Introduction
Colorectal cancer (CRC) is the third most common
cause of cancer-associated mortality (1). Despite advances in the diagnosis and
therapy, CRC remains a major global health problem (2). Thus, it is necessary to explore the
molecular mechanisms underlying the progression of CRC.
Sirtuins (SIRTs) 1–7 are a class of nicotine adenine
dinucleotide (NAD)+-dependent deacetylases (3), involved in cellular processes (4), including metabolism (5), stress response (6,7), genomic
stability (8,9) and longevity (10). Among SIRTs, SIRT1, SIRT6 and SIRT7 are
localized in the nucleus (11–13), SIRT2
resides within the cytoplasm (11),
whereas SIRT3, SIRT4 and SIRT5 are mitochondrial (14,15).
Aberrant expression of SIRTs is associated with the pathogenesis of
several human diseases, including cancer (16). SIRT4 does not have a NAD+-dependent
deacetylase activity but downregulates glutamate dehydrogenase
activity (17), an enzyme that
converts glutamate to α-ketoglutarate and regulates the metabolism
of glutamine and glutamate to ultimately produce ATP (18). SIRT4 regulates the balance between
fatty acid oxidation and lipid synthesis in skeletal muscle cells
and adipocytes (19,20), and regulates insulin secretion in
human pancreatic β cells (17,21).
Additionally, SIRT4 serves a role in the metabolism of amino acids
(22). In tumor cells, SIRT4 may
negatively regulate cell proliferation by inhibiting the uptake of
glutamine (23). Jeong et al
(24) reported that SIRT4 regulates
the cellular metabolism in response to DNA damage by inhibiting
mitochondrial glutamine metabolism. Additionally, decreased protein
levels of SIRT4 have been detected in endometrial adenocarcinoma
tissues and are associated with advanced American Joint Committee
on Cancer (AJCC) stages (25).
Downregulation of SIRT4 is associated with poor prognosis in
esophageal squamous cell carcinoma (26). Numerous studies demonstrated that
SIRT4 is downregulated in several types of human cancer, including
breast (27), gastric (28), liver (29), colon (30) and bladder cancer (31) and leukemia (32), however, primarily in lung cancer
(33,34), Although previous studies suggested
that SIRT4 may act as a tumor suppressor (35), the function of SIRT4 in human cancer
remains unclear.
In the present study, the expression of SIRT4 was
detected in CRC tissues. Additionally, the effect of SIRT4
downregulation on colon cancer proliferation, migration and
invasion was investigated. The effects of SIRT4 on the
chemotherapeutic sensitivity of CRC cells and the underlying
molecular mechanisms were also explored. The present study provides
insights on the role of SIRT4 in CRC progression.
Materials and methods
Cell lines and cell culture
All cell lines were provided by the Shanghai
Institute of Cell Biology (Shanghai, China). The human CRC cell
lines HCT116, SW1116, SW620 and DLD1 were maintained in RPMI-1640
(Biological Industries, Kibbutz Beit Haemek, Israel) and the normal
colorectal cell line FHC were maintained in Dulbecco's modified
Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin. Cells were cultured at 37°C
in a humidified atmosphere containing 5% CO2.
Tissue samples
A total of 30 tissues (15 CRC and 15 matched
adjacent normal tissues of male patients) were obtained from the
Department of Gastrointestinal Surgical Oncology at Harbin Medical
University Cancer Hospital (Heilongjiang, China) between 2011 and
2012. The range years was 38–76 years and the mean age of patients
was 52 years. None of the patients received chemotherapy before
surgery. All patients signed written informed consent, and the
study was approved by the Ethics Committee of Harbin Medical
University (Heilongjiang, China).
The Cancer Genome Atlas (TCGA)
database
The SIRT4 expression data of 174 samples were
downloaded from TCGA data portal on August 2016 (https://cancergenome.nih.gov/). According to the TCGA
barcode, the expression data was divided into CRC data and normal
data, including 155 CRC tissues and 19 normal colon tissues. The
statistical analyses and figures were performed by GraphPad Prism
software 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) based on
two sets of expression data.
Immunohistochemistry and evaluation of
staining
Tissue sections (3.5 µm thick) were prepared from
paraffin-embedded tissues. Briefly, sections were deparaffinized in
xylene for 5 min (four times) and rehydrated in a descending
ethanol series at room temperature. Antigen retrieval was performed
using a pressure cooker for 3 min at 121°C in Tris-EDTA buffer.
Endogenous peroxidase activity was then blocked by incubation in 3%
hydrogen peroxide for 10 min at room temperature. The sections were
incubated with primary antibody against SIRT4 (ab10140; 1:100;
Abcam, Cambridge, UK) overnight at 4°C. Following the primary
incubation, sections were incubated with secondary antibody
solution (ZB-2306; 1:5,000; ZSGB-BIO; OriGene Technologies, Inc.,
Beijing, China) for 1 h at room temperature. The sections were
stained with 3,3-diaminobenzidine tetrahydrochloride (DAB;
ZSGB-BIO; OriGene Technologies, Inc.) and counterstained with
hematoxylin. Tissues stained with PBS instead of primary antibody,
served as negative controls.
