Introduction
MicroRNAs (miRNAs/miRs) exist in the majority of
eukaryotes and consist of 21–25 nucleotides. miRNAs regulate gene
expression via binding to target mRNAs, resulting in mRNA
degradation or inhibition of translation (1–4). mRNAs
acts as oncogenes or suppressor genes to affect tumor development
(5–8).
In 2013, Takahashi et al reported that hepatocellular
carcinoma (HCC) is the most common primary malignancy of the liver
globally (9), and numerous miRNAs are
abnormally expressed in HCC (9).
miR-21 is one of the most prominently expressed miRNAs in a number
of human cancer types, including pancreas, breast, prostate, colon,
lung and stomach (10). The
expression of miR-21 is increased in HCC tissues, compared with
normal tissues (11). However,
miR-146a has decreased expression in HCC tissues, compared with
normal liver tissues (12). miR-34a
is a member of the miR-34 family and has been demonstrated to
modulate critical gene transcripts involved in tumorigenesis, but
its role in tumorigenesis remains unknown. miR-34a may be activated
by p53 to induce apoptosis and inhibit tumor growth (13–16).
However, it often dysfunctions or mutates in tumors (17–21). Luo
et al (22) has demonstrated
that miR-34a was able to suppress the migration of breast cancer
cells via targeting Fra-1. Wang et al (23) observed an inverse association between
programmed death-ligand 1 (PD-L1) and miR-34a expression in a
number of acute myeloid leukemia (AML) samples. miR-34a is a
putative binder of the PD-L1-3′ untranslated region (UTR), and
overexpression of miR-34a in HL-60 and Kasumi-1 cells could block
PD-L1 expression (23). The aim of
the present study was to verify the effect of miR-34a on metastasis
of liver cancer cells.
Materials and methods
Materials
The miRNeasy Mini kit was purchased from Qiagen GmbH
(Hilden, Germany). Real-time PCR detection kit was from
GeneCopoeia, Inc., (Rockville, MD, USA). 5-fluorouracil (5-FU) and
MTT were from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). The
concentration of 5-FU in the present study was 5 µg/ml, and cells
were incubated at 37°C for 48 h. Lipofectamine® 2000 was
purchased from Thermo Fisher Scientific, Inc., (Waltham, MA, USA).
Minimum Essential Medium (MEM) was from Hyclone (GE Healthcare,
Chicago, IL, USA), and fetal bovine serum was from Gibco (Thermo
Fisher Scientific, Inc.).
Cell lines and cell culture
MHCC97H liver cancer cells were purchased from the
Resource Center of Shanghai Institutes of Biological Sciences
(Shanghai, China) were maintained in a monolayer culture at 37°C
and 5% CO2 in MEM that was supplemented with 10% fetal
bovine serum.
Cell transfection
MHCC97H cells were transfected with 5 mg/ml miR-NC
and 5 mg/ml miR-34a mimic (Shanghai GenePharma Co., Ltd., Shanghai,
China) using Lipofectamine® 2000 as follows. The cells
(4–5×104 cells/ml) were plated in a 6-well plate at 37°C
for 24 h. Prior to transfection, the miRNA-Lipofectamine solution
was prepared by mixing MEM separately with Lipofectamine or miRNA
and then mixing the solutions together. Finally, the
miRNA-Lipofectamine solution was added to each well, and the cells
were incubated at 37°C for 24–72 h for subsequent analyses.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
All independent experiments were conducted in
triplicate. Negative control [no complementary DNA (cDNA)] and RT
control (no reverse transcriptase). Total RNA (including miRNA) was
extracted using the miRNeasy Mini kit (Qiagen GmbH), and DNA was
removed using DNase I. Reverse transcription was conducted with
cDNA Synthesis kit (cat. no. 6130; Takara Bio. Inc., Otsu, Japan).
Reverse transcription was conducted with 2 µg total RNA, according
to the manufacturer's instructions, and the negative control used
diethyl pyrocarbonate (DEPC)-treated water instead of the total
RNA, the positive control used the control RNA (contained within
the cDNA Synthesis kit) instead of the total RNA. In brief, the
following reagents were added in turn, 2 µl 10X reaction buffer, 1
µl RiboLock™ Ribonuclease Inhibitor (40 U/µl), DEPC-treated water
up to 10 µl and incubated at 37°C for 30 min, and then 1 µl reverse
transcriptase (1 U/µl) was added and incubated at 72°C for 5 min.
