Introduction
Ovarian cancer is a common malignant tumor in female
genital system and its incidence rate ranks second in the malignant
tumors of female genital system. Moreover, its mortality has been
ranked as first in female malignant tumors. Approximately 85% of
the patient has been reported to be suffering from epithelial
ovarian cancer (1). At present,
ovarian cancer is mainly treated with comprehensive treatment
methods including surgery, chemotherapy, radiotherapy, biological
therapy, etc., clinically. Although the methods for diagnosis and
treatment of ovarian cancer is being developed continuously, the
5-year survival rate of the patients is still just 30%. Therefore,
it is of great clinical significance to search molecular markers
for early diagnosis of ovarian cancer and for the judgment of
prognosis (2).
MicroRNA (miRNA/miR) is a small highly-conserved,
single-stranded and non-coding RNA, which could be specifically
associated with its target mRNA, as it affects directly the
transcription or translation processes of the target genes
(3,4).
It has been confirmed in an earlier study that the abnormal
expression of miRNA in tumor cells is responsible for the
development of various malignant tumors (5). miR-23 has two subtypes: miR-23a and
miR-23b. They only differ by one nucleotide and are coded by
different genes (6). miR-23a is
located on chromosome 19. It not only regulates the growth and
differentiation of normal cells, but also controls the
proliferation as well as metastasis of a variety of malignant tumor
cells (7,8). miR-23b is located on chromosome 9 and
its abnormal expression has been noticed in prostate cancer,
pancreatic cancer, liver cancer and other malignant tumors.
Moreover, miR-23b has an influence on the pathology and prognosis
of the patients (9).
Currently, there is neither any study on the
correlation between the expressions of miR-23a and miR-23b during
ovarian cancer nor the any study about their influence on the
pathological parameters and prognosis of ovarian cancer. Therefore,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) was adopted in this study to detect the expressions of
miR-23a and miR-23b in tumor tissues of patients with ovarian
cancer. The prime aim of the study was to investigate the
correlation between the expressions of the two items. Meanwhile,
the influences of miR-23a and miR-23b in tumor tissues of the
patients on the pathological parameters and prognosis of patients
with ovarian cancer were studied in combination with the analysis
of clinical data.
Materials and methods
Study design
A total of 50 patients who were treated in surgical
department of our hospital from June 2008 to December 2011 were
enrolled as study subjects in this study. The study subjects aged
28–76 years old and had an average age of 54.8. Tumor tissues and
normal tissues adjacent to the cancer of the patients with ovarian
cancer were taken during surgery and were preserved in liquid
nitrogen. Inclusion criteria for the patients: Patients who were
treated for the first time without radiotherapy or chemotherapy,
and patients confirmed with ovarian epithelial cancer by pathology
after surgery Exclusion criteria: patients who had a history of
cardiovascular diseases, neurological diseases, and diabetes,
accompanied with malignant tumors in other parts during
preoperative examinations. The clinical ethics committee of our
hospital had approved the present study. Further, all patients
approved for the study or their families had signed the informed
consent.
TRIzol kits, reverse transcription kits and RT-qPCR
kits (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA); primer synthesis (Takara Biotechnology Co., Ltd., Dalian,
China).
Detection of the expressions of
miR-23a and miR-23b in tumor tissues of the ovarian cancer patients
with RT-qPCR
Approximately 200 mg of samples was taken from
frozen tumor tissues and normal tissues adjacent to the cancer
respectively. The total RNA in the specimens of the tissues was
extracted as per the method described in the package insert of
TRIzol kit to detect the absorbance of RNA at 260 and 280 nm.
Reverse transcription was conducted for the samples with A260/A280
between 1.8–2.0. The operation was carried out as per the package
insert of reverse transcription kit. The-obtained cDNA was used as
the template and PCR operation was carried out as per the
recommended method of the kit. The total volume of the PCR reaction
system was 20 µl and the primer sequence was indicated in Table I. Reaction conditions: 94°C for 4 min
(initial denaturation); 95°C for 1 min; 60°C for 1 min and 72°C for
1 min; amplification for 40 cycles. Ct value was output from the
instrument. The results were analyzed by 2−∆∆Cq method
using U6 RNA as control.
 | Table I.Primer sequence of reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequence of reverse
transcription-quantitative polymerase chain reaction.
| Genes | Direction | Primer sequence |
|---|
| miR-23a | Forward primer |
5′-ATCACATTGCCAGGGATTTCC-3′ |
|
| Reverse primer |
5′-CCAGTGCAGGGTCCGAGGT-3′ |
| miR-23b | Forward primer |
5′-CGCGGCCGCTAGTATTATGTT-3′ |
|
| Reverse primer |
5′-CACATTTTAAAAAACATA-3′ |
| U6 | Forward primer |
5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
|
| Reverse primer |
5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
Analysis of the correlation between t miR-23a and
miR-23b expressions in tumor tissues during ovarian cancer and the
pathological/prognosis parameters. The patients were divided into
high-expression group and low-expression group with the median of
the expressions of miR-23a (5.35) and miR-23b (2.87) in tumor
tissues of the patients with ovarian cancer as the benchmark.
χ2 test was conducted to analyze the correlation between
the expressions of miR-23a and miR-23b and the pathological
parameters of the patients. Follow-up visit was conducted once
every month from the first day after surgery for 5 years.
Statistical analysis
SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA)
was used for data analyses. Further, χ2 test was
conducted for the comparison of enumeration data between groups.
Enumeration data were expressed as mean ± standard deviation and
t-test was conducted for the comparison between two groups. Pearson
analysis was performed to analyze correlation, Kaplan-Meier
analysis was adopted for survival analysis and log-rank method was
used for the comparison between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Detection of the expressions of
miR-23a and miR-23b in tumor tissues of ovary ancer patients with
RT-qPCR
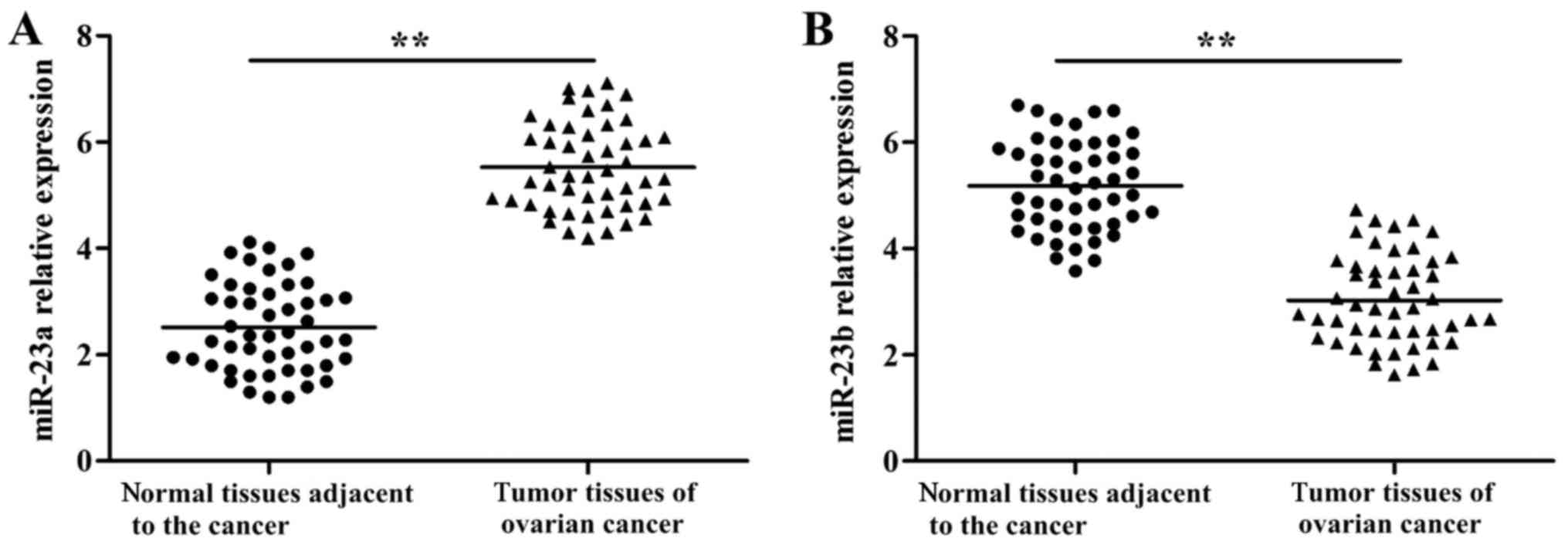
RT-qPCR results are indicated in Fig. 1. miR-23a showed significantly higher
expression in tumor tissues (P<0.01) in comparison to adjacent
normal tissues. On the other hand, miR-23b revealed significantly
lower expression (P<0.01) in tumor tissues when compared with
adjacent normal tissues.
Correlation analysis on the
expressions of miR-23a and miR-23b in tumor tissues of patients
with ovarian cancer
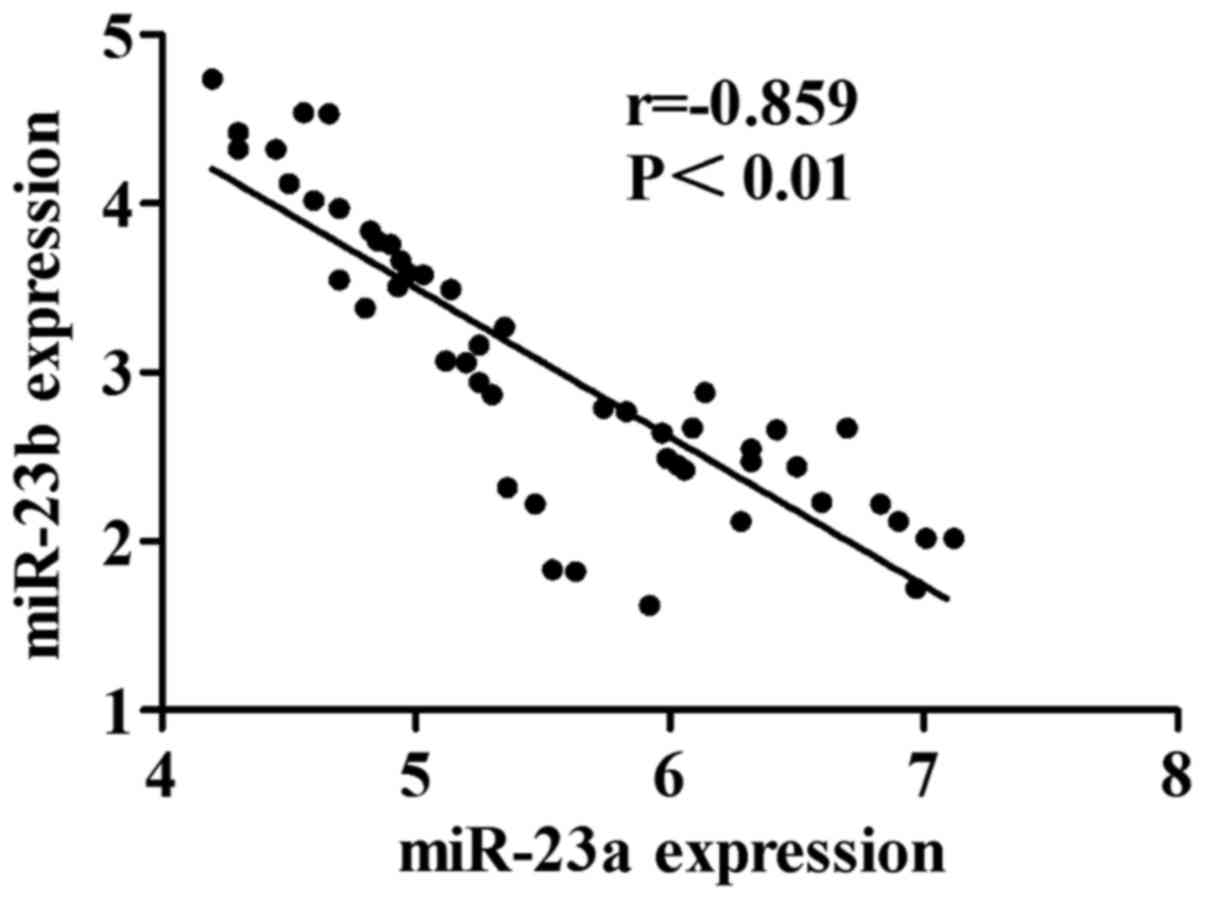
Pearson correlation analysis was conducted for the
expressions of miR-23a and miR-23b in tumor tissues of patients
with ovarian cancer. The results are shown in Fig. 2. The expression amount of miR-23a is
negatively correlated with that of miR-23b in tumor tissues of
patients with ovarian cancer (r=−0.859, P<0.01).
Relationship between expressions of
miR-23a and miR-23b in ovarian tumors tissues with clinical
pathology parameters
The results are shown in Table II. χ2 test indicated that
the high expression of miR-23a and the low expression of miR-23b in
tumor tissues of the patients were related with the degree of tumor
differentiation, metastasis of lymph nodes and clinical
staging.
 | Table II.Association between abnormal
expression levels of miR-23a and miR-23b and the
clinicopathological parameters of ovarian cancer. |
Table II.
Association between abnormal
expression levels of miR-23a and miR-23b and the
clinicopathological parameters of ovarian cancer.
|
|
| miR-23a | miR-23b |
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameters | n | High expression (n,
%) | χ2 | P-value | Low expression (n,
%) | χ2 | P-value |
|---|
| Age |
|
| 0.25 | >0.05 |
| 0.09 | >0.05 |
| ≥50 years
old | 33 | 18 (54.55) |
|
| 16 (48.48) |
|
|
| <50
years old | 17 | 8 (47.06) |
|
| 9 (52.94) |
|
|
| Differentiation
degree |
|
| 7.96 | <0.01 |
| 13.88 | <0.01 |
| Low
differentiation | 21 | 6 (28.57) |
|
| 4 (19.05) |
|
|
|
Medium/high
differentiation | 29 | 20 (68.97) |
|
| 21 (72.41) |
|
|
| Metastasis of lymph
nodes |
|
| 11.76 | <0.01 |
| 19.10 | <0.01 |
| Yes | 31 | 22 (70.97) |
|
| 23 (74.19) |
|
|
| No | 19 | 4 (21.05) |
|
| 2 (10.53) |
|
|
| Clinical staging |
|
| 6.61 | <0.05 |
| 5.56 | <0.05 |
| I–II | 18 | 5 (27.78) |
|
| 5 (27.78) |
|
|
|
III–IV | 32 | 21 (65.63) |
|
| 20 (62.50) |
|
|
Analysis of survival condition and
prognosis of the patients
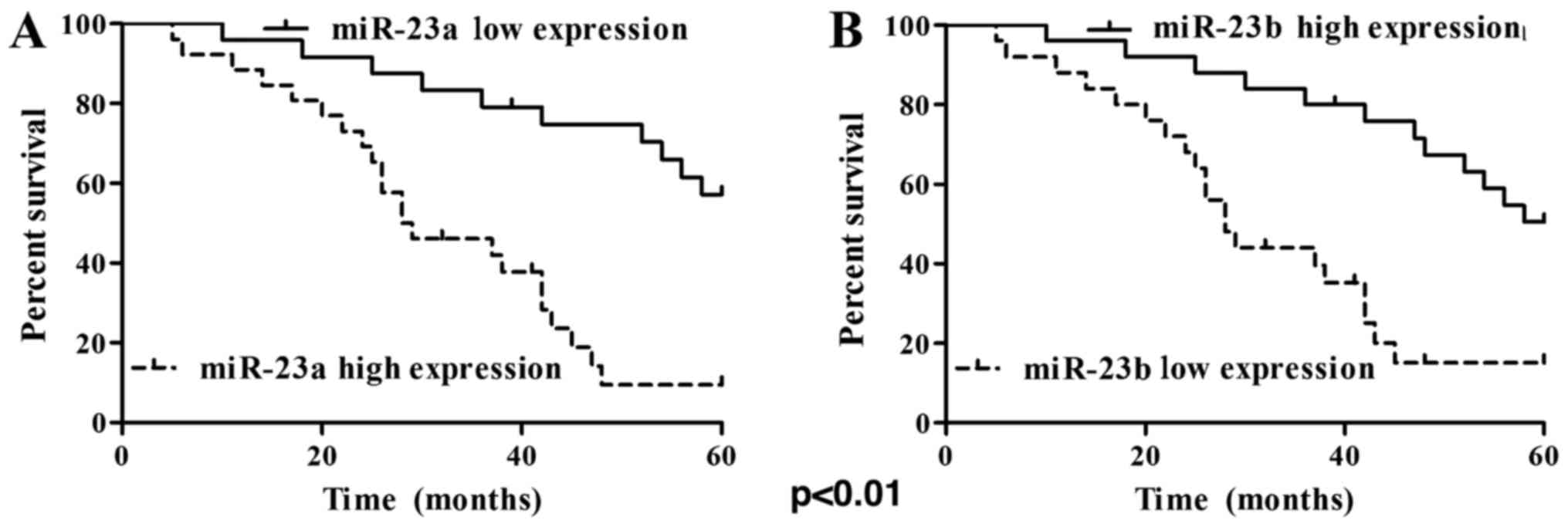
The results of 5-year follow-up visit showed that 18
cases survived and 32 cases died among the 50 cases. The 5-year
overall survival rate was 36% (18/50) and the death rate was 64%
(32/50). The Kaplan-Meier survival curve of patients with ovarian
cancer is shown in Fig. 3A and B. The
results clearly confirmed that the patients with high expression of
miR-23a or low expression of miR-23b had relatively poor prognosis.
Results of univariate survival analyses are shown in Table III, which showed that both miR-23a
and miR-23b had impacts on the overall survival rate of patients
with ovarian cancer (P<0.01).
 | Table III.Univariate analysis on the correlation
of the expression levels of miR-23a and miR-23b with the overall
survival rate of patients with ovarian cancer. |
Table III.
Univariate analysis on the correlation
of the expression levels of miR-23a and miR-23b with the overall
survival rate of patients with ovarian cancer.
| Groups | n | Number of survived
case in 5 years | 5-year survival rate
(%) | Wald (log-rank) | P-value |
|---|
| miR-23a |
|
|
|
|
|
| High
expression | 26 | 4 | 15.38 | 16.11 | <0.01 |
| Low
expression | 24 | 14 | 58.33 |
|
|
| miR-23b |
|
|
|
|
|
| High
expression | 25 | 13 | 52.00 | 12.86 | <0.01 |
| Low
expression | 25 | 5 | 20.00 |
|
|
Discussion
The pathogenesis of ovarian cancer is obscured.
Further, it is quite difficult to ascertain and eradicate the
cancer effectively by surgery at an early stage. In addition,
ovarian cancer cells easily develop tolerance to radiotherapy and
chemotherapy, leading to tumor recurrence and treatment failure.
Therefore, discovering tumor markers of ovarian cancer is of great
significance for the diagnosis, treatment and prognosis of ovarian
cancer (10,11).
The normal expression of miRNA could regulate
proliferation and differentiation of cells, while its abnormal
expression would result in development of tumors (12). It is a confirmed fact that miRNAs had
an effect on the expression of target genes as they pair with the
target genes, thereby, regulate various physiological processes
(13). Further, it was observed in an
earlier study that miRNA could regulate the expression of
approximately 60% protein (14). The
expression of miRNA was closely related with the growth,
differentiation, multiplication, transfer and apoptosis of cells
(15,16).
The expressions of miR-23a and miR-23b are closely
related with the proliferation, differentiation and apoptosis,
etc., of the cells. Our study observed higher expression ofmir-23a
in ovarian tumor tissues. miR-23a has significantly higher
expression in breast cancer patients with lymph node metastasis
than those without lymph node metastasis (17). MiR-23a is highly expressed in liver
cancer cells, which promotes the growth and proliferation of liver
cancer cells by regulating Samds signaling pathway (18). miR-23a is highly expressed in
cisplatin-resistant A2780 cells of ovarian cancer. An earlier study
showed that inhibition of miR-23a expression reduced the expression
of P-gp protein in A2780 cells leading to elevation in sensitivity
of cells to cisplatin (19). Studies
have shown that miR-23b is highly expressed in the serum of
patients with gastric cancer, which is negatively correlated with
the prognosis of the patients (20).
miR-23b showed lower expression in the tissues of ovarian cancer.
When miR-23b is excessively expressed in ovarian cancer cells, the
expressions of protein such as CCNG1, Survivin, Bcl-xL, P70S6K and
MMP9 are decreased, thus the proliferation and migration of tumor
cells are reduced (21).
In order to investigate the expressions of miR-23a
and miR-23b in tumor tissues of patients with ovarian cancer and
its influence on the pathological parameters and prognosis of
patients with ovarian cancer, RT-qPCR was firstly adopted in this
study to detect the expressions of miR-23a and miR-23b in tumor
tissues of patients with ovarian cancer. The results showed that
compared with normal tissues adjacent to the cancer, miR-23a had
significantly higher expression in tissues of ovarian cancer, while
the expression of miR-23b was reduced significantly. Meanwhile,
Pearson correlation analysis indicated that the expression of
miR-23a was negatively correlated with that of miR-23b in tumor
tissues of patients with ovarian cancer. In order to further
confirm the relationship between expressions of miR-23a and miR-23b
and ovarian cancer, the correlation between the expressions of
miR-23a and miR-23b and the clinical pathology/prognosis parameters
was performed. The results showed that the high expression of
miR-23a and the low expression of miR-23b were correlated with the
degree of tumor differentiation, metastasis of lymph nodes and
clinical staging. Further, survival analysis indicated that
patients with high expression of miR-23a and the low expression of
miR-23b had relatively poor prognosis. The univariate survival
analysis showed that both miR-23a and miR-23b had a significant
influence on the overall survival rate of patients with ovarian
cancer. As, both miR-23a and miR-23b were negatively correlated
with each other. So, there is no need of observing individual
parameter with survival rate. Moreover, if in an established tumor,
mir-23a was observed to be higher, and then its negative
counterpart mi23b would be automatically down as a result of
negative correlation. Therefore, we have studied overall survival
correlation with thee two parameters collectively. Further,
increase in mi23a could be called as independent prognostic marker
of ovarian cancer. The above study results showed that miR-23a and
miR-23b could be used as the important indicators for predicting
the prognosis of patients with ovarian cancer.
In conclusion, the high expression of miR-23a and
the low expression of miR-23b confirmed presence of ovarian cancer
in patients. So, miR-23a and miR-23b might be utilized as important
reference indicators for timely diagnosis of ovarian cancer.
However, similar studies with large sample size are essential for
concrete conclusion.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and material
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LS wrote the manuscript and assisted with RT-qPCR.
ML analyzed the survival condition and prognosis of the patients.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the ethics committee of
Longnan Hospital and informed consent was signed by the patients
and/or guardians.
Consent for publication
Informed consents were signed by the patients and/or
guardians.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Panera N, Gnani D, Crudele A, Ceccarelli
S, Nobili V and Alisi A: MicroRNAs as controlled systems and
controllers in non-alcoholic fatty liver disease. World J
Gastroenterol. 20:15079–15086. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: microRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siomi H and Siomi MC: On the road to
reading the RNA-interference code. Nature. 457:396–404. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang Z, Wang XL, Bai R, Liu WY, Li X, Liu
M and Tang H: miR-23a promotes IKKα expression but suppresses ST7L
expression to contribute to the malignancy of epithelial ovarian
cancer cells. Br J Cancer. 115:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aghaee-Bakhtiari SH, Arefian E, Naderi M,
Noorbakhsh F, Nodouzi V, Asgari M, Fard-Esfahani P, Mahdian R and
Soleimani M: MAPK and JAK/STAT pathways targeted by miR-23a and
miR-23b in prostate cancer: Computational and in vitro approaches.
Tumour Biol. 36:4203–4212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wen YC, Lee WJ, Tan P, Yang SF, Hsiao M,
Lee LM and Chien MH: By inhibiting snail signaling and miR-23a-3p,
osthole suppresses the EMT-mediated metastatic ability in prostate
cancer. Oncotarget. 6:21120–21136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Donadelli M, Dando I, Fiorini C and
Palmieri M: Regulation of miR-23b expression and its dual role on
ROS production and tumour development. Cancer Lett. 349:107–113.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bookman MA: Optimal primary therapy of
ovarian cancer. Ann Oncol. 27 Suppl 1:i58–i62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suh DH, Kim HS, Chang SJ and Bristow RE:
Surgical management of recurrent ovarian cancer. Gynecol Oncol.
142:357–367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blower PE, Chung JH, Verducci JS, Lin S,
Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et
al: MicroRNAs modulate the chemosensitivity of tumor cells. Mol
Cancer Ther. 7:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du T and Zamore PD: microPrimer: The
biogenesis and function of microRNA. Development. 132:4645–4652.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang S and Olson EN: AngiomiRs-key
regulators of angiogenesis. Curr Opin Genet Dev. 19:205–211. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gounaris-Shannon S and Chevassut T: The
role of miRNA in haematological malignancy. Bone Marrow Res.
2013:2691072013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Liu X, Xu W, Zhou P, Gao P, Jiang S,
Lobie PE and Zhu T: c-MYC-regulated miR-23a/24-2/27a cluster
promotes mammary carcinoma cell invasion and hepatic metastasis by
targeting sprouty2. J Biol Chem. 288:18121–18133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang S, He X, Ding J, Liang L, Zhao Y,
Zhang Z, Yao X, Pan Z, Zhang P, Li J, et al: Upregulation of
miR-23a approximately 27a approximately 24 decreases transforming
growth factor-beta-induced tumor-suppressive activities in human
hepatocellular carcinoma cells. Int J Cancer. 123:972–978. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin AH, Zhou XP and Zhou FZ: Inhibition of
microRNA-23a increases cisplatin sensitivity of ovarian cancer
cells: The possible molecular mechanisms. Nan Fang Yi Ke Da Xue Xue
Bao. 35:125–128. 2015.(In Chinese). PubMed/NCBI
|
|
20
|
Zhuang K, Han K, Tang H, Yin X, Zhang J,
Zhang X and Zhang L: Up-regulation of plasma miR-23b is associated
with poor prognosis of gastric cancer. Med Sci Monit. 22:356–361.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan J, Jiang JY, Meng XN, Xiu YL and Zong
ZH: MiR-23b targets cyclin G1 and suppresses ovarian cancer
tumorigenesis and progression. J Exp Clin Cancer Res. 35:312016.
View Article : Google Scholar : PubMed/NCBI
|