Introduction
The incidence and mortality of lung cancer is the
highest among all malignant tumors worldwide; it is a disease that
severely threatens human health and affects quality of life
(1,2).
Non-small cell lung cancer (NSCLC) accounts for ~80% of all lung
cancer cases. The efficacy of surgical resection, adjuvant
radiochemotherapy, immunotherapy and individualized targeted
therapy have improved in recent years; however, the 5-year survival
rate of NSCLC patients remains ~15% (3). The occurrence and development of lung
cancer is closely associated with the biological behavior of lung
cancer cells, although the molecular mechanism that drives the
disease remains unclear (4–6). It is widely accepted that the
proliferation, invasion and metastasis of tumor cells, and the
alteration of intrinsic cellular processes such as apoptosis and
the cell cycle, promotes the occurrence and development of NSCLC
(7,8).
The molecular regulatory mechanism of the biological behavior of
lung cancer cells is extremely complex. Therefore, identification
of and screening for tumor-associated genes, together with the
study of their molecular mechanisms, have considerable clinical
value in the diagnosis and treatment of NSCLC.
Homeobox-containing genes are highly evolutionarily
conserved; proteins encoded by homeobox-containing genes are
important transcription factors that precisely regulate the
differentiation of cells by specifically activating downstream
genes (9,10). Studies have indicated that
homeobox-containing genes serve key roles in the generation and
differentiation of embryos and organs of organisms. In addition,
homeobox-containing genes are important for the regulation of the
development and differentiation of adult tissues (11). In recent years, numerous studies have
confirmed that HOX genes, a subclass of homeobox-containing genes,
also perform important biological functions in the occurrence and
development of a variety of tumors (12,13). For
example, Zhong et al (14)
reported that the methylation of the HOXD13 gene is an important
marker in the early diagnosis of breast cancer (14). In addition, Aquino et al
(15) revealed that 13
homeobox-containing genes exhibit abnormal expression in oral
squamous cell carcinoma (15). The
members of homeobox-containing genes are numerous and have complex
structures. Their functions and molecular mechanisms of action in
tumors require further investigation. The engrailed homeobox (EN)
family of genes includes EN1 and EN2. EN2 has been reported to
serve an important role in the development of embryos and the
nervous system (16). It has been
demonstrated that EN2 is abnormally expressed in prostate, breast
and bladder cancer, and is closely associated with the procession
of tumors (17,18). However, the expression level and role
of EN2 in NSCLC remains unclear. In the present study, the
expression and mechanism of action of EN2 in NSCLC was investigated
using reverse transcription-quantitative polymerase chain reaction
(RT-qPCR), western blot analysis, and a Transwell assay. The
present study provided an experimental basis for screening
biological targets for the diagnosis and treatment of NSCLC.
Materials and methods
Patients
A total of 42 patients (31 males and 11 females) who
received surgical resection of NSCLC tissues between January 2014
and January 2015 were included in the present study. All patients
were clearly diagnosed with NSCLC by pathologists. NSCLC and
tumor-adjacent normal tissues were collected from all NSCLC
patients. The age range of the patients was 27–65 years and the
mean age was 41.7±2.4 years. Patients were included if they were
without: Any other tumors; a long history of drug intake;
autoimmune diseases; and adjuvant therapy prior to surgery. Among
the 42 patients, 25 had lymphatic metastasis and 17 did not. The
Tumor-Node-Metastasis (TNM) staging followed NSCLC TNM staging
criteria of American Joint Committee on Cancer 2003 edition
(19). Of the 42 patients, 11 had
stage I disease, 18 had stage II disease and 13 had stage III
disease. All procedures were approved by the ethics committee of
Jining No. 1 People's Hospital (Jining, China). Written informed
consent was obtained from all patients or their families.
Cells and transfection
The lung cancer A549 cell line was purchased from
the Type Culture Collection Chinese Academy of Sciences (Shanghai,
China). A549 cells were defrosted at 37°C and cultured in 10 ml
fresh Dulbecco's Modified Eagle's Medium (DMEM; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) medium at 37°C and 5%
CO2 for 24 h. The old medium was discarded; 5 ml fresh
high-glucose DMEM medium supplemented with 10% fetal bovine serum
was added (Hyclone; GE Healthcare Life Sciences). The medium was
changed every 2 days and the cells were passaged at 90%
confluence.
A549 cells were cultured in DMEM medium and divided
into negative control (NC) and pcDNA-3.1-EN2 groups. At 70–90%
confluence, 0.5 µg NC or pcDNA-3.1-EN2 plasmids (Hanbio
Biotechnology Co., Ltd., Shanghai, China) were mixed with 1 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and the mixture was added into
two individual vials containing 50 µl OptiMemi medium (Hyclone; GE
Healthcare Life Sciences). After 5 min, the liquids in the two
vials were mixed prior to standing for another 15 min. Next, the
mixture was added to the cells, which were incubated for 6 h.
Following replacement of the DMEM medium supplemented with 10%
fetal bovine serum, the cells were cultured under normal conditions
for 48 h prior to use.