The histological evaluation was performed by two
pathologists from Harbin Medical University Cancer Hospital
(Harbin, China) using a light microscope. The staining score was
defined by using intensity (0, negative; 1, weak; 2, moderate; 3,
strong) and area (0, <5%; 1, 5–25%; 2, 25–50%; 3, 50–75%; 4,
>75%). The final score was generated by assessing the percentage
of area stained multiplied by the intensity of staining.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues and cells with
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. RNA was
reverse-transcribed into cDNA using TransScript Reverse
Transcriptase (AT101-02; Beijing Transgen Biotech Co., Ltd.,
Beijing, China). qPCR was performed using the SYBR Green PCR Mix
(AQ141-01; Beijing Transgen Biotech, Beijing, China) and the
Bio-Rad CFX96TM Real-Time PCR system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The primers used were as follows: SIRT4,
5′-ATGAAGATGAGCTTTGCGT-3′ (forward) and 5′-TCAGCATGGGTCTATCAAAGG-3′
(reverse); GAPDH, 5′-ATGGGGAAGGTGAAGGTCG-3′ (forward) and
5′-GGGGTCATTGATGGCAACAATA-3′ (reverse). GAPDH was used as an
endogenous control. The thermocycling conditions were as follows:
95°C for 10 min, followed by 40 cycles at 95°C for 10 sec and 61°C
for 30 sec. The analysis of RT-qPCR results according to the
2−ΔΔCq method (36).
Western blot analysis
Total protein was extracted from cells and tissues
using a radioimmunoprecipitation assay buffer (Beyotime Institute
of Biotechnology, Haimen, China) supplemented with 1 mM
phenylmethanesulfonyl fluoride (PMSF, Beyotime Institute of
Biotechnology, Shanghai, China). Protein determination was
performed using the BCA Protein assay kit (Takara Bio, Inc., Otsu,
Japan) and proteins (25 µg) were separated by SDS-PAGE (10% gels)
and then transferred onto polyvinylidene difluoride (PVDF)
membranes (EMD Millipore, Billerica, MA, USA) and blocked in 5%
bovine serum albumin (Beyotime Institute of Biotechnology) was
diluted in PBS for 2 h at room temperature. Membranes were
incubated with anti-SIRT4 (catalog no. ab10140; 1:300, Abcam) or
mouse anti-β-actin (catalog no. sc-58673, 1:5,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) antibodies at 4°C overnight,
followed by incubation with Rabbit Anti-goat IgG H&L (catalog
no. ab6697; 1:5,000; Abcam) or goat anti-mouse IgG H&L (catalog
no. ab6708; 1:5,000; Abcam) for 1 h at room temperature. The
protein bands were visualized by enhanced chemiluminescence (ECL;
GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) via FluorChem M
imaging system (FCM, FM0422; ProteinSimple, San Jose, CA, USA) and
analyzed using the Image J software (https://imagej.nih.gov/ij/).
Apoptosis analysis
Cell apoptosis was evaluated by using Annexin
V-phycoerythrin (PE)/7-aminoactinomycin D (7-AAD) Apoptosis
Detection kit (BD Biosciences, Franklin Lakes, NJ, USA), according
to the manufacturer's protocol. Briefly, a total of
1×106 HCT116 cells were collected and washed with PBS,
and then the cells were resuspended with 1-ml Binding Buffer. 100
µl solution (1×105), 5 µl PE Annexin V and 5 µl 7-ADD
were added in a 5 ml tube. The cells were vortexed gently and then
incubated for 10 min in the dark at room temperature. At last, 400
µl Binding Buffer was added in each tube before analyzed by flow
cytometry. The flow cytometry data were analyzed by software
(FlowJo 7.6.1; FlowJo LLC, Ashland, OR, USA).
Cell viability assay
Cellular viability was evaluated using CellTiter-Glo
assay (Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. Briefly, HCT116 cells in RPMI-1640
(Biological Industries, Kibbutz Beit Haemek, Israel) were plated in
96-well plates at a density of 1×104 cells/well at room
temperature. At 0, 24, 48 and 72 h, 100 µl CellTiter-Glo reagent
was added into each well and mixed completely using an orbital
shaker at room temperature. Following incubation for 10 min at room
temperature, the luminescence signal was assessed using a
luminometer.
Clustered Regularly Interspaced Short
Palindromic Repeats (CRISPR)/CRISPR-associated protein-9 nuclease
(Cas9)-mediated knockout of SIRT4 in HCT116 cells
In the present study a CRISPR-Cas9 vector, targeting
specific region of SIRT4 (5′-CCGAATCGGGGATACCAGAC-3′) was used.
Guide RNA sequence for CRISPR/Cas9 was designed using the CRISPR
Design Tool (http://tools.genome-engineering.org) on August 2016.
HCT116 cells were grown to 80–90% confluence and transfected with
CRISPR plasmids (pSpCas9) by using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at room temperature. Cells were
treated with 2 µg/ml puromycin (ST551, Beyotime Institute of
Biotechnology, Shanghai, China) for 24 h. Transfected cells were
sorted to obtain single cell clones. Western blot analysis was used
to confirm the knockout of SIRT4 in HCT116 cells.
Invasion and migration assay
Cell migration was evaluated using a Transwell
chamber without (3422, Corning, USA) according to the
manufacturer's protocol. Cell invasion was evaluated using a
Transwell chamber with Matrigel (Corning Incorporated, Corning, NY,
USA) according to the manufacturer's protocol. For the upper
chambers, 3×105 cells/ml were suspended in 0.5 ml of
serum-free DMEM. The lower chamber contained DMEM supplemented with
10% FBS. Plates were incubated at 37°C in a humidified atmosphere
containing 5% CO2. At 48 h, cells remaining on the upper
membrane surface were removed and the invasive cells on the lower
surface were fixed with 1% crystal violet for 30 min and counted at
room temperature. Images were captured using an Olympus inverted
microscope (Olympus Corporation, Tokyo, Japan). The number of
invaded cells was counted using ImageJ software (National
Institutes of Health, MD, Bethesda, USA).
5-FU treatment
5-fluorouracil 0.25 g/10 ml or oxaliplatin 50 mg was
purchased from Harbin Medical University Cancer Hospital and
storage at 4°C. HCT116 cells were treated with 5-FU for 36 h at
37°C and treated HCT116 cells with oxaliplatin for 48 h at
37°C.
Statistical analysis
Data were analyzed using GraphPad Prism (version
5.0; GraphPad Software, Inc.). Data are expressed as the mean ±
standard deviation. Three individual experiments were performed.
Results were analyzed using Student's t-test or one-way ANOVA
analysis, following the analysis of variance. P<0.05 was
considered to indicate a statistically significant difference.
Results
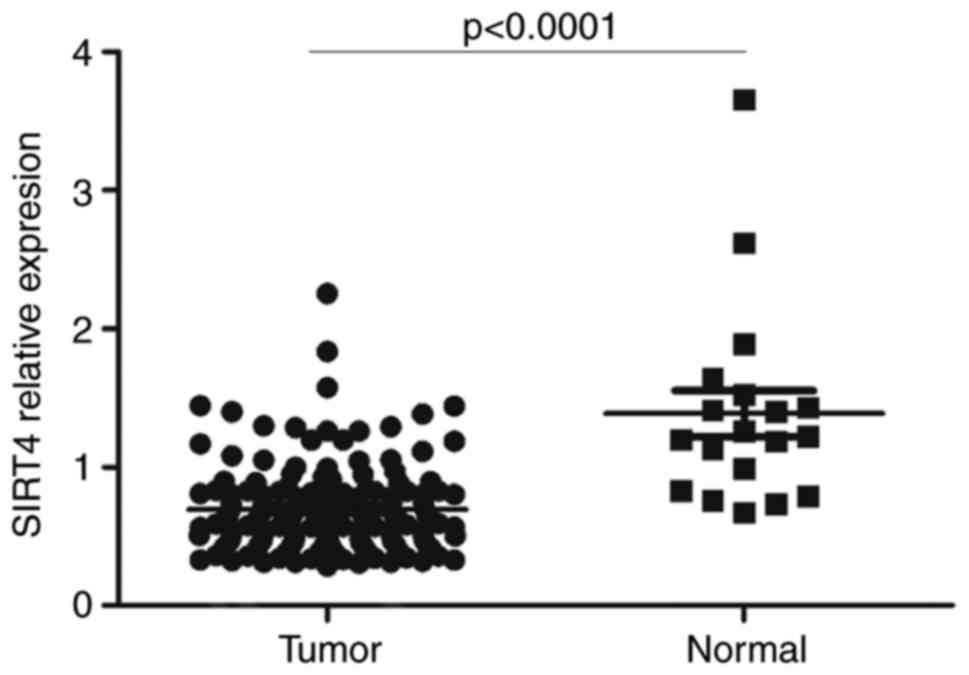
Expression of SIRT4 in CRC based on
data obtained from TCGA
The SIRT4 expression data from 174 samples
(including 19 normal colon tissues and 155 CRC tissues) were
downloaded from TCGA data portal. The results demonstrated that the
expression of SIRT4 was significantly decreased in colon
adenocarcinoma tissues compared with normal colon tissues (Fig. 1).
SIRT4 is downregulated in CRC cell
lines and tissues
mRNA and protein expression levels of SIRT4 were
examined in CRC cell lines and tissues. The results confirmed that
mRNA expression levels of SIRT4 were significantly decreased
compared with that in adjacent normal tissues (Fig. 2A). Additionally, mRNA expression
levels of SIRT4 were decreased in CRC cell lines (HCT116, SW1116,
SW620 and DLD1) compared with that in the normal colorectal cell
line FHC (Fig. 2B). Protein
expression of SIRT4 was evaluated in tissues using western blot
analysis and immunohistochemistry. The results demonstrated that
the expression level of SIRT4 in CRC tissues was decreased compared
with that in adjacent normal tissues (Fig. 2C-E). Thus, SIRT4 is downregulated in
CRC tissues and cells, suggesting that SIRT4 may be involved in the
pathogenesis of CRC.
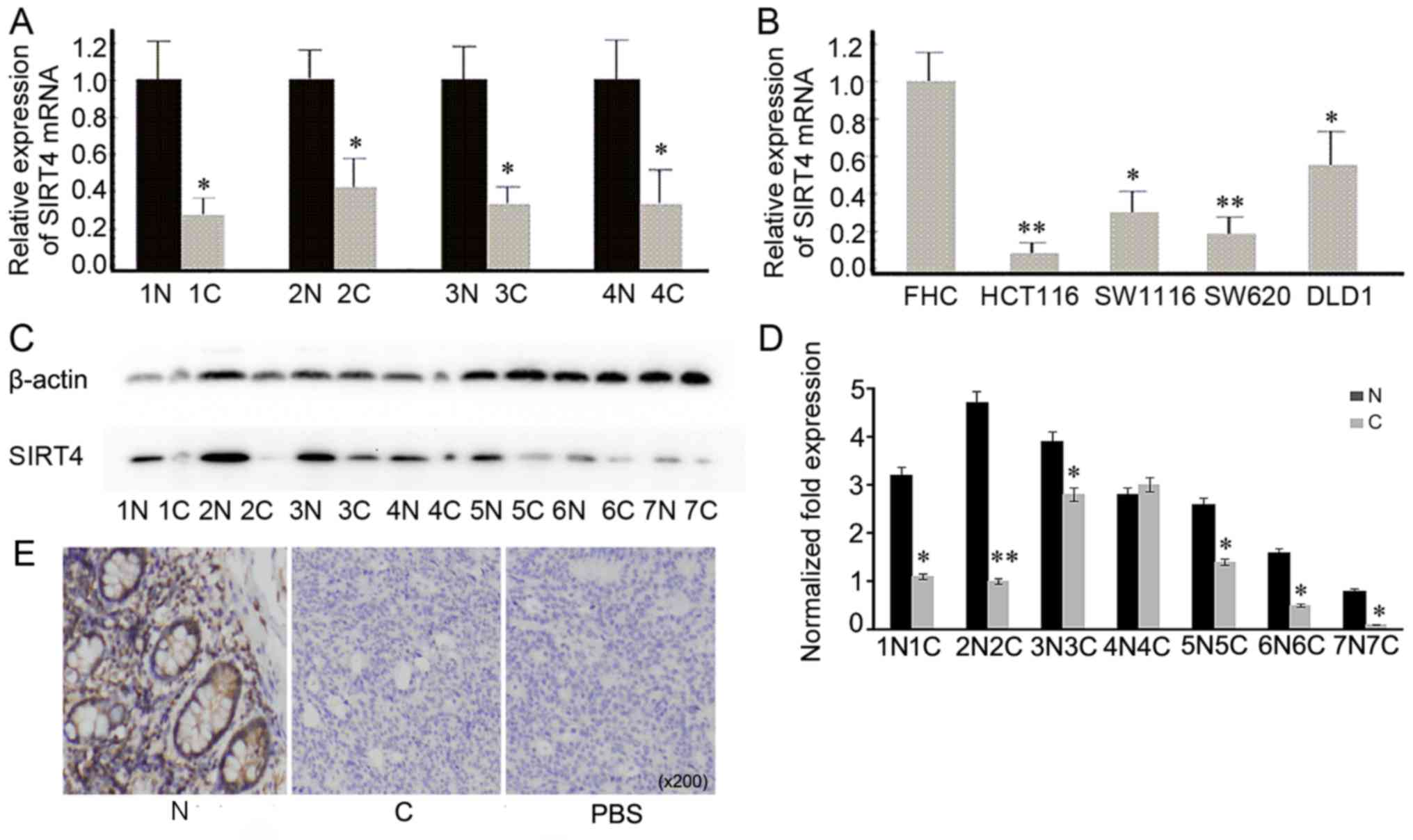 | Figure 2.Detection of SIRT4 expression in CRC
tissues and cell lines. (A) mRNA expression levels of SIRT4 in
representative CRC tissues and adjacent normal tissues was
examined. *P<0.05 vs. N. (B) mRNA expression levels of SIRT4 in
CRC cell lines (HCT116, SW1116, SW620 and DLD1) was decreased
compared with that in the normal cell line FHC. **P<0.01,
*P<0.05 vs. FHC. (C) The expression of SIRT4 in seven CRC
tissues and adjacent normal tissues was evaluated using western
blot analysis. (D) The densitometric analysis of SIRT4 expression
protein was downregulated in CRC tissues compared with that in
normal tissues. **P<0.01, *P<0.05 vs. N. (E) Representative
immunohistochemical staining of SIRT4 in human CRC and adjacent
normal tissues. SIRT4 was mainly expressed in the cytoplasm, and
its expression was decreased in CRC tissues compared with that in
adjacent normal tissue. SIRT, sirtuin; CRC, colorectal cancer; N,
normal tissue; C, CRC tissue; PBS, negative control. |
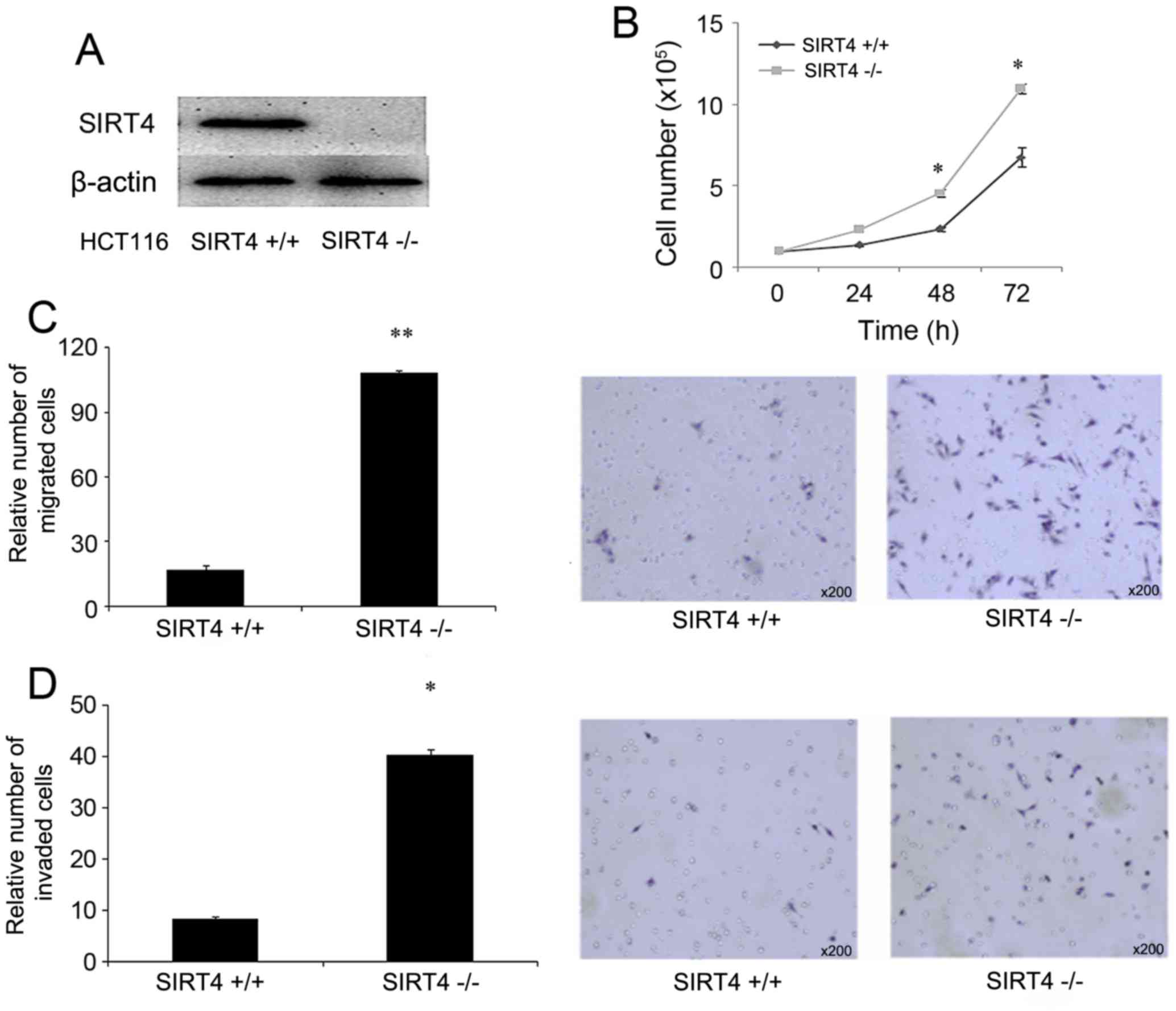
SIRT4 functions as a potential tumor
suppressor by repressing the proliferation, and migration of CRC
cells
To determine the function of SIRT4 in colorectal
cancer, CRISPR/Cas9-mediated knockout of SIRT4 was performed in
HCT116 cells. The knockout of SIRT4 was confirmed using western
blot analysis (Fig. 3A). The
proliferative ability of control cells (HCT116 SIRT4+/+)
and SIRT4 knockout cells (HCT116 SIRT4−/−) was
evaluated. The results demonstrated that HCT116 SIRT4−/−
cells grew faster compared with HCT116 SIRT4+/+ cells
(Fig. 3B). Additionally, the effect
of SIRT4 knockout on the migratory abilities was determined in
HCT116 cells. The results demonstrated that HCT116
SIRT4−/− significantly enhanced the migratory ability
(Fig. 3C) and invasive ability
(Fig. 3D). Therefore, the knockout of
SIRT4 promoted the proliferation, migration and invasion of HCT116
cells.
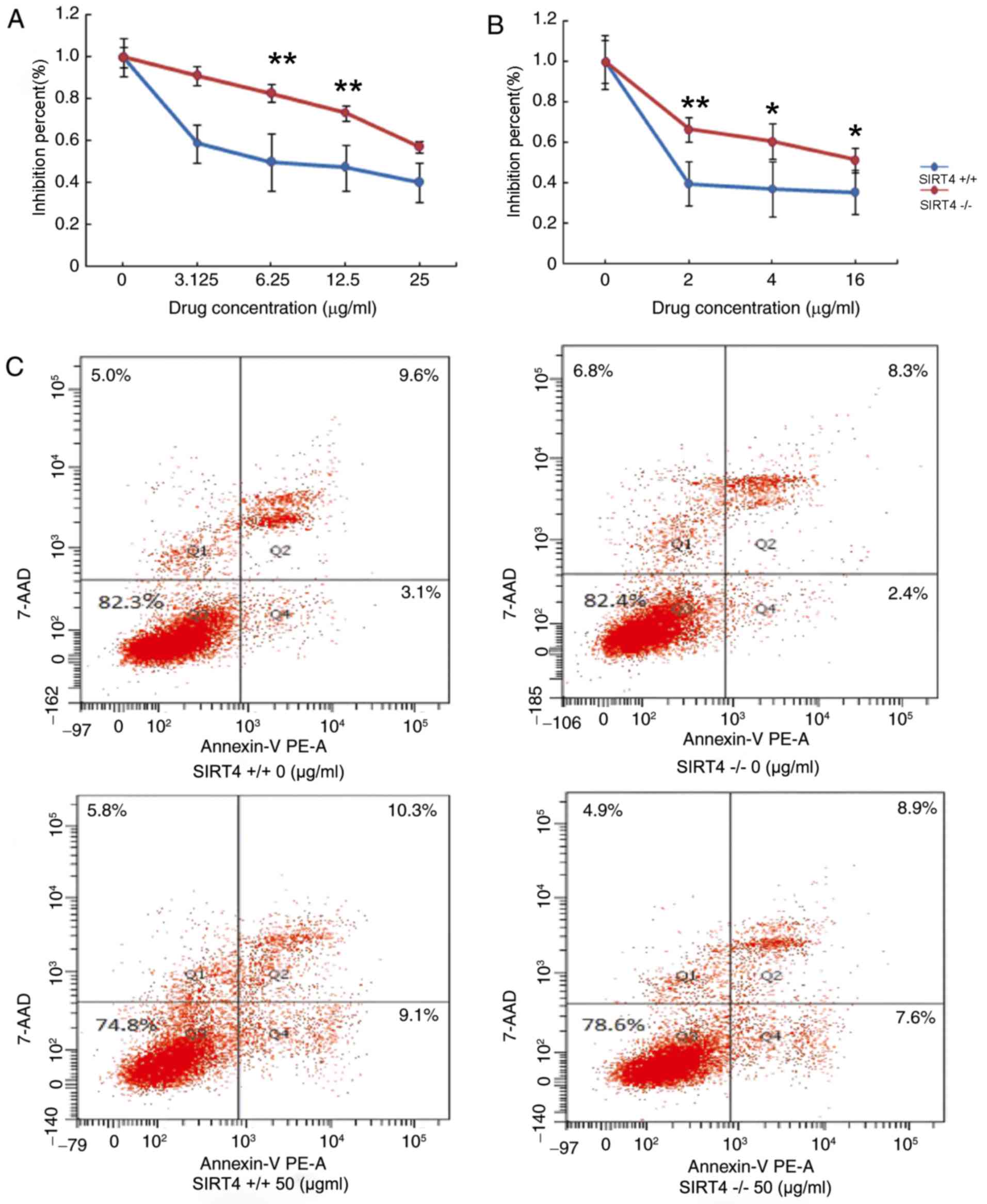
Knockout of SIRT4 decreases
chemosensitivity in CRC cells by inhibiting apoptosis
The effects of SIRT4 knockout in regulating
chemotherapy in CRC were also evaluated. HCT116 SIRT4−/−
and HCT116 SIRT4+/+ were treated with various
concentrations of 5-fluorouracil (5-FU) or oxaliplatin for 48 h at
37°C. CellTiter-Glo Luminescent assay was employed to determine the
effect of SIRT4 on the chemosensitivity of CRC cells. The results
demonstrated that the viability of HCT116 SIRT4−/− cells
was increased compared with that in SIRT4+/+ in response
to treatment with 5-FU or oxaliplatin by inhibition percent (%)
(Fig. 4A and B). These results
suggest that knockout of SIRT4 may decrease the chemosensitivity of
CRC cells. Next, the effect of SIRT4 knockout on cells apoptosis
was evaluated. Due to the high drug concentration required for
apoptosis, HCT116 SIRT4−/− cells and HCT116
SIRT4+/+ cells were treated with 50 µg/ml 5-FU and cell
apoptosis was evaluated using flow cytometry. The results
demonstrated that the apoptosis rate was decreased in HCT116
SIRT4−/− cells compared with that of HCT116
SIRT4+/+ cells (Fig. 4C),
suggesting that knockout of SIRT4 may decrease sensitivity to
chemotherapy by inhibiting apoptosis in CRC cells.
Discussion
Previous studies demonstrated that SIRT4 is
downregulated in various cancer types (23,24) and is
associated with multiple cancer biological behaviors (23). However, the function of SIRT4 in CRC
remains unclear. The results of the present study revealed that the
expression of SIRT4 was significantly decreased in CRC tissues and
cell lines at mRNA and protein levels, which is consistent with
previous studies. Previous studies revealed that SIRT4 might
function as a tumor suppressor. Jeong et al (24) demonstrated that SIRT4 may suppress
tumor formation by inhibiting glutamine metabolism, and
overexpression of SIRT4 may prevent the growth of HeLa cells.
Additionally, Csibi et al (23) revealed that overexpression of SIRT4
may prevent the growth of the prostate cancer cell line DU145 and
the colon cancer cell line DLD-1. Jeong et al (32) revealed that SIRT4 might inhibit the
growth of Myc-induced B lymphoma cell by inhibiting glutamine
metabolism. In the present study, stable human SIRT4 KO cells were
established and the effects of knockout of SIRT4 on proliferation,
migration and invasion of CRC cells were examined. The results
demonstrated that knockout of SIRT4 increased the proliferation,
migration and invasion of CRC cells, thus confirming the findings
of previous studies (30).
Targeting SIRT4 may be a novel therapeutic approach
in CRC. Chemotherapy is considered as one of the most effective
treatment approaches in CRC (37).
5-FU and oxaliplatin are widely used for the treatment of CRC
(38). 5-FU-based chemotherapy in
combination with oxaliplatin or irinotecan is used for the
treatment of metastatic CRC (39).
5-FU metabolites are incorporated into DNA to suppress cell growth
(40). In tumor cells, 5-FU is
metabolized to cytotoxic compounds, which bind to thymidylate
synthase in order to repress DNA synthesis (41). Oxaliplatin, a platinum-based
chemotherapeutic drug, containing 1,2-diaminocyclohexane carrier
ligand, forms platinum-DNA adducts, which block DNA replication. In
addition, oxaliplatin has demonstrated high efficacy against tumor
growth both in vitro and in vivo (42). Oxaliplatin treatment combined with
5-FU increases progression-free survival and overall survival of
CRC (43). Oxaliplatin and 5-FU may
act synergistically to regulate thymidylate synthase (44). In the present study, the effects SIRT4
on the chemosensitivity of CRC cells were evaluated. To the best of
our knowledge, this is the first study to examine the role of SIRT4
in cancer chemotherapy. The results demonstrated that SIRT4 KO
cells exhibited increased viability compared with control cells,
suggesting that SIRT4 may enhance the chemosensitivity of CRC cells
to 5-FU and oxaliplatin. Next, the underlying molecular mechanism
was investigated. A number of events involved in apoptosis are
closely associated with mitochondria, including the release of
caspase activators, loss of mitochondrial transmembrane potential,
altering electron transport, and participation of Bcl-2 family
proteins (45,46). SIRT4 is located in the mitochondria.
Previous studies have demonstrated that SIRT4 serves an important
function in hypoxia-induced apoptosis in H9c2 cardiomyoblast cells
through affecting Bcl-2-associated-X protein (Bax) translocation
(47). Therefore, it was hypothesized
that SIRT4 may affect the chemotherapeutic sensitivity of CRC cells
through regulating apoptosis. The results revealed that SIRT4
knockout prevents the apoptosis of CRC cells in response to 5-FU.
These results suggest that SIRT4 may act as a tumor suppressor and
may be used as a novel therapeutic target in CRC.
In conclusion, the present study demonstrated that
the expression of SIRT4 is significantly decreased in CRC tissues
at mRNA and protein levels, and that SIRT4 knockout increases tumor
proliferation, migration and invasion, thus suggesting that SIRT4
may act as a tumor suppressor. Additionally, SIRT4 knockout affects
the chemotherapeutic sensitivity of CRC cells by regulating
apoptosis. The present study provides a promising target for CRC
therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81172265).
Availability of data and materials
The data used and analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
YaZ conceived and designed the experiments. YuZ, GW,
XL, TW and MW performed the experiments and analyzed the data. YuZ
and YaZ wrote the paper. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent prior
to their inclusion and the present study was approved by the Ethics
Committee of Harbin Medical University (Heilongjiang, China).
Consent for publication
This research was completed in compliance with the
Helsinki Declaration. The patients provided written informed
consent for the publication of the data. The data collection and
analysis were carried out without disclosing the identities of the
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Argmann C and Auwerx J: Insulin secretion:
SIRT4 gets in on the act. Cell. 126:837–839. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Finkel T, Deng CX and Mostoslavsky R:
Recent progress in the biology and physiology of sirtuins. Nature.
460:587–591. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kleszcz R, Paluszczak J and Baer-Dubowska
W: Targeting aberrant cancer metabolism-The role of sirtuins.
Pharmacol Rep. 67:1068–1080. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeong SM, Hwang S and Seong RH: SIRT4
regulates cancer cell survival and growth after stress. Biochem
Biophys Res Commun. 470:251–256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng J, Yan PF, Zhao HY, Zhang FC, Zhao WH
and Feng M: SIRT6 suppresses glioma cell growth via induction of
apoptosis, inhibition of oxidative stress and suppression of
JAK2/STAT3 signaling pathway activation. Oncol Rep. 35:1395–1402.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kiran S, Oddi V and Ramakrishna G: Sirtuin
7 promotes cellular survival following genomic stress by
attenuation of DNA damage, SAPK activation and p53 response. Exp
Cell Res. 331:123–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim HS, Vassilopoulos A, Wang RH, Lahusen
T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, et al: SIRT2
maintains genome integrity and suppresses tumorigenesis through
regulating APC/C activity. Cancer Cell. 20:487–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Benigni A, Perico L and Macconi D:
Mitochondrial dynamics is linked to longevity and protects from
End-organ injury: The emerging role of Sirtuin 3. Antioxid Redox
Signal. 25:185–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Singh S, Kumar PU, Thakur S, Kiran S, Sen
B, Sharma S, Rao VV, Poongothai AR and Ramakrishna G:
Expression/localization patterns of sirtuins (SIRT1, SIRT2, and
SIRT7) during progression of cervical cancer and effects of sirtuin
inhibitors on growth of cervical cancer cells. Tumour Biol.
36:6159–6171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Azuma Y, Yokobori T, Mogi A, Altan B,
Yajima T, Kosaka T, Onozato R, Yamaki E, Asao T, Nishiyama M and
Kuwano H: SIRT6 expression is associated with poor prognosis and
chemosensitivity in patients with non-small cell lung cancer. J
Surg Oncol. 112:231–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang PY, Li G, Deng ZJ, Liu LY, Chen L,
Tang JZ, Wang YQ, Cao ST, Fang YX, Wen F, et al: Dicer interacts
with SIRT7 and regulates H3K18 deacetylation in response to DNA
damaging agents. Nucleic Acids Res. 44:3629–3642. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Padmaja Divya S, Pratheeshkumar P, Son YO,
Vinod Roy R, Andrew Hitron J, Kim D, Dai J, Wang L, Asha P, Huang
B, et al: Arsenic induces insulin resistance in mouse adipocytes
and myotubes via oxidative stress-regulated mitochondrial
Sirt3-FOXO3a signaling pathway. Toxicol Sci. 146:290–300. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pacella-Ince L, Zander-Fox DL and Lane M:
Mitochondrial SIRT5 is present in follicular cells and is altered
by reduced ovarian reserve and advanced maternal age. Reprod Fertil
Dev. 26:1072–1083. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yuan H, Su L and Chen WY: The emerging and
diverse roles of sirtuins in cancer: A clinical perspective. Onco
Targets Ther. 6:1399–1416. 2013.PubMed/NCBI
|
|
17
|
Ahuja N, Schwer B, Carobbio S, Waltregny
D, North BJ, Castronovo V, Maechler P and Verdin E: Regulation of
insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase.
J Biol Chem. 282:33583–33592. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ho L, Titus AS, Banerjee KK, George S, Lin
W, Deota S, Saha AK, Nakamura K, Gut P, Verdin E and
Kolthur-Seetharam U: SIRT4 regulates ATP homeostasis and mediates a
retrograde signaling via AMPK. Aging (Albany NY). 5:835–849. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Laurent G, German NJ, Saha AK, de Boer VC,
Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran
B, et al: SIRT4 coordinates the balance between lipid synthesis and
catabolism by repressing malonyl CoA decarboxylase. Mol Cell.
50:686–698. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nasrin N, Wu X, Fortier E, Feng Y, Bare'
OC, Chen S, Ren X, Wu Z, Streeper RS and Bordone L: SIRT4 regulates
fatty acid oxidation and mitochondrial gene expression in liver and
muscle cells. J Biol Chem. 285:31995–32002. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Haigis MC, Mostoslavsky R, Haigis KM,
Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos
GD, Karow M, Blander G, et al: SIRT4 inhibits glutamate
dehydrogenase and opposes the effects of calorie restriction in
pancreatic beta cells. Cell. 126:941–954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Houtkooper RH, Pirinen E and Auwerx J:
Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol
Cell Biol. 13:225–238. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Csibi A, Fendt SM, Li C, Poulogiannis G,
Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T,
et al: The mTORC1 pathway stimulates glutamine metabolism and cell
proliferation by repressing SIRT4. Cell. 153:840–854. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jeong SM, Xiao C, Finley LW, Lahusen T,
Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al:
SIRT4 has tumor-suppressive activity and regulates the cellular
metabolic response to DNA damage by inhibiting mitochondrial
glutamine metabolism. Cancer Cell. 23:450–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen X, Lai X, Wu C, Tian Q, Lei T, Pan J
and Huang G: Decreased SIRT4 protein levels in endometrioid
adenocarcinoma tissues are associated with advanced AJCC stage.
Cancer Biomark. 19:419–424. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakahara Y, Yamasaki M, Sawada G, Miyazaki
Y, Makino T, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S,
Mimori K, et al: Downregulation of SIRT4 expression is associated
with poor prognosis in esophageal squamous cell carcinoma.
Oncology. 90:347–355. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Igci M, Kalender ME, Borazan E, Bozgeyik
I, Bayraktar R, Bozgeyik E, Camci C and Arslan A: High-throughput
screening of Sirtuin family of genes in breast cancer. Gene.
586:123–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang G, Cui F, Yu F, Lu H, Zhang M, Tang
H and Peng Z: Sirtuin-4 (SIRT4) is downregulated and associated
with some clinicopathological features in gastric adenocarcinoma.
Biomed Pharmacother. 72:135–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang JX, Yi Y, Li YW, Cai XY, He HW, Ni
XC, Zhou J, Cheng YF, Jin JJ, Fan J and Qiu SJ: Down-regulation of
sirtuin 3 is associated with poor prognosis in hepatocellular
carcinoma after resection. BMC Cancer. 14:2972014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miyo M, Yamamoto H, Konno M, Colvin H,
Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, et
al: Tumour-suppressive function of SIRT4 in human colorectal
cancer. Br J Cancer. 113:492–499. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Blaveri E, Simko JP, Korkola JE, Brewer
JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P
and Waldman FM: Bladder cancer outcome and subtype classification
by gene expression. Clin Cancer Res. 11:4044–4055. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jeong SM, Lee A, Lee J and Haigis MC:
SIRT4 protein suppresses tumor formation in genetic models of
Myc-induced B cell lymphoma. J Biol Chem. 289:4135–4144. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu L, Dong Q, He J, Wang X, Xing J, Wang
E, Qiu X and Li Q: SIRT4 inhibits malignancy progression of NSCLCs,
through mitochondrial dynamics mediated by the ERK-Drp1 pathway.
Oncogene. 36:2724–2736. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu Y, Yan Y, Principe DR, Zou X,
Vassilopoulos A and Gius D: SIRT3 and SIRT4 are mitochondrial tumor
suppressor proteins that connect mitochondrial metabolism and
carcinogenesis. Cancer Metab. 2:152014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lombardi L, Morelli F, Cinieri S, Santini
D, Silvestris N, Fazio N, Orlando L, Tonini G, Colucci G and
Maiello E: Adjuvant colon cancer chemotherapy: Where we are and
where we'll go. Cancer Treat Rev. 36(Suppl 3): S34–S41. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
AlShamaileh H, Wang T, Xiang D, Yin W,
Tran PH, Barrero RA, Zhang PZ, Li Y, Kong L, Liu K, et al:
Aptamer-mediated survivin RNAi enables 5-fluorouracil to eliminate
colorectal cancer stem cells. Sci Rep. 7:58982017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Siesing C, Sorbye H, Dragomir A, Pfeiffer
P, Qvortrup C, Pontén F, Jirström K, Glimelius B and Eberhard J:
High RBM3 expression is associated with an improved survival and
oxaliplatin response in patients with metastatic colorectal cancer.
PLoS One. 12:e01825122017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Noordhuis P, Holwerda U, van Laar JA, van
der Wilt CL and Peters GJ: A non-radioactive sensitive assay to
measure 5-fluorouracil incorporation into DNA of solid tumors.
Nucleosides Nucleotides Nucleic Acids. 23:1481–1484. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Q, Meng F, Wang L, Mao Y, Zhou H, Hua
D, Zhang H and Wang W: A polymorphism in ABCC4 is related to
efficacy of 5-FU/capecitabine-based chemotherapy in colorectal
cancer patients. Sci Rep. 7:70592017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Raymond E, Faivre S, Woynarowski JM and
Chaney SG: Oxaliplatin: Mechanism of action and antineoplastic
activity. Semin Onco. 25(2 Suppl 5): S4–S12. 1998.
|
|
43
|
Douillard JY, Cunningham D, Roth AD,
Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J,
Alakl M, et al: Irinotecan combined with fluorouracil compared with
fluorouracil alone as first-line treatment for metastatic
colorectal cancer: A multicentre randomised trial. Lancet.
355:1041–1047. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Raymond E, Faivre S, Chaney S, Woynarowski
J and Cvitkovic E: Cellular and molecular pharmacology of
oxaliplatin. Mol Cancer Ther. 1:227–235. 2002.PubMed/NCBI
|
|
45
|
Wang L, Zhou H, Wang Y, Cui G and Di LJ:
CtBP maintains cancer cell growth and metabolic homeostasis via
regulating SIRT4. Cell Death Dis. 6:e16202015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Verdin E, Hirschey MD, Finley LW and
Haigis MC: Sirtuin regulation of mitochondria: Energy production,
apoptosis, and signaling. Trends Biochem Sci. 35:669–675. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu B, Che W, Xue J, Zheng C, Tang K,
Zhang J, Wen J and Xu Y: SIRT4 prevents hypoxia-induced apoptosis
in H9c2 cardiomyoblast cells. Cell Physiol Biochem. 32:655–662.
2013. View Article : Google Scholar : PubMed/NCBI
|