All reagents were contained within the cDNA synthesis kit. qPCR was
performed using a SYBR® Premix Ex Taq™ kit
(Takara Bio, Inc., Otsu, Japan) on the QuantStudio™ 5 Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.), and
with the following primers: miR-34a forward, 5′-TGGCAGTGTCTTAGCT-3′
(10 µM) and reverse, 5′-TGGTGTCGTGGAGTCG-3′ (10 µM); and U6 primer
was used as the positive control, U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ (10 µM) and reverse,
5′-AACGCTTCACGAATTTGCGT-3′ (10 µM; all BioSune, Shanghai, China).
The final concentration of the primers was 0.4 µM. A 3-step PCR was
performed using the SYBR Select Master Mix kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocols, and conducted under the following conditions: Initial
denaturation at 95°C for 30 sec, followed by a total of 40 cycles
were run with a initial denaturation at 95°C for 5 sec, and an
annealing temperature at 60°C for 30 sec. Finally, 95°C for 5 sec,
and 60°C for 1 min, and 95°C for 5 sec. Relative quantification was
analyzed using the 2−∆∆Cq method (24).
Invasion assay
Each Transwell insert was coated with diluted BD
Matrigel matrix coating solution (BD Biosciences, Franklin Lakes,
NJ, USA) and then incubated at 37°C for 30 min. Cell suspension
(5×104 cells/ml) in Opti-MEM® I culture media
(Thermo Fisher Scientific, Inc.) with no FBS was prepared. The
negative control was cells transfected with miR-NC. Cell suspension
(1 ml) was added to each 6-well invasion chamber. Culture media
with 20% FBS was placed in each Transwell chamber and incubated in
a humidified incubator (37°C, 5% CO2). Matrix and
non-invading cells were gently scraped off following 24 h. The
cells were fixed with 95% ethanol for 20 min and subsequently
stained with hematoxylin at room temperature for 10 min. The
membrane was washed with PBS and observed under a fluorescent
microscope (Nikon Corporation, Tokyo, Japan). A total of 5 fields
(×20) were selected randomly and the number of invaded cells were
counted.
Statistical analyses
Experimental data are presented as the mean ±
standard deviation. Multiple groups were compared using one-way
analysis of variance with SPSS (version 17.0, SPSS, Inc., Chicago,
IL, USA) statistical software. P<0.05 was used to indicate
significant difference. All independent experiments were conducted
in triplicate.
Results
miR-34a expression is promoted by
5-FU
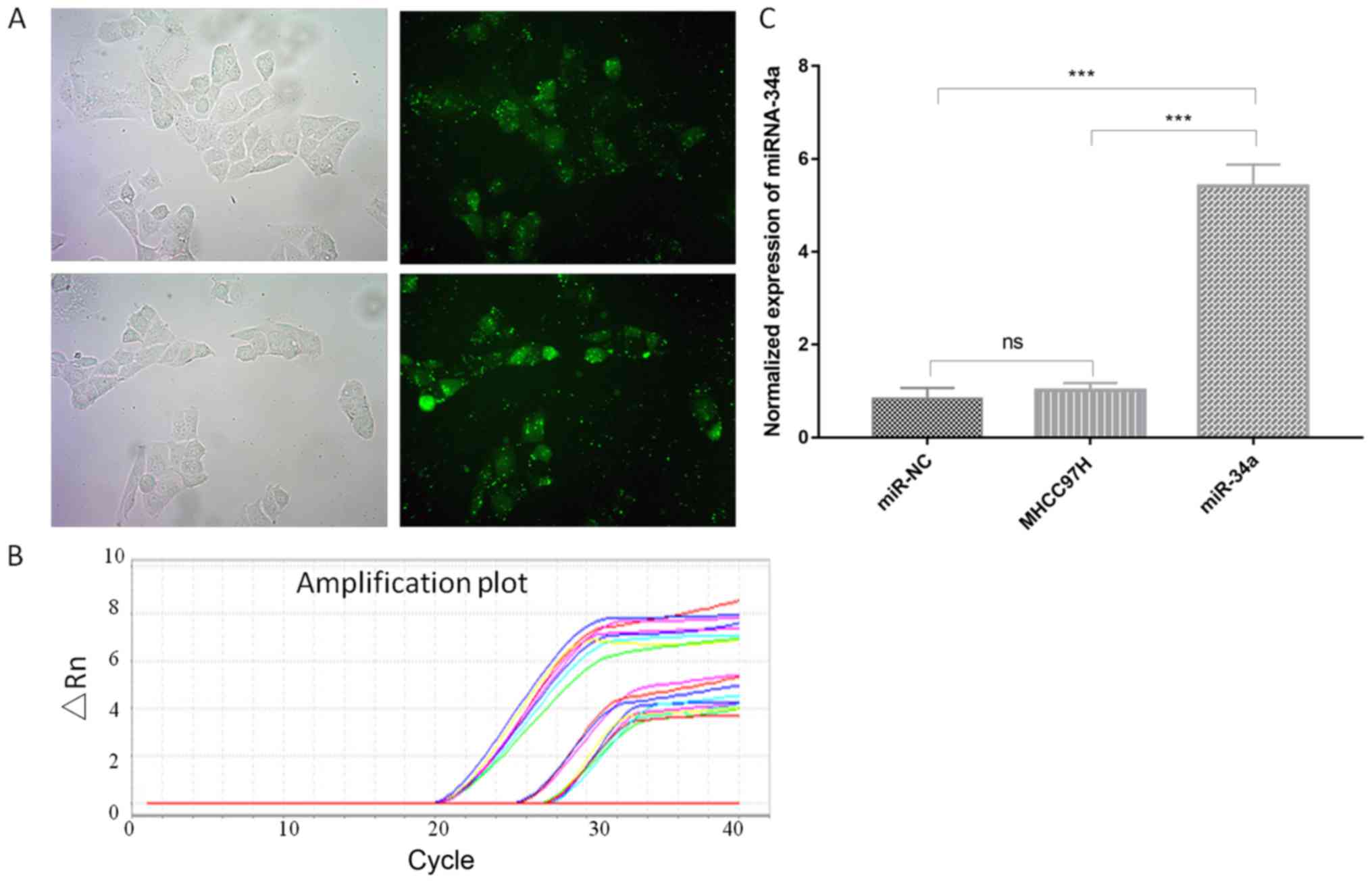
To study the effect of miR-34a on cancer cells, cell
lines were transfected with negative miR control (NC) and miR-34a
vector. The transfected cells were green (Fig. 1A). The cells that were transfected
with miR-34a exhibited a markedly increased miR-34a expression
(5.35-fold) compared with non-transfected cells according to
RT-qPCR analysis (Fig. 1B and C;
Table I). The transfected cells were
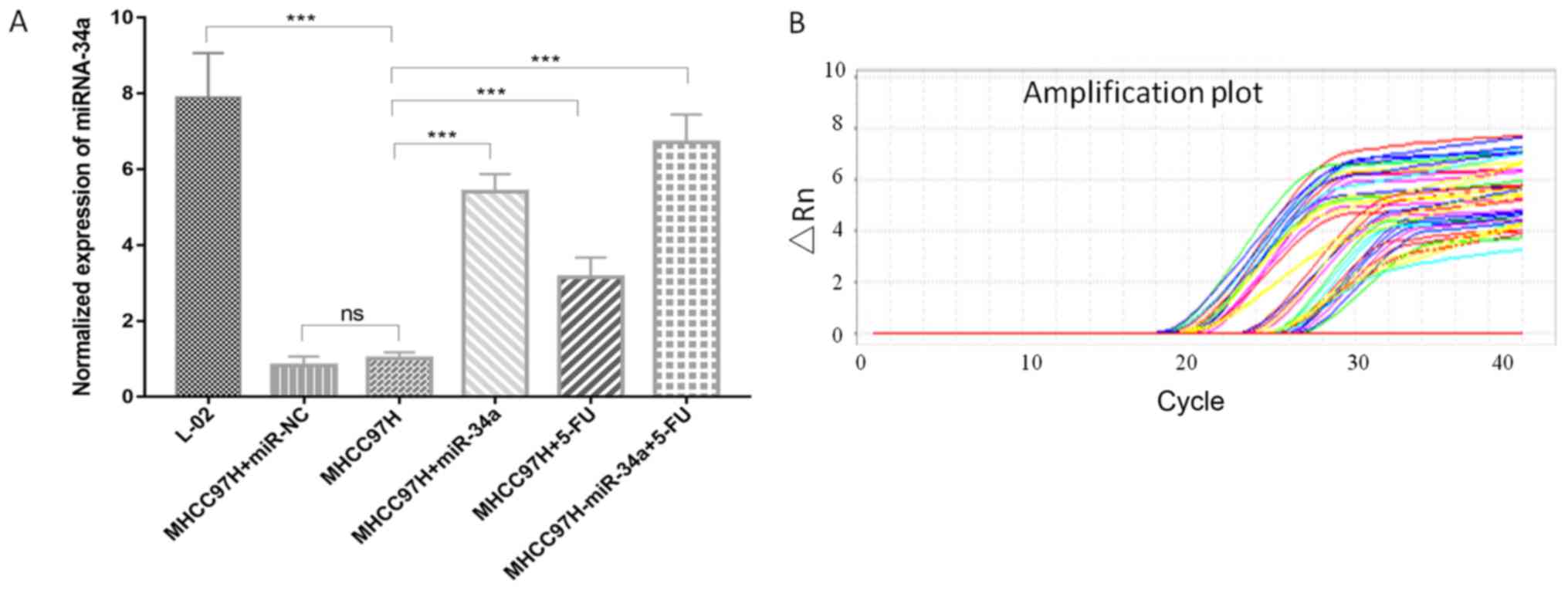
treated with 5 µg/ml 5-FU to observe miR-34a expression. 5-FU was
able to promote miR-34a expression. The cells that were transfected
with miR-34a and then treated with 5-FU (MHCC97H-miR-34a+5-FU) were
able to express a relatively increased level of miR-34a, compared
with the control group (MHCC97H-miR-34a) (Fig. 2 and Table
I).
 | Table I.Relative quantification of miR-34a
expression in transfected and 5-FU-treated cells. |
Table I.
Relative quantification of miR-34a
expression in transfected and 5-FU-treated cells.
|
| U6 | miR-34a |
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Groups | Cq | Mean Cq | Cq | Mean Cq | ∆Cq | ∆∆Cq |
2−ΔΔCq |
|---|
| L-02 | 20.13 | 20.02 | 25.01 | 24.93 | 4.91 | −2.97 | 7.84 |
|
| 20.12 |
| 25.02 |
|
|
|
|
|
| 20.14 |
| 25.01 |
|
|
|
|
|
| 20.21 |
| 24.71 |
|
|
|
|
|
| 19.63 |
| 24.82 |
|
|
|
|
|
| 19.58 |
| 24.70 |
|
|
|
|
|
| 20.03 |
| 25.01 |
|
|
|
|
|
| 20.00 |
| 25.03 |
|
|
|
|
|
| 20.31 |
| 25.06 |
|
|
|
|
| MHCC97H | 21.01 | 20.88 | 28.76 | 28.76 | 7.88 | 0.00 | 1.00 |
|
| 21.03 |
| 28.82 |
|
|
|
|
|
| 21.00 |
| 28.79 |
|
|
|
|
|
| 20.53 |
| 28.69 |
|
|
|
|
|
| 20.47 |
| 28.80 |
|
|
|
|
|
| 20.70 |
| 28.77 |
|
|
|
|
|
| 21.13 |
| 28.70 |
|
|
|
|
|
| 21.02 |
| 28.77 |
|
|
|
|
|
| 21.00 |
| 28.71 |
|
|
|
|
| MHCC97H-miR-NC | 20.14 | 20.49 | 28.67 | 28.67 | 8.18 | 0.30 | 0.81 |
|
| 20.31 |
| 28.69 |
|
|
|
|
|
| 20.22