RT-qPCR
Tissues (100 mg) were ground into powder following
freezing with liquid nitrogen prior to the addition of 1 ml TRIzol
(Thermo Fisher Scientific, Inc.) for lysis. Subsequently, total RNA
was extracted using phenol chloroform method (20). The purity of RNA was determined at
A260/A280 using ultraviolet spectrophotometry
(Nanodrop ND1000, Thermo Fisher Scientific, Inc., Wilmington, DE,
USA). Next, cDNA was obtained by RT using a PrimeScript RT reagent
kit (Takara Biotechnology Co., Ltd., Dalian, China) from 1 µg RNA
and stored at −20°C. The RT reaction system RNA constituted 5 µl
RNA template, 2 µl Oligo (dT), 2 µl super pure dNTP, and 5.5 µl
ddH2O. The reaction system was first heated to 70°C and
then kept on ice for 2 min. Following centrifugation at 1,000 × g
at 4°C for 30 sec, 4 µl 5 X first-strand buffer and 0.5 µl RNasin
(Tiangen Biotech Co., Ltd., Beijing, China) were added, followed by
addition of 1 µl TIANScript M-MLV (Tiangen Biotech Co., Ltd.). The
mixture was then incubated at 25°C for 10 min and 42°C for 50 min;
at 95°C for 5 min the stop reaction was conducted. Subsequently,
the samples were kept on ice and 5 µl was used for RT-qPCR.
To determine the expression of EN2 in tumor and
tumor-adjacent tissues, the Quant One Step qRT-PCR (SYBR-Green) kit
(Tiangen Biotech Co., Ltd.) was used, with GAPDH serving as the
internal reference. The RT-qPCR reaction system (30 µl) contained 5
µl cDNA, 10 µl mix, 1 µl upstream primer, 1 µl downstream primer,
and 13 µl ddH2O. The primers used were: EN2 forward,
5′-CTACTGTACGCGCTACTCGG-3′ and reverse, 5′-CCCGTGGCCTTCTTGATCTT-3′;
GAPDH forward, 5′-AGGTGAAGGTCGGAGTCAAC-3′ and reverse,
5′-CGCTCCTGGAAGATGGTGAT-3′. The PCR protocol was as follows:
Initial denaturation at 95°C for 10 min; 40 cycles of denaturation
at 95°C for 30 sec and annealing at 60°C for 30 sec (iQ5; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The 2−ΔΔCq
method (21) was used to calculate
the relative expression of EN2 mRNA against GAPDH; each sample was
tested in triplicate.
Cell Counting kit-8 (CCK-8) assay
For the growth curve assay, A549 cells were seeded
into 96-well plates at a density of 5,000 cells per well in
triplicate. Every 24 h, the cells were incubated with CCK-8 reagent
(Beyotime Institute of Biotechnology, Shanghai, China) at 37°C for
30 min. Absorbance at 490 nm was read on a microplate reader
(168–1,000; Model 680, Bio-Rad Laboratories, Inc.) at 24, 48 and 72
h, and proliferation curves were plotted using absorbance values at
each time point.
Transwell assay
Transwell chambers (8 µm diameter and 24 wells;
Corning Inc., Corning, NY, USA) were used to evaluate the migration
ability of A549 cells. Transfected cells were collected by trypsin
digestion and resuspended to a density of 2×105 cells/ml
using serum-free DMEM. The cell suspension (200 µl per well) was
added into the upper chamber. In the lower chamber, 500 µl DMEM
medium supplemented with 10% fetal bovine serum was added.
Following incubation for 24 h, cells in the upper chamber were
removed with a cotton swab. Next, the chamber was fixed using 4%
formaldehyde for 10 min at room temperature and then subjected to
Giemsa staining at room temperature for 1 min. Following three
washes of the cells that migrated to the other side of the chamber
with phosphate-buffered saline (PBS), these cells were counted in 5
individual fields under a light microscope (magnification, ×200) to
evaluate migration ability.
Matrigel invasion chambers (BD Biosciences, Franklin
Lakes, NJ, USA) were used to determine the invasive ability of
cells. Matrigel was first diluted with serum-free DMEM medium at a
ratio of 1:2. In the upper chamber, 50 µl diluted Matrigel was
added and maintained at 37°C for 1 h. In the lower chamber, 500 µl
DMEM medium supplemented with 10% fetal bovine serum was added.
Following incubation at 37°C for 24 h, the cells (1×105)
in upper chamber were removed with a cotton swab. Next, cells in
the chamber was fixed using 4% formaldehyde for 10 min at room
temperature and then subjected to Giemsa staining at room
temperature for 1 min. Following three washes of the cells that
moved to the other side of the chamber with PBS, these cells in
five fields were counted under a light microscope (magnification,
×200) to evaluate invasion ability.
Flow cytometry
At 24 h post-transfection, 1×106 cells of
each group were washed twice with precooled PBS. A Cell Cycle Assay
kit (BD Biosciences) was used to determine the cell cycle according
to the manufacturer's protocols. The cells were incubated at room
temperature with 200 µl liquid A for 10 min, and 150 µl liquid B
for another 10 min. Then, the cells were incubated at room
temperature with 120 µl liquid C in the dark for 10 min prior to
flow cytometry analysis using FACSVerse™ flow cytometer
(BD Biosciences). The results were analyzed using ModFit software
version 3.2 (Verity Software House, Inc., Topsham, ME, USA).
Western blotting
At 48 h after transfection, A549 cells were
trypsinized and collected. Then, precooled radioimmunoprecipitation
assay lysis buffer (600 µl; 50 mM Tris-base, 1 mM EDTA, 150 mM
NaCl, 0.1% SDS, 1% Triton X-100, and 1% sodium deoxycholate;
Beyotime Institute of Biotechnology) and phenylmethylsulfonyl
fluoride were added to the samples. Following lysis for 5 min on
ice, the mixture was centrifuged at 12,000 × g and 4°C for 10 min.
Protein samples (50 µg per lane) were then mixed with equal volume
of 2 X SDS loading buffer prior to denaturation in boiling water
bath for 10 min. Subsequently, the samples (5 µl) were subject to
10% SDS-PAGE at 100 V. The resolved proteins were transferred to
polyvinylidene difluoride membranes on ice (300 mA, 2 h) and
blocked with 50 g/l skimmed milk at room temperature for 1 h. Next,
the membranes were incubated with rabbit anti-human EN2 polyclonal
primary antibody (1:1,000; cat. no. ab28731; Abcam, Cambridge, UK)
and rabbit anti-human GAPDH primary antibody (1:5,000; cat. no.
ab9485; Abcam) at 4°C overnight. Following five washes with PBS
with 0.1% Tween-20 for 5 min, the membranes were incubated with
goat anti-mouse (1:5,000; cat. no. ab6789) and goat anti-rabbit
(1:2,000; cat. no. ab205718; both Abcam) horseradish
peroxidase-conjugated secondary antibodies, respectively, for 1 h
at room temperature prior to five washes with PBS with Tween-20 for
5 min. Next, the membrane was developed with an enhanced
chemiluminescence detection kit (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for imaging. Image lab v3.0 software (Bio-Rad
Laboratories, Inc.) was used to acquire and analyze imaging
signals. The relative content of EN2 protein was expressed as the
EN2/GAPDH ratio. For the measurement of relative β-Catenin
expression, polyclonal rabbit anti-human β-Catenin primary antibody
(1:1,000; cat. no. AF0066) and polyclonal rabbit anti-human H3
histone primary antibody (1:3,000; cat. no. AF0009; both Beyotime
Institute of Biotechnology) were incubated according to the western
blotting protocol used for EN2, and H3 histone was used as internal
reference.
Statistical analysis
The results were analyzed using SPSS 16.0
statistical software (SPSS, Inc., Chicago, IL, USA). Each test was
performed in triplicate. All measurement data were expressed as the
mean ± standard deviation. Data were tested for normality;
comparisons between two groups were performed using paired t-test.
Multigroup measurement data were analyzed using one-way analysis of
variance. In case of homogeneity of variance, the least significant
difference and Student-Newman-Keuls post hoc methods were used; in
case of heterogeneity of variance, Tamhane's T2 or Dunnett's T3
post hoc methods were used. P<0.05 was considered to indicate a
statistically significant difference.
Results
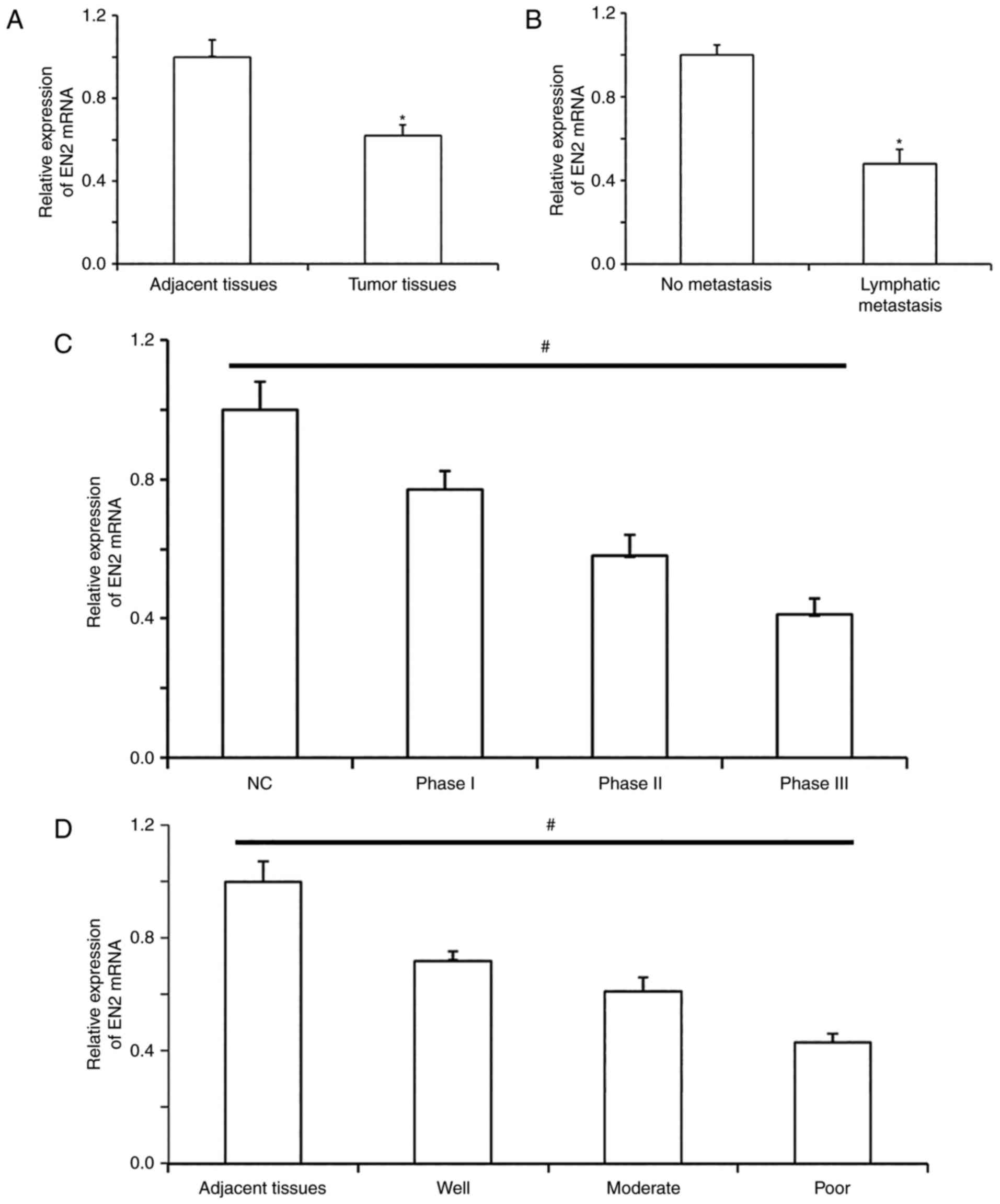
EN2 mRNA expression levels in NSCLC
tissues are lower than those in normal tissues and are associated
with the metastasis, clinical staging and differentiation degrees
of NSCLC
To measure the expression levels of EN2 mRNA in
NSCLC tissues, RT-qPCR was utilized. EN2 mRNA expression levels in
NSCLC tissues were significantly lower than those in tumor-adjacent
tissues (Fig. 1A). In addition, the
expression levels of EN2 mRNA in patients with lymphatic metastasis
were significantly lower than those in patients without lymphatic
metastasis (Fig. 1B). When assessing
the association between E2 expression and clinicopathological
characteristics, EN2 expression gradually decreased with the
increases in TNM staging (Fig. 1C)
and differentiation degrees (Fig.
1D). The results indicated that EN2 mRNA expression levels in
NSCLC tissues were lower than those in normal tissues and may be
associated with the metastasis, clinical staging and the degree of
differentiation of NSCLC.
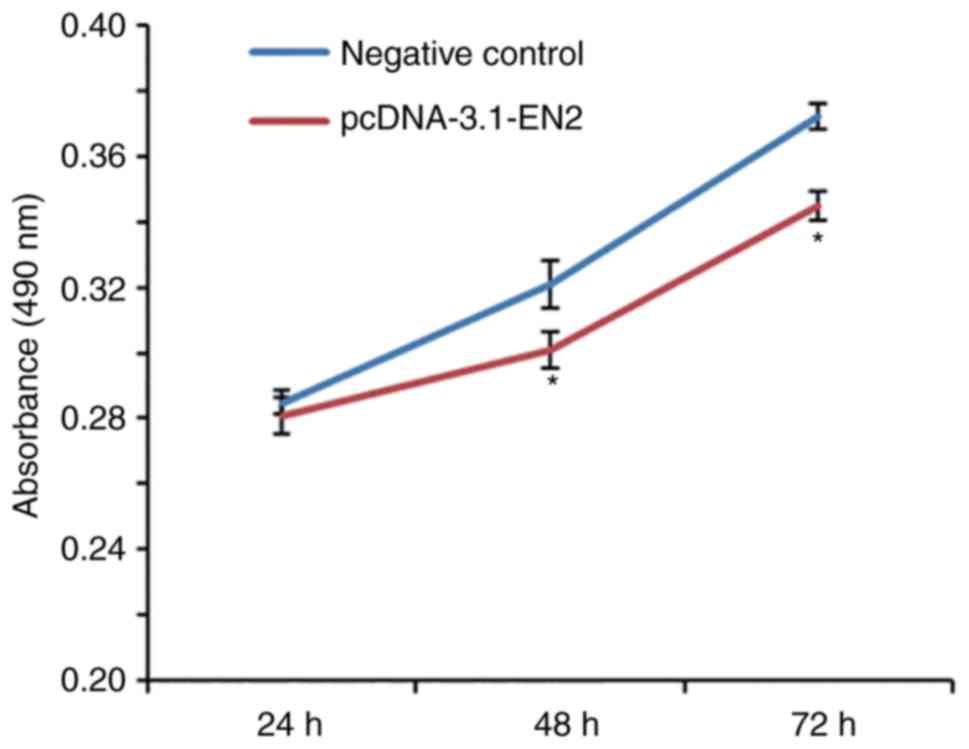
Increased expression of EN2 inhibits
the proliferation of A549 cells in vitro
To investigate the effect of EN2 on the
proliferation of A549 cells, a CCK-8 assay was performed. The data
revealed that the absorbance of cells transfected with
pcDNA-3.1-EN2 was significantly lower than that of cells in NC
group at 48 and 72 h (Fig. 2),
indicative of a reduced cellular proliferation. The results
indicated that increased expression of EN2 inhibited the
proliferation of A549 cells in vitro.
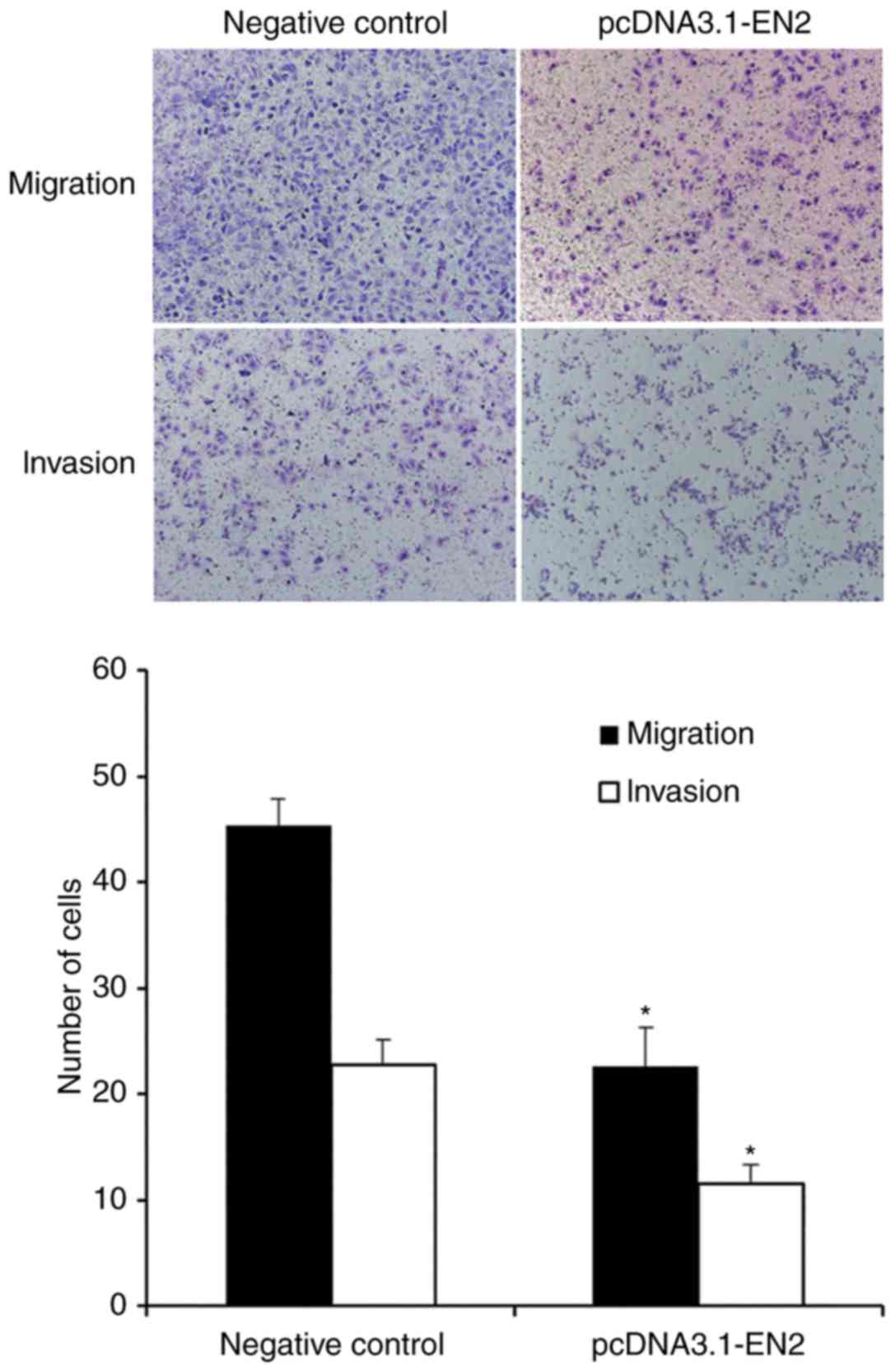
Overexpression of EN2 suppresses the
migration and invasion of A549 cells
To study the migration and invasive abilities of
A549 cells, a Transwell assay was performed. Transwell migration
and invasion assays demonstrated that the number of A549 cells
transfected with pcDNA-3.1-EN2 that crossed the membrane was lower
than the number of cells in the negative control group (Fig. 3). These results indicated that the
overexpression of EN2 may suppress the migration and invasion of
A549 cells.
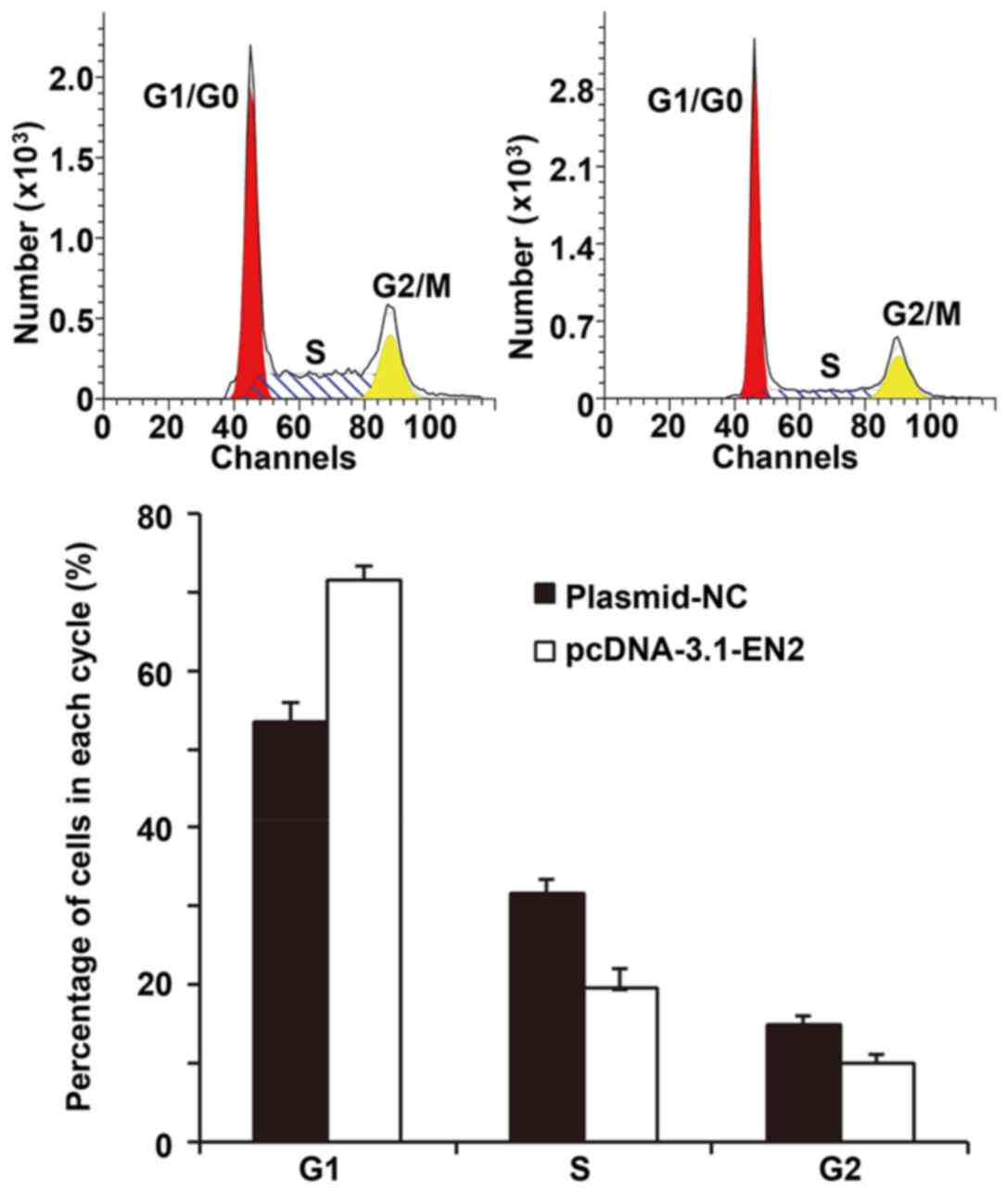
Elevated EN2 expression inhibits the
proliferation of A549 cells by regulating the G1/S phase
transition
To investigate the effect of EN2 on the A549 cell
cycle, flow cytometry was conducted. The data revealed that A549
cells overexpressing EN2 exhibited G1/S phase arrest,
unlike the negative control cells (Fig.
4). The results indicated that elevated EN2 expression might
inhibit the proliferation of A549 cells by regulating
G1/S phase transition.
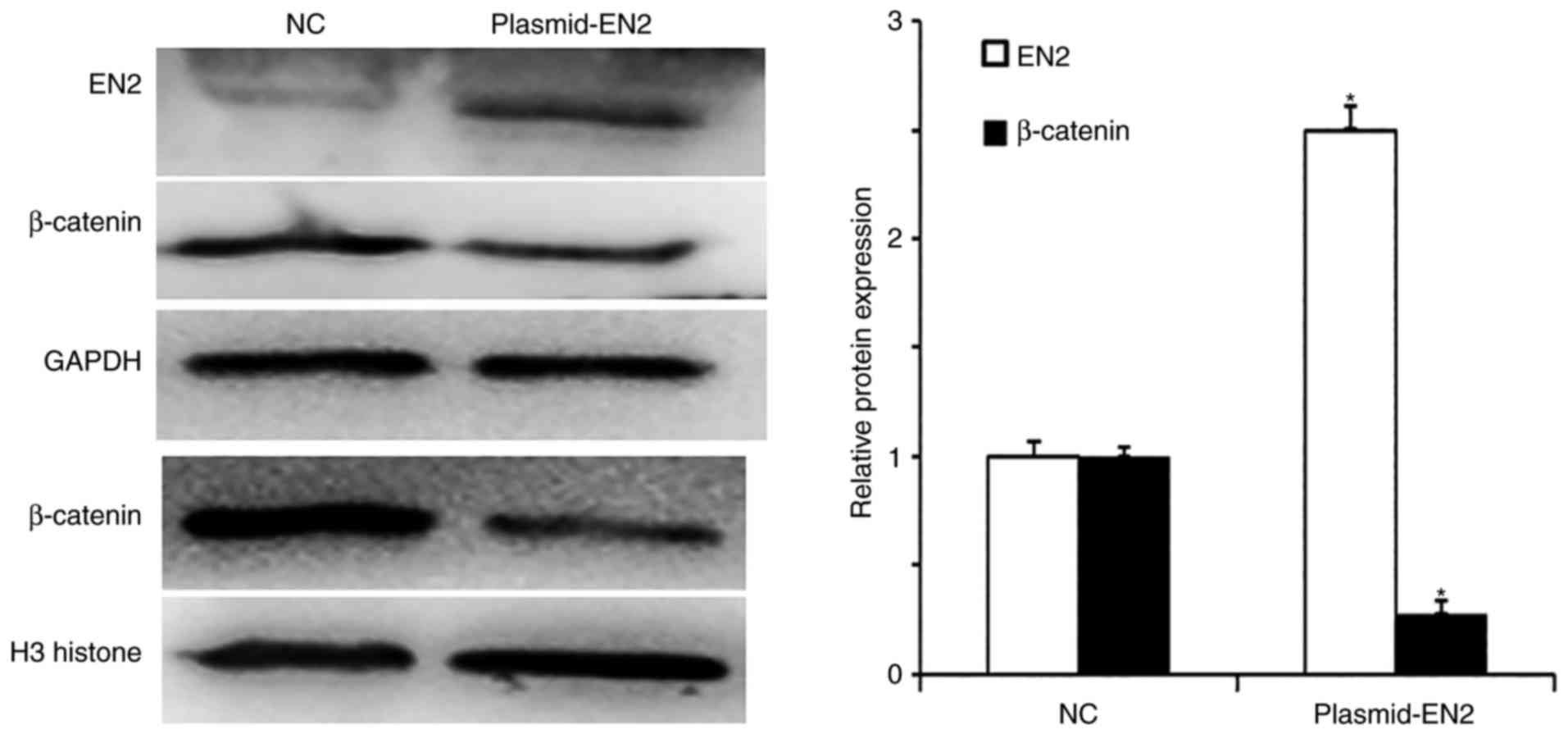
β-catenin protein expression and
nuclear translocation in A549 cells are inhibited by EN2
overexpression
To determine protein expression in A549 cells,
western blotting was conducted. Following transfection with the
pcDNA3.1-EN2 plasmid, EN2 protein expression levels in A549 cells
were significantly enhanced. In addition, β-catenin protein
expression levels were downregulated as EN2 expression levels
increased (Fig. 5). The results
indicated that β-catenin protein expression levels and nuclear
translocation in A549 cells might be inhibited by EN2
overexpression.
Discussion
The results of the present study demonstrated that
EN2 may act as a tumor-suppressor gene in NSCLC and that reduced
expression of EN2 may be closely associated with the occurrence and
development of NSCLC. It has been reported that EN2 may serve a
notable role in the early development of embryos and the
maintenance of the function of nervous system, and is closely
associated with the development of the nervous system and autism
(22). EN2 has also been demonstrated
to be closely associated with the occurrence and development of
tumors, potentially acting as a tumor-suppressor gene in certain
types of cancer. Martin et al (23) reported that the expression of EN2 in
breast cancer cells was upregulated and might have promoted the
proliferation of tumor cells. Bose et al (24) revealed that EN2 expression levels were
upregulated in prostate cancer cell lines and that the silencing of
EN2 expression may inhibit the proliferation of prostate cancer
cells via the downregulation of PAX2 gene expression. Morgan et
al (25) demonstrated that EN2
content in urine from bladder cancer patients was elevated to
levels that were of clinical diagnostic value. In addition, Lai
et al (26) reported that EN2
expression was downregulated in renal clear cell carcinoma and was
negatively associated with clinicopathological staging, further
indicating that EN2 may be a tumor-suppressor gene. These studies
indicated that the biological functions of EN2 might be distinct in
different types of tumors.
At present, there is one report on EN2 expression in
NSCLC (27). The results of the
present study revealed that EN2 expression levels were
downregulated in NSCLC, and were negatively associated with
lymphatic metastasis, degree of differentiation and TNM staging,
indicating that EN2 may have tumor-suppressor gene functions in
NSCLC. In vitro experiments demonstrated that the
overexpression of EN2 in A549 cells might inhibit the
proliferation, migration and invasion of A549 cells. Flow cytometry
conducted in the present study revealed that EN2 inhibited the
G1/S transition of A549 cells, indicating that the
downregulation of EN2 facilitates the occurrence and development of
NSCLC.
The Wnt signaling pathway is closely associated with
cell proliferation, differentiation, migration and invasion. As a
key protein in the Wnt signaling pathway, β-catenin aggregates in
the cytoplasm, enters the cell nucleus, exerts its function as a
transcription factor and activates the transcription and expression
of downstream genes (28). In
addition, β-catenin can also bind with epithelial marker E-cadherin
and mediate the migration of cells (29). Western blotting data reported in the
present study demonstrated that EN2 overexpression led to the
downregulation of β-catenin protein expression levels in A549
cells; β-catenin expression levels in the nucleus were
significantly reduced, indicating that the upregulation of EN2 may
inhibit the activity of the Wnt signaling pathway, in addition to
the migration and invasion of A549 cells.
In conclusion, the present study demonstrated that
downregulated EN2 expression might be closely associated with the
occurrence and development of NSCLC. Overexpression of EN2 may
inhibit the proliferation, migration and invasion of NSCLC,
possibly due to the inhibition of Wnt signaling pathway by EN2.
Therefore, EN2 has to potential to be considered to be a potential
diagnostic marker and therapeutic target for NSCLC.
Acknowledgements
The authors would like to thank Dr Cheng Zhang of
the Affiliated Hospital of Jining Medical University (Jining,
China) for their instructions.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XXL, XCL and CQG designed the study; XXL and XCL
performed the experiments; XXL, XCL and CQG analyzed the data. All
authors collaborated to interpret results and develop the
manuscript. The final version of the manuscript has been read and
approved by all authors.
Ethics approval and consent to
participate
All procedures were approved by the ethics committee
of Jining No. 1 People's Hospital (Jining, China). Written informed
consent was obtained from all patients or their families.
Consent for publication
Written informed consent was obtained from all
patients or their families.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang YZ, Chen X, Fan XX, He JX, Huang J,
Xiao DK, Zhou YL, Zheng SY, Xu JH, Yao XJ, et al: Compound library
screening identified cardiac glycoside digitoxin as an effective
growth inhibitor of gefitinib-resistant non-small cell lung cancer
via downregulation of α-tubulin and inhibition of microtubule
formation. Molecules. 21:3742016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng H, Wang M, Wu J, Wang ZM, Nan HJ and
Sun H: Inhibition of mTOR enhances radiosensitivity of lung cancer
cells and protects normal lung cells against radiation. Biochem
Cell Biol. 94:213–220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trinidad López C, Bayarri Souto M, Pernas
Oca R, Sánchez-Gracián Delgado C, Vázquez González M, Liste
Vaamonde A, De La Fuente Tardáguila G and De La Fuente Aguado J:
Characteristics of computed tomography perfusion parameters in
non-small-cell-lung-cancer and its relationship to histology, size,
stage an treatment response. Clin Imaging. 50:5–12. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei H, Su M, Lin R, Li H and Zou C:
Prognostic factors analysis in EGFR mutation-positive non-small
cell lung cancer with brain metastases treated with whole
brain-radiotherapy and EGFR-tyrosine kinase inhibitors. Oncol Lett.
11:2249–2254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao T, Gao Z, Wu W, He W and Yang YI:
Effect of synchronous solitary bone metastasectomy and lung cancer
resection on non-small cell lung cancer patients. Oncol Lett.
11:2266–2270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee P, Leung CC, Restrepo MI, Takahashi K,
Song Y and Porcel JM: Year in review 2015: Lung cancer, pleural
diseases, respiratory infections, bronchiectasis and tuberculosis,
bronchoscopic intervention and imaging. Respirology. 21:961–967.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matsumoto M, Nakajima W, Seike M, Gemma A
and Tanaka N: Cisplatin-induced apoptosis in non-small-cell lung
cancer cells is dependent on Bax- and Bak-induction pathway and
synergistically activated by BH3-mimetic ABT-263 in p53 wild-type
and mutant cells. Biochem Biophys Res Commun. 473:490–496. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rocco G, Nason K, Brunelli A, Varela G,
Waddell T and Jones DR: Management of stage IIIA (N2) non-small
cell lung cancer: A transatlantic perspective. J Thorac Cardiovasc
Surg. 151:1235–1238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Parrillo L, Costa V, Raciti GA, Longo M,
Spinelli R, Esposito R, Nigro C, Vastolo V, Desiderio A, Zatterale
F, et al: Hoxa5 undergoes dynamic DNA methylation and
transcriptional repression in the adipose tissue of mice exposed to
high-fat diet. Int J Obes (Lond). 40:929–937. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morgan R and El-Tanani M: Hox genes as
potential markers of circulating tumour cells. Curr Mol Med.
16:322–327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sandberg ES, Calikoglu AS, Loechner KJ and
Snyder LL: Short stature homeobox-containing haploinsufficiency in
seven siblings with short stature. Case Rep Endocrinol.
2017:72873512017.PubMed/NCBI
|
|
12
|
Catela C, Shin MM, Lee DH, Liu JP and
Dasen JS: Hox proteins coordinate motor neuron differentiation and
connectivity programs through ret/gfralpha genes. Cell Rep.
14:1901–1915. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krumlauf R: Hox genes and the hindbrain: A
study in segments. Curr Top Dev Biol. 116:581–596. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhong Z, Shan M, Wang J, Liu T, Xia B, Niu
M, Ren Y and Pang D: HOXD13 methylation status is a prognostic
indicator in breast cancer. Int J Clin Exp Pathol. 8:10716–10724.
2015.PubMed/NCBI
|
|
15
|
Aquino G, Franco R, Sabatino R, Mantia EL,
Scognamiglio G, Collina F, Longo F, Ionna F, Losito NS, Liguori G,
et al: Deregulation of paralogous 13 HOX genes in oral squamous
cell carcinoma. Am J Cancer Res. 5:3042–3055. 2015.PubMed/NCBI
|
|
16
|
DiCicco-Bloom E, Lord C, Zwaigenbaum L,
Courchesne E, Dager SR, Schmitz C, Schultz RT, Crawley J and Young
LJ: The developmental neurobiology of aut-ism spectrum disorder. J
Neurosci. 26:6897–6906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ranzi AD, da Silva JN, Graziottin TM,
Annels N and Bica CG: Immunohistochemistry biomarkers in nonmuscle
invasive bladder cancer. Appl Immunohistochem Mol Morphol.
25:178–183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McGrath SE, Michael A, Morgan R and Pandha
H: EN2 in prostate cancer. Adv Clin Chem. 71:47–76. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kaur H, Sehgal IS, Bal A, Gupta N, Behera
D, Das A and Singh N: Evolving epidemiology of lung cancer in
India: Reducing non-small cell lung cancer-not otherwise specified
and quantifying tobacco smoke exposure are the key. Indian J
Cancer. 54:285–290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cepollaro S, Della Bella E, de Biase D,
Visani M and Fini M: Evaluation of RNA from human trabecular bone
and identification of stable reference genes. J Cell Physiol.
233:4401–4407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-(Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Omi M, Harada H, Watanabe Y, Funahashi J
and Nakamura H: Role of En2 in the tectal laminar formation of
chick embryos. Development. 141:2131–2138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martin NL, Saba-El-Leil MK, Sadekova S,
Meloche S and Sauvageau G: EN2 is a candidate oncogene in human
breast cancer. Oncogene. 24:6890–6901. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bose SK, Bullard RS and Donald CD:
Oncogenic role of engrailed-2 (en-2) in prostate cancer cell growth
and survival. Transl Oncogenomics. 3:37–43. 2008.PubMed/NCBI
|
|
25
|
Morgan R, Bryan RT, Javed S, Launchbury F,
Zeegers MP, Cheng KK, James ND, Wallace DM, Hurst CD, Ward DG, et
al: Expression of Engrailed-2 (EN2) protein in bladder cancer and
its potential utility as a urinary diagnostic biomarker. Eur J
Cancer. 49:2214–2222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lai CY, Pan B, Luo Y, Liang WB, Chen J, Ye
DM, Guo JN, Li L and Su ZX: Engrailed-2 is down-regulated but also
ectopically expressed in clear cell renal cell carcinoma. Mol Biol
Rep. 41:3651–3657. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu W, Banerji S, Davie JR, Kassie F, Yee D
and Kratzke R: Yin yang gene expression ratio signature for lung
cancer prognosis. PLoS One. 8:e687422013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Togel L, Nightingale R, Chueh AC,
Jayachandran A, Tran H, Phesse T, Wu R, Sieber OM, Arango D,
Dhillon AS, et al: Dual targeting of bromodomain and extra-terminal
domain proteins, and WNT or MAPK signaling, inhibits c-MYC
expression and proliferation of colorectal cancer cells. Mol Cancer
Ther. 15:1217–1226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sempou E, Biasini E, Pinzon-Olejua A,
Harris DA and Malaga-Trillo E: Activation of zebrafish Src family
kinases by the prion protein is an amyloid-β-sensitive signal that
prevents the endocytosis and degradation of E-cadherin/β-catenin
complexes in vivo. Mol Neurodegener. 11:182016. View Article : Google Scholar : PubMed/NCBI
|