Introduction
Although stem cells comprise only a very small
percentage of a tumor, they have a critical role in promoting
disease pathogenesis (1). This is
largely due to their self-renewal capabilities, which allow these
cells to survive for a long period of time, during which, they may
acquire mutations that confer a growth advantage (2). Cancer stem cells have also been shown to
be associated with therapeutic resistance (3). Genes involved in stem cell biology
during normal development, such as the transcription factor
octamer-binding transcription factor 4 (OCT4), may also contribute
to tumorigenesis (4).
OCT4 is a homeodomain transcription factor with an
essential role in embryonic stem (ES) cell self-renewal (5,6). This was
shown by genetic ablation of the OCT4 gene, since the loss of OCT4
suppressed a large number of the stem characteristics of an ES cell
line (7). OCT4 has also been detected
in adult cells possessing stem cell characteristics, such as
mammary stem cells (8). In addition
to its role in maintaining stem cell pluripotency, a role for OCT4
in cancer stem cells is also emerging (4). While the role of OCT4 in cancer is
unknown, a previous study identified high expression of OCT4 in
cluster of differentiation (CD)44+/CD24-breast cancer stem cells
(4). Importantly, the authors
determined that OCT4 was more highly expressed in tumor tissue than
in normal adjacent tissue, and that individuals with high OCT4
expression had worse disease-free survival than those with low
expression (4). The human OCT4 gene
can generate three transcript variants (OCT4A, OCT4B and OCT4B1)
and four protein isoforms via a combination of alternative splicing
and use of multiple translation initiation sites (9,10). While,
in ES cells, self-renewal is conferred by OCT4A, OCT4B appears to
serve a role in responding to cellular stress (11–14).
However, the exact roles of each of these OCT4 variants in normal
physiology and in cancer remain unclear.
The deregulation of cell cycle progression and
apoptosis may contribute to tumorigenesis. Two important cell cycle
inhibitors, p16INK4a and p14ARF, are encoded by the
cyclin-dependent kinase inhibitor 2A (CDKN2A) (INK4a/ARF)
locus (15). p16INK4a regulates
retinoblastoma protein-dependent G1 arrest (16). By contrast, p14ARF can be activated by
oncogenic and hyperproliferative stimuli (including Myc, Ras and
E1A), resulting in activation of the p53-p21 pathway and causing
p53-dependent G1 cell cycle arrest (17–19). p53
is a tumor-suppressor protein with essential roles in apoptosis and
cell cycle arrest (20). Activated
p53 mediates apoptosis via regulating the expression of B-cell
lymphoma (Bcl)-2 gene family members or by directly binding to
anti-apoptotic Bcl-2 family proteins at the mitochondrial surface,
thereby activating Bcl-2 associated X protein (Bax) and initiating
the apoptosis cascade (21,22). Although numerous studies concerning
p16INK4a, p14ARF and p53 have been published, little is known about
the genes and networks that co-regulate the p16INK4a and p53
pathways.
Recent studies have started to elucidate the
association between OCT4 and pathways regulating cell proliferation
and apoptosis (23–25). These associations are important, since
they aid to strengthen the link between OCT4 and cancer. One group
recently demonstrated that OCT4B is upregulated by p53 under
genotoxic stress, functioning in the stress response in ES cells
(14). Another group recently
published that OCT4 suppresses p53 expression (26). This raises the possibility that, in
certain cellular contexts, these factors may be part of a negative
regulatory loop. Despite these facts, the precise nature of the
association between these proteins remains unclear.
Previous studies by the present authors confirmed
the presence of OCT4B expression in MCF-7 cells, and it was
speculated that the transcription of this gene in this cancer cell
line may be driven by an independent promoter (27). In the present study, we build upon
this initial finding by showing that OCT4B is widely expressed
across a panel of human breast cancer cell lines. It was also
observed that OCT4 regulates the expression of factors such as p53
and p16, which control essential cell regulatory pathways. Finally,
the reciprocal regulation of OCT4A and OCT4B in cancer cells was
explored. In conclusion, the present data will aid to elucidate the
role of OCT4 in the regulation of proliferation and apoptosis of
breast cancer cells. These findings could be clinically important
for patients with breast tumors in which OCT4 becomes
re-expressed.
Materials and methods
Cell culture
The human breast adenocarcinoma cell lines MCF-7,
MDA-MB-231 (mm231) and MDA-MB-468 (mm468) were cultured in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
calf serum (FCS; HyClone; GE Healthcare Life Sciences, Logan, UT,
USA), 100 U/ml of penicillin and 100 µg/ml of streptomycin solution
(HyClone; GE Healthcare Life Sciences) at 37°C and under 5%
CO2. The T-47D (human ductal breast epithelial tumor
cell line) cell line was cultured in RPMI 1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FCS (HyClone;
GE Healthcare Life Sciences), 100 U/ml of penicillin and 100 µg/ml
of streptomycin solution (HyClone; GE Healthcare Life Sciences).
All the cell lines were purchased from Peking Union Medical College
(Beijing, China). Similar cell densities were maintained across
different experiments. All experiments and protocols were reviewed
by and approved by the Institutional Review Board of Peking Union
Medical College.
Construction of overexpression vector
and RNA interference (RNAi) vector
The open reading frames (ORFs) of OCT4A and OCT4B
were amplified from the messenger RNA (mRNA) of MCF-7 cells, and
were subcloned into the pDsRed-ex-C1 vector [containing
Discosoma sp. red fluorescent protein (DsRed); Clontech
Laboratories, Inc., Mountainview, CA, USA] or the pEGFP-N1 vector
[containing enhanced green fluorescent protein (EGFP); Clontech
Laboratories, Inc.] to generate the fusion expression vectors
pDsRed-ex-C1-OCT4A and pEGFP-N1-OCT4B, respectively (which are
referred to as pC1-4A and pN1-4B, respectively, throughout the
present manuscript). An additional two OCT4 vectors lacking
fluorescent protein fusions were also generated. For both OCT4A and
OCT4B, these were generated by inserting the complementary DNAs
(cDNAs) into pEGFP-N1, thus deleting EGFP. The resulting constructs
were named pN1-4As and pN1-4Bs for OCT4A and OCT4B, respectively.
The sequences of the primers used for subcloning the OCT4A ORF with
a mutation at the stop codon were
5′-ACGTAAGCTTATGGCGGGACACCTGGCTTC-3′ (forward) and
5′-TTTTGAATTCCGGGCAGGCACCACAGTTT-3′ (reverse). The primers for
subcloning the OCT4B ORF were 5′-ACTGAAGCTTATGCACTTCTACAGAC-3′
(forward) and 5′-TTTTGAATTCCGGGCAGGCACCACAGTTT-3′ (reverse). The
primers used for OCT4A ORF with a stop codon were
5′-ACGTAAGCTTATGGCGGGACACCTGGCTTC-3′ (forward) and
5′-TTTTGAATTCCGGGCAGGCACCTCAGTTT-3′ (reverse). The primers for
OCT4B ORF with a stop codon were 5′-ACTGAAGCTTATGCACTTCTACAGAC-3′
(forward) and 5′-TTTTGAATTCCGGGCAGGCACCTCAGTTT-3′ (reverse). RNAi
expression vectors were generated by cloning small hairpin RNA
(shRNA) oligonucleotides into the BbsI site of the
pFIV-h1/U6-copGFP vector (Clontech Laboratories, Inc.). The
sequences of the oligonucleotides used were
5′-AAAGAAGAGAAAGCGAACCAGTATTCCAGTCTATACTGGTTCGCTTTCTCT-3′ (sense)
and 5′-AAAAAGAGAAAGCGAACCAGTATTAGCGATTATACTGGTTCGCTTTCTCTT-3′
(antisense). The sequence targets a common exon (exon 5) of OCT4A
and OCT4B.
Transient expression and RNAi
Transfection of shRNA vectors was performed with
VigoFect (Vigorous, Beijing, China), according to the
manufacturer's recommendations. Cells grown to 50–60% confluence in
24-well plates were transfected with 4 µg of the small interfering
RNA plasmid pFIV-4B. After 24 h, the cells were harvested for
isolation of RNA and protein, as well as for flow cytometry
analysis.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was prepared from cultured cells using the
RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) with on-column DNase
(Promega Corporation, Madison, WI, USA) treatment. Next, cDNA was
synthesized using the SuperScript® First-Strand
Synthesis System (Promega Corporation). RT was performed in a 20-µl
sample volume containing 2 µg RNA. The reaction included 60 min of
RT at 42°C and 15 min of inactivation at 70°C. Negative controls
were performed without reverse transcriptase. Amplification was
performed in a 25-µl sample volume according to a standard
procedure. The primer sets used in the experiment were as follows:
OCT4A, 5′-ATGGCGGGACACCTGGCTTC-3′ (forward) and
5′-AAGGGCAGGCACCTCAGTTT-3′ (reverse); OCT4B,
5′-ATGCACTTCTACAGACTATTCCTTGGGGCC-3′ (forward) and
5′-AAGGGCAGGCACCTCAGTTT-3′ (reverse); p53, 5′-TCAGATCCGTGGGCGTGA-3′
(forward) and 5′-GTCAGTGGGGAACAAGAAG-3′ (reverse); p21,
5′-ACCATGTGGACCTGTCACTGTCTTG-3′ (forward) and
5′-AGGCTTCCTGTGGGCGGATTAG-3′ (reverse); p14ARF,
5′-TTTTCGTGGTTCACATCCC-3′ (forward) and 5′-TCTAAGTTTCCCGAGGTTTCT-3′
(reverse); p16INK4a, 5′-TGCCCAACGCACCGAATAGTTA-3′ (forward) and
5′-GTGCAGCACCACCAGCGTGTCC-3′ (reverse); Bcl-2,
5′-AGATTGATGGGATCGTTGCCTTAT-3′ (forward) and
5′-CCAATTCCTTTCGGATCTTTATTTC-3′ (reverse); Bax,
5′-GGGAGCGGCGGTGATGGA-3′ (forward) and 5′-GCAAAAGGGCCCCTGTCTTC-3′
(reverse); and β-actin, 5′-TCATCACTATTGGCAACGAGC-3′ (forward) and
5′-AACAGTCCGCCTAGAAGCAC-3 (reverse). PCR products were visualized
and semiquantified by agarose gel electrophoresis and
densitometry.
Immunocytochemistry detection of
OCT4
Human breast cancer cells were cultured on
coverslips and grown to 80% confluence. Cells were fixed in 4%
paraformaldehyde for 30 min at room temperature and then incubated
with 0.3% Triton X-100 in PBS at 4°C for 20 min. After blocking
with 10% FCS in PBS, cells were incubated with an anti-OCT4
antibody that recognizes both OCT4A and OCT4B (1:50 dilution;
sc-8629; Santa Cruz Biotechnology Inc., Dallas, TX, USA) for 1 h at
room temperature. Fluorescein isothiocyanate (FITC)-conjugated
anti-goat immunoglobulin G (1:100 dilution; sc-2489; Santa Cruz
Biotechnology Inc.) was used as a secondary antibody. Propidium
iodide (PI) staining of the nucleus was performed at room
temperature for 5 min. Fluorescent images were photographed using a
confocal laser scanning microscope (OLS3000; Nikon Corporation,
Tokyo, Japan).
Western blotting
Western blotting followed by chemiluminescence
detection was performed by a conventional method. A polyclonal
antibody (ab19857; Abcam, Cambridge, UK) was used to detect the
C-termini of both OCT4A and OCT4B. Detection of GAPDH with an
anti-GAPDH antibody (1:200 dilution; 2251-1; Epitomics, Burlingame,
CA, USA) served as a loading control.
RT-quantitative PCR (RT-qPCR)
RT-q PCR for quantification of cDNA was
performed using SYBR Green II PCR Master Mix (DRR081S; Takara
Biotechnology Co., Ltd., Dalian, China) and the ABI 7500 Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. RT-qPCR was performed in
triplicate. The cycling conditions were 40 cycles of 95°C for 5
sec, 55°C for 10 sec and 72°C for 10 sec. The primer sets used in
the study were as follows: OCT4B, 5′-CAGGGAATGGGTGAATGAC-3′
(forward) and 5′-CGTGTGGCCCCAAGGA-3′ (reverse); p53,
5′-TGCTTGCAATAGGTGTGCGTCA-3′ (forward) and
5′-AAACACCAGTGCAGGCCAACTT-3′ (reverse); p21,
5′-ATGTGTCCTGGTTCCCGTTTCT-3′ (forward) and
5′-TTGTGGGAGGAGCTGTGAAAGA-3′ (reverse); Bcl-2,
5′-TAATTGCCAAGCACCGCTTCGT-3′ (forward) and
5′-TGATCCGGCCAACAACATGGAA-3′ (reverse); p16INK4a,
5′-ACCCCGCTTTCGTAGTTTTC-3′ (forward) and 5′-GCAGAAGCGGTGTTTTTCTT-3′
(reverse); p14ARF, 5′-AGAACATGGTGCGCAGGTTCTTG-3′ (forward) and
5′-TAGACGCTGGCTCCTCAGTAGCAT-3′ (reverse); Bax,
5′-GGGAGCGGCGGTGATGGA-3′ (forward) and 5′-GCAAAAGGGCCCCTGTCTTC-3′
(reverse); and GAPDH, 5′-TCAAGAAGGTGGTGAAGCA-3′ (forward) and
5′-AAAGGTGGAGGAGTGGGT-3′ (reverse). Relative gene expression levels
were quantified using the 2−ΔΔCq method (28).
Analysis of cell apoptosis
For each sample, ~105 cells were prepared
for the apoptosis assay. An Annexin V-FITC and PI double staining
kit (Vigorous) was used according to the manufacturer's protocol.
Stained cells were observed using a confocal laser scanning
microscope (OLS3000; Nikon Corporation) and analyzed by flow
cytometry (FACSCalibur; BD Biosciences, San Jose, CA, USA).
Statistical analysis
Data were presented as means ± standard deviation,
and statistical comparisons between data groups were conducted
using the Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
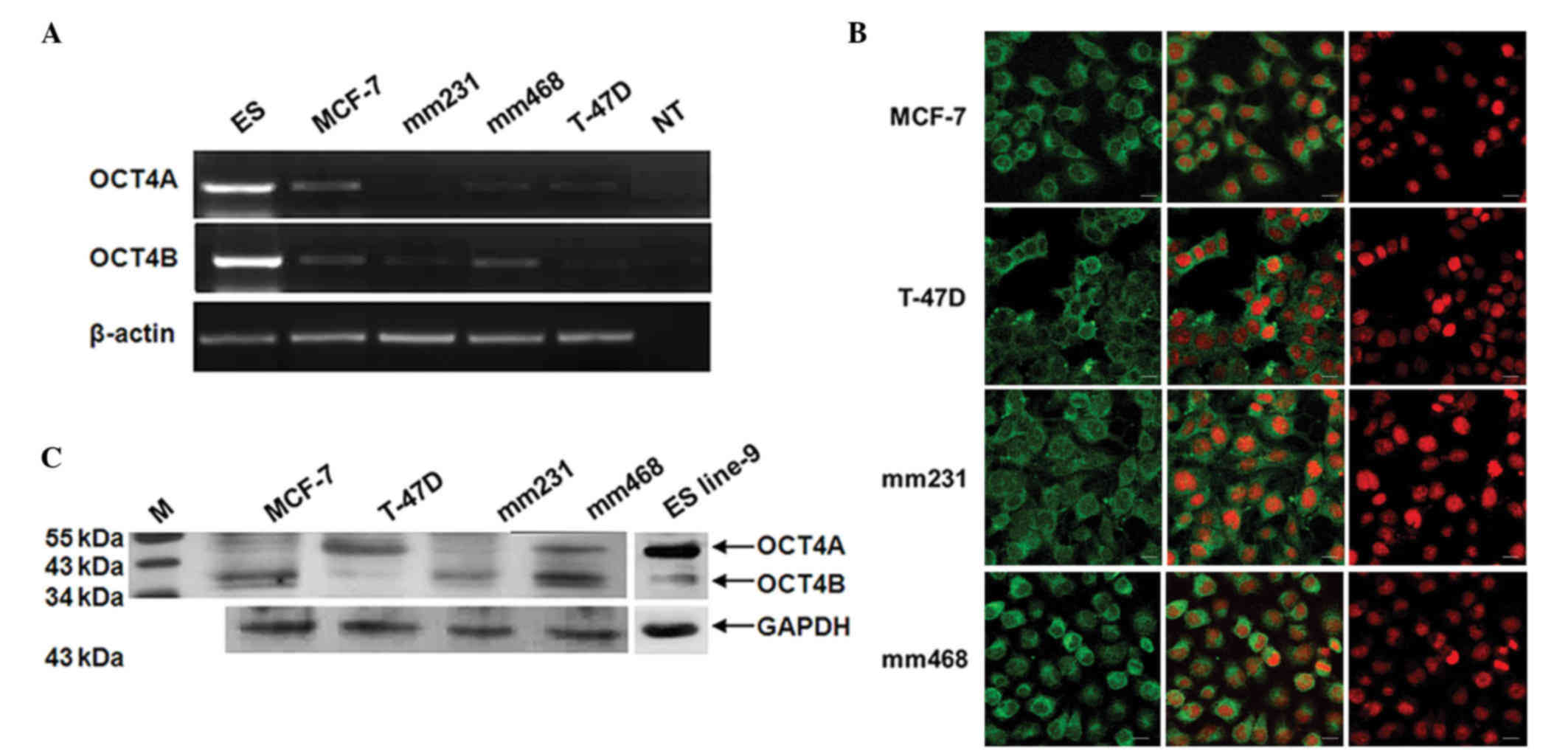
OCT4 is expressed in a panel of breast
cancer cell lines
OCT4 gene expression was examined in a panel of four
breast cancer cells, including two estrogen receptor (ER)-positive
lines (MCF-7 and T-47D) and two ER-negative lines [MDA-MB-231
(mm231) and MDA-MB-468 (mm468)](Fig.
1). Semiquantitative RT-PCR was performed using primers capable
of distinguishing OCT4A from OCT4B. PCR for each OCT4 variant
produced a single band, which was then verified by cloning and
sequencing. While transcripts for both OCT4A and OCT4B were
detected in each breast cancer cell line, their levels of
expression were significantly lower than their levels in ES cells
(Fig. 1A). It should be noted that
the transcript levels of OCT4A and OCT4B fluctuated with
differences in cell density.
Next, the protein expression of OCT4A and OCT4B was
examined in breast cancer cell lines by western blotting and
immunocytochemistry. By western blot analysis, it was observed that
MCF-7 and mm231 cells express very low levels of OCT4A (45 kDa),
and that the expression of OCT4A was higher in T-47D and mm468
cells (Fig. 1B). Differences in OCT4B
(35 kDa) expression were more apparent among the four cell lines,
with mm468 showing the highest level of OCT4B expression and T-47D
showing the lowest (Fig. 1B).
Immunocytochemistry for OCT4 was next performed using an antibody
that fails to distinguish OCT4A from OCT4B. Prominent cytoplasmic
staining of OCT4 was observed in each of the four breast cancer
cell lines (Fig. 1C). Taken together,
our data show that OCT4 is commonly re-expressed in multiple breast
cancer cell lines and is predominantly located in the
cytoplasm.
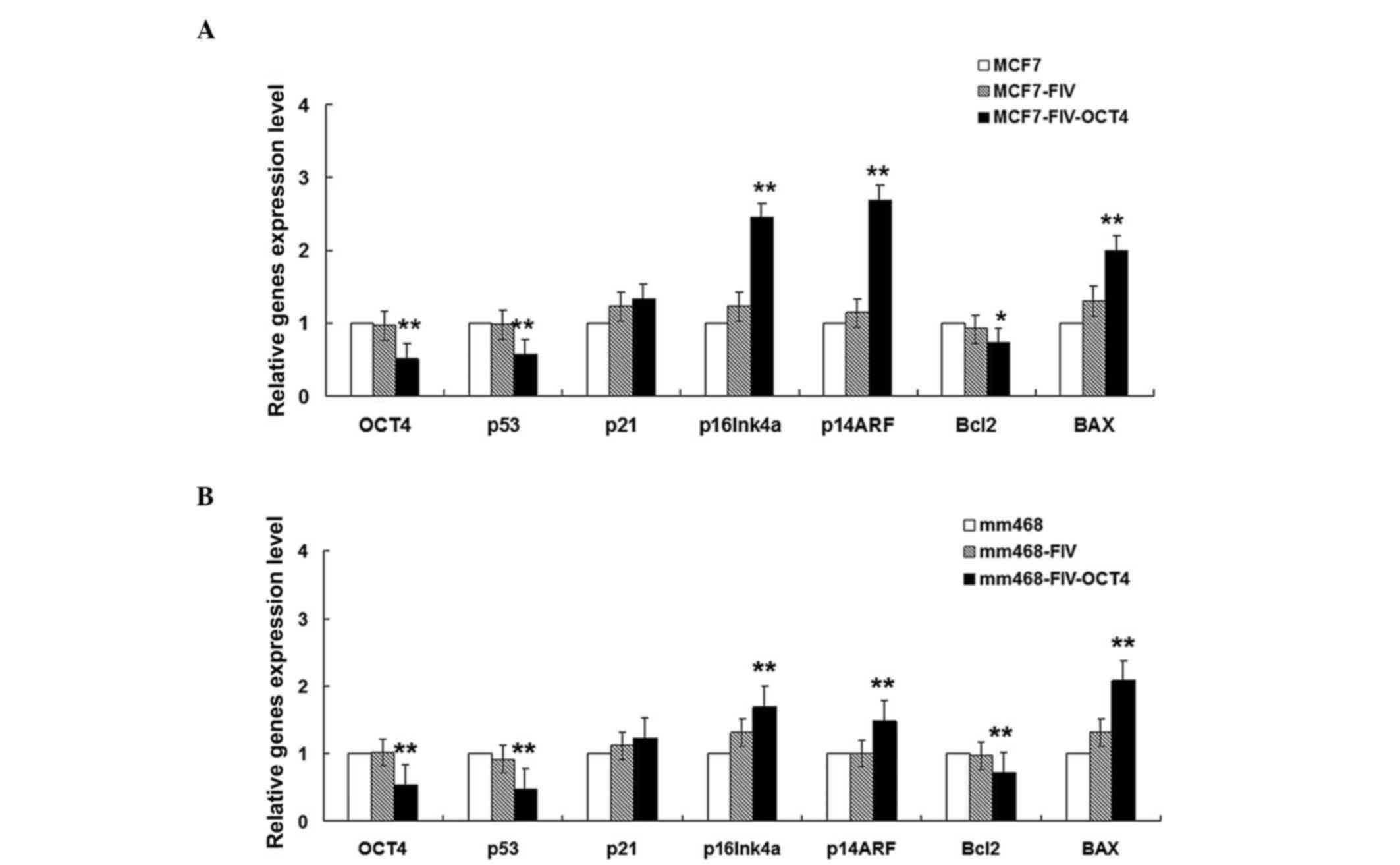
OCT4 regulates p53 and p16INK4a/p14ARF
gene transcription
The possible role of OCT4 in breast cancer cells was
examined by assessing its ability to influence the transcription of
genes downstream of the p53 and p16 pathways. Since the MCF-7 and
mm468 cell lines expressed the highest levels of OCT4B, these were
selected for further study. Each of these two cell lines was
transfected with OCT4-RNAi, which efficiently depleted the mRNA
levels of OCT4 in both cell lines (Fig.
2A and B).
Transcript levels of the cell cycle inhibitor genes
p16INK4a and p14ARF were significantly increased in OCT4-RNAi cells
compared with control cells transfected with empty pFIV vector
(Fig. 2A). These results indicate
that OCT4 modulates p16INK4a and p14ARF transcription, and may
regulate cell cycle progression in MCF-7 cells.
Following OCT4 knockdown, mRNA expression of the
anti-apoptotic gene Bcl-2 was significantly downregulated, while
mRNA expression of the pro-apoptotic gene Bax was upregulated.
These results indicate that OCT4B can influence the Bcl-2/Bax ratio
and may regulate cell apoptosis.
Activated p53 is important in promoting apoptosis
via its negative regulation of Bcl-2 and positive regulation of Bax
(21,29). However, the results from our OCT4-RNAi
cell experiments revealed that p53 was positively regulated by OCT4
(Fig. 2). Additionally, expression of
p21, a downstream target gene of p53 functioning in cell cycle
arrest (18), was unchanged following
OCT4B interference.
The RT-qPCR results in MCF-7 cells (ER-positive),
together with the results from flow cytometry (Fig. 3), suggest that OCT4 may induce
p53-independent apoptosis. Furthermore, the similar transcriptional
changes induced by OCT4 in both the ER-negative mammary epithelial
cancer cell line mm468 (Fig. 2B) and
the ER-positive line MCF-7 (Fig. 2A)
suggest that OCT4 is a likely regulator of apoptosis in multiple
breast cancer cell lines regardless of their ER status.
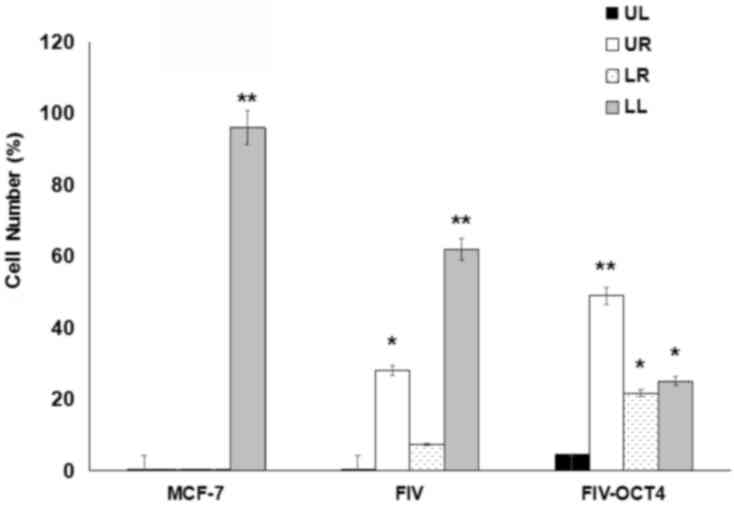
OCT4 inhibits apoptosis in MCF-7
cells
The current study next explored whether OCT4
regulates cell cycle progression or apoptosis in MCF-7 cells. To do
so, MCF-7 cells were transfected with OCT4 shRNA and then analyzed
by flow cytometry. Transfection of empty vector served as control.
Although no significant change in cell cycle distribution following
OCT4 knockdown was observed (data not shown), a significant change
in the rate of apoptosis was noticed. Cells transfected with OCT4
RNAi exhibited a greater degree of apoptosis (35.2%) than control
cells (Fig. 3 and Table I). Notably, it was possible to
distinguish early vs. late apoptotic cells in our analysis. These
results suggest that endogenous OCT4 may be involved in preventing
apoptosis in MCF-7 cells.
 | Table I.Apoptosis analysis of MCF-7 cells by
flow cytometry following OCT4 knockdown. |
Table I.
Apoptosis analysis of MCF-7 cells by
flow cytometry following OCT4 knockdown.
| Cells | MCF-7 | pFIV | OCT4 |
|---|
| UR (%) | 0.1 | 28.1 | 48.9 |
| LR (%) | 0.1 | 7.3 | 21.7 |
| Apoptosis (%) | 0.2 | 35.4 | 70.6 |
OCT4B and OCT4A regulate each other,
but OCT4A is not involved in OCT4B function
The observation that a large number of the breast
cancer cell lines examined in the present study, including MCF-7,
express both OCT4A and OCT4B raises the possibility that these two
OCT4 variants may cooperate at some level to regulate target gene
expression, cell proliferation and apoptosis. To examine this
possibility, overexpression vectors for both OCT4A and OCT4B were
constructed. It was observed that OCT4B overexpression increased
endogenous transcription of OCT4A; similarly, overexpression of
OCT4A increased endogenous transcription of OCT4B (Fig. 4). Despite this mutual regulation,
overexpression of each OCT4 variant had different effects on the
expression of p16INK4a and p53 pathway genes. While overexpression
of OCT4B significantly altered the levels of p53, p14ARF, p16INK4a,
Bcl-2 and Bax, overexpression of OCT4A was unable to do so
(Fig. 4C). These findings suggest
that OCT4B may serve a dominant role compared with OCT4A, or that
OCT4B may function via an independent pathway to regulate genes
involved in cell cycle progression and apoptosis in MCF-7
cells.
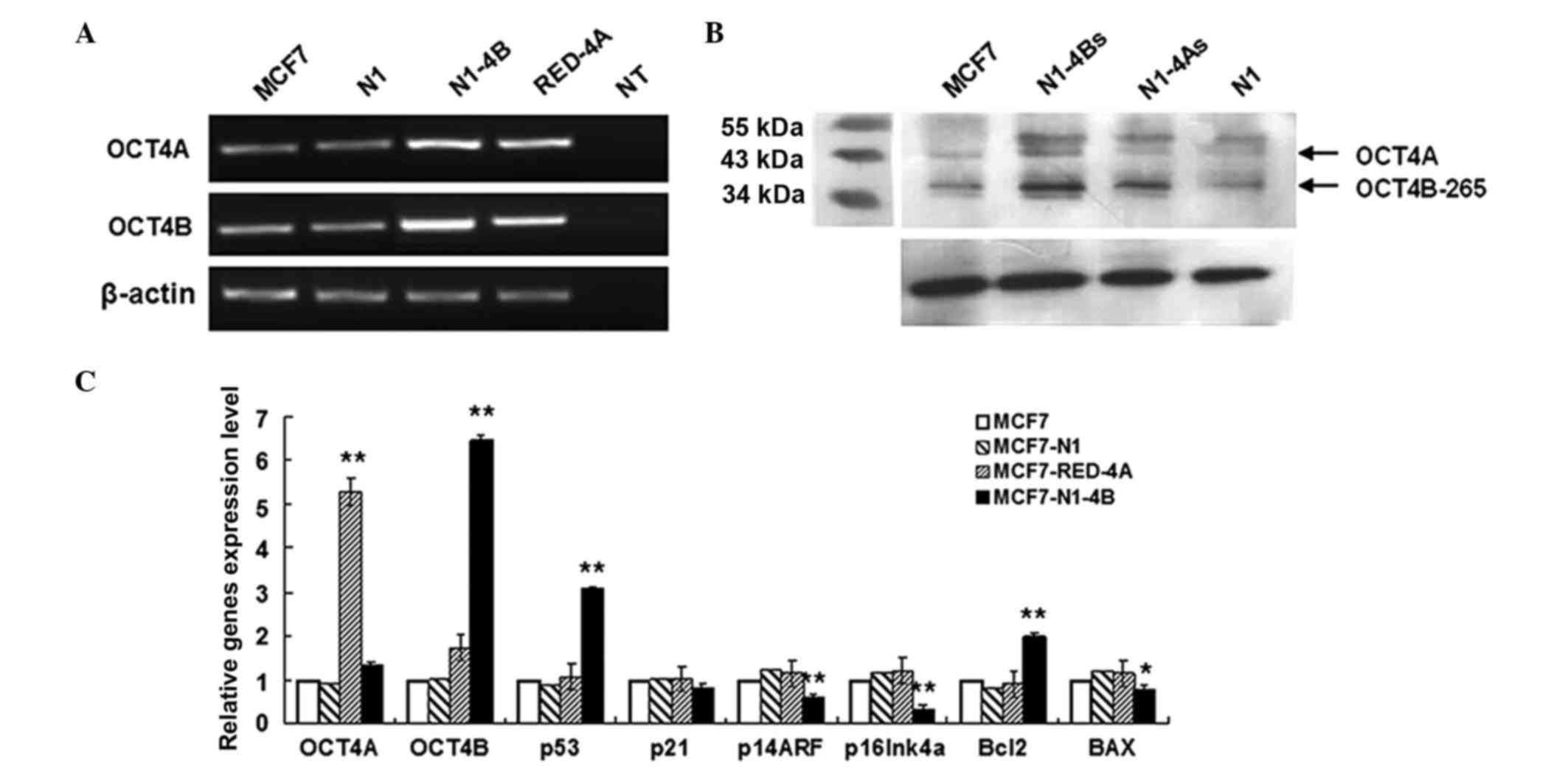 | Figure 4.Comparison of OCT4B and OCT4A in the
regulation of gene expression. (A) Reverse transcription-qPCR
analysis of the mutual regulation between OCT4B and OCT4A in MCF-7
cells. Overexpression of OCT4A led to increased expression of OCT4B
and vice versa. (B) Western blot analysis of the mutual regulation
between OCT4B and OCT4A in MCF-7 cells. (C) qPCR transcriptional
analysis of genes involved in cell cycle arrest and apoptosis
following OCT4A and OCT4B overexpression in MCF-7 cells. Of note,
cyclin-dependent kinase inhibitor 2A codes for two proteins: p16
(or p16INK4a) and p14ARF. Error bars represent the standard
deviation from three replicates. *P<0.05. **P<0.01. OCT4,
octamer-binding transcription factor 4; Bcl, B-cell lymphoma; Bax,
Bcl-2-associated X protein; qPCR, quantitative polymerase chain
reaction; N1, exogenous transcription of empty vector; N1-4B,
overexpression vector for OCT4B; Red-4A, overexpression vector for
OCT4A; NT, negative control; N1-4Bs, overexpression vector for
OCT4B; N1-4As, overexpression vector for OCT4A. |
Discussion
OCT4 is a key transcription factor for stem cell
self-renewal (5). As cells
differentiate, OCT4 is downregulated (6,30,31). However, it can become aberrantly
re-expressed in several cancers, including breast cancer (8,32). It was
observed that OCT4B is widely expressed across a panel of human
breast cancer cell lines, and that OCT4 regulates the expression of
factors such as p53 and p16, which control essential cell
regulatory pathways. The present data suggest that re-expressed
OCT4 may function as an oncogene in human breast cancer.
The expression of both OCT4A and OCT4B was examined
in a panel of breast cancer cell lines, and evidence for
re-expression of both variants was detected. However, the ratio of
OCT4A to OCT4B was observed to vary among different cell lines.
This could be an important factor for cancer cell biology, since
the two variants may influence one another's function; the data
provided in the present manuscript supports this claim, as does
recent work from other groups (33).
Although OCT4A and OCT4B may influence one another, the exact
mechanisms governing this interaction and the biological
consequence of this interaction require additional investigation.
It should also be noted that OCT4 was predominantly located in the
cytoplasm of the four breast cancer cell lines examined. This is of
interest, given its canonical function as a transcription factor,
and raises the possibility that OCT4 may have functions outside the
nucleus as well.
The present study revealed that OCT4 regulates the
expression of several factors, which control essential cell
regulatory pathways such as proliferation and apoptosis. For
example, RNAi-mediated knockdown of OCT4 in breast cancer cells
results in upregulation of the cell cycle inhibitors p16INK4a and
p14ARF. OCT4 knockdown also results in upregulation of the
pro-apoptotic factor Bax and downregulation of the anti-apoptotic
factor Bcl-2. Notably, the gene expression changes in
apoptosis-related genes correlate with the biological outcome. That
is, OCT4 knockdown cells exhibit increased rates of apoptosis. In
contrast, despite transcriptional changes in the cell cycle
regulators p16INK4a and p14ARF, no alteration was observed in cell
cycle progression in breast cancer cells depleted of OCT4. This
suggests that OCT4 may cooperate with other factors to control cell
cycle and proliferation, and that this cooperation may exert a more
robust effect on apoptosis than OCT4 alone. This influence on
apoptosis may be p53-independent, since OCT4 knockdown results in
decreased mRNA expression of p53. In addition, the p53-target gene
p21 is unaltered following OCT4 knockdown. Although the exact
mechanisms governing the influence of OCT4 on apoptosis require
additional research, these findings suggest that it may occur in a
p53-independent manner.
Additionally, the hyperactivity of mitogenic
oncogenes, such as c-Myc, adenovirus E1A and Ras, triggers a common
pathway of p53 accumulation via induction of the p14ARF
tumor-suppressor gene (34). Thus,
the inhibitory role of OCT4 in p14ARF transcription suggests that
OCT4 can antagonize oncogene-induced apoptosis.
p16INK4a and p14ARF are both encoded by the CDKN2A
locus but are under the control of independent promoters (15). Despite the fact that under certain
circumstances each gene can be independently regulated (16), various authors have shown that
p16INK4a and p14ARF can also be coordinately regulated in a process
that largely depends on chromatin remodeling events (35). In the present study, concomitant
transcriptional regulation of p16INK4a and p14ARF was detected in
cells with OCT4B overexpression or silencing, suggesting that OCT4B
may be involved in a chromatin remodeling event at this locus.
As aforementioned, OCT4A and OCT4B may cooperate at
certain level to regulate target gene expression, cell
proliferation and apoptosis. The present study demonstrated that
OCT4B overexpression increases the endogenous transcription of
OCT4A; similarly, overexpression of OCT4A increases the endogenous
transcription of OCT4B. Despite this mutual regulation,
overexpression of each OCT4 variant had markedly different effects
on the expression of p16INK4a and p53 pathway genes. While
overexpression of OCT4B significantly altered the levels of p53,
p14ARF, p16INK4a, Bcl-2 and Bax, overexpression of OCT4A was unable
to do so. These observations suggest that there is a certain
threshold of OCT4B that must be attained to elicit these specific
biological effects, and that this level is not achieved by OCT4A
overexpression. Thus, although overexpression of OCT4A increases
the levels of OCT4B, it does not cause a sufficient increase in
OCT4B levels and subsequently does not affect the transcription of
p16 or p53 target genes.
In conclusion, the OCT4B isoform serves a role in
the regulation of p53 and p16 pathway genes in breast cancer cells.
Although OCT4A and OCT4B may influence one another to regulate
breast cancer cell biology, OCT4B appears to be the dominant
player. OCT4B may serve as a tumor marker with potential
diagnostic, prognostic and therapeutic value. In conclusion, the
present results aid to elucidate a role for OCT4 in regulating the
proliferation and apoptosis of breast cancer cells. These findings
could be clinically important for patients with breast tumors in
which OCT4 becomes re-expressed.
References
|
1
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions: Nature Rev Cancer. 8:755–768. 2008.
|
|
2
|
Pece S, Tosoni D, Confalonieri S, Mazzarol
G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG and Di Fiore
PP: Biological and molecular heterogeneity of breast cancers
correlates with their cancer stem cell content. Cell. 140:62–73.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu R, Wang X, Chen GY, Dalerba P, Gurney
A, Hoey T, Sherlock G, Lewicki J, Shedden K and Clarke MF: The
prognostic role of a gene signature from tumorigenic breast-cancer
cells. N Engl J Med. 356:217–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu CG, Lu Y, Wang BB, Zhang YJ, Zhang RS,
Lu Y, Chen B, Xu H, Jin F and Lu P: Clinical implications of stem
cell gene Oct-4 expression in breast cancer. Ann Surg.
253:1165–1171. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Niwa HJ, Miyazaki J and Smith AG:
Quantitative expression of Oct-3/4 defines differentiation,
dedifferentiation or self-renewal of ES cells. Nat Genet.
24:372–376. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Okamoto K, Okazawa H, Okuda A, Sakai M,
Muramatsu M and Hamada H: A novel octamer binding transcription
factor is differentially expressed in mouse embryonic cells. Cell.
60:461–472. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Monsef N, Soller M, Isaksson M,
Abrahamsson PA and Panagopoulos I: The expression of pluripotency
marker Oct 3/4 in prostate cancer and benign prostate hyperplasia.
Prostate. 69:909–916. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Atlasi Y, Mowla SJ, Ziaee SA and Bahrami
AR: OCT-4, an embryonic stem cell marker, is highly expressed in
bladder cancer. Int J Cancer. 120:1598–1602. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ
and Andrews PW: OCT4 spliced variants are differentially expressed
in human pluripotent and nonpluripotent cells. Stem Cells.
26:3068–3074. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Papamichos SI, Kotoula V, Tarlatzis BC,
Agorastos T, Papazisis K and Lambropoulos AF: OCT4B1 isoform: The
novel OCT4 alternative spliced variant as a putative marker of
stemness. Mol Hum Reprod. 15:269–270. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X and Dai J: Concise review: Isoforms
of OCT4 contribute to the confusing diversity in stem cell biology.
Stem Cells. 28:885–893. 2010.PubMed/NCBI
|
|
12
|
Lee J, Kim HK, Rho JY, Han YM and Kim J:
The human OCT-4 isoforms differ in their ability to confer
self-renewal. J Biol Chem. 281:33554–33565. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kotoula V, Papamichos SI and Lambropoulos
AF: Revisiting OCT4 expression in peripheral blood mononuclear
cells. Stem Cells. 26:290–291. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Y, Wei J, Han J, Wang X, Su G, Zhao Y,
Chen B, Xiao Z, Cao J and Dai J: The novel function of OCT4B
isoform-265 in genotoxic stress. Stem Cells. 30:665–672. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xing EP, Nie Y, Song Y, Yang GY, Cai YC,
Wang LD and Yang CS: Mechanisms of inactivation of p14ARF,
p15INK4b, and p16INK4a genes in human esophageal squamous cell
carcinoma. Clin Cancer Res. 5:2704–2713. 1999.PubMed/NCBI
|
|
16
|
Li J, Poi MJ and Tsai MD: Regulatory
mechanisms of tumor suppressor P16(INK4A) and their relevance to
cancer. Biochemistry. 50:5566–5582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sherr CJ: Autophagy by ARF: A short story.
Mol Cell. 22:436–437. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sherr CJ: Divorcing ARF and p53: An
unsettled case. Nat Rev Cancer. 6:663–673. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agrawal A, Yang J, Murphy RF and Agrawal
DK: Regulation of the p14ARF-Mdm2-p53 pathway: An overview in
breast cancer. Exp Mol Pathol. 81:115–122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fisher DE: The p53 tumor suppressor:
critical regulator of life & death in cancer. Apoptosis.
6:7–15. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haupt S, Berger M, Goldberg Z and Haupt Y:
Apoptosis-the p53 network. J Cell Sci. 116:4077–4085. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chipuk JE, Kuwana T, Bouchier-Hayes L,
Droin NM, Newmeyer DD, Schuler M and Green DR: Direct activation of
Bax by p53 mediates mitochondrial membrane permeabilization and
apoptosis. Science. 303:1010–1014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao Y, Siegel D and Knöchel W:
Xenopus POU factors of subclass V inhibit activin/nodal
signaling during gastrulation. Mech Dev. 123:614–625. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao Y, Knöchel S, Donow C, Miethe J,
Kaufmann E and Knöchel W: The POU factor Oct-25 regulates the
Xvent-2B gene and counteracts terminal differentiation in
Xenopus embryos. J Biol Chem. 279:43735–43743. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hinkley CS, Martin JF, Leibham D and Perry
M: Sequential expression of multiple POU proteins during amphibian
early development. Mol Cell Biol. 12:638–649. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ng WL, Chen G, Wang M, Wang H, Story M,
Shay JW, Zhang X, Wang J, Amin AR, Hu B, et al: OCT4 as a target of
miR-34a stimulates p63 but inhibits p53 to promote human cell
transformation. Cell Death Dis. 5:e10242014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Meng L, Hu H, Zhang Y, Zhao C, Li
Q, Shi F, Wang X and Lin A: Oct-4B isoform is differentially
expressed in breast cancer cells: Hypermethylation of regulatory
elements of Oct-4A suggests an alternative promoter and
transcriptional start site for Oct-4B transcription. Biosci Rep.
31:109–115. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agarwal ML, Taylor WR, Chernov MV,
Chernova OB and Stark GR: The p53 network. J Biol Chem. 273:1–4.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pesce M, Gross MK and Schöler HR: In line
with our ancestors: Oct-4 and the mammalian germ. Bioessays.
20:722–732. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schöler HR, Ruppert S, Suzuki N, Chowdhury
K and Gruss P: New type of POU domain in germ line-specific protein
Oct-4. Nature. 344:435–439. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang P, Branch DR, Bali M, Schultz GA,
Goss PE and Jin T: The POU homeodomain protein OCT3 as a potential
transcriptional activator for fibroblast growth factor-4 (FGF-4) in
human breast cancer cells. Biochem J. 375:199–205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li D, Yang ZK, Bu JY, Xu CY, Sun H, Tang
JB, Lin P, Cheng W, Huang N, Cui RJ, et al: OCT4B modulates OCT4A
expression as ceRNA in tumor cells. Oncol Rep. 33:2622–2630. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sherr CJ and Weber JD: The ARF/p53
pathway. Curr Opin Genet Dev. 10:94–99. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim WY and Sharpless NE: The regulation of
INK4/ARF in cancer and aging. Cell. 127:265–275. 2006. View Article : Google Scholar : PubMed/NCBI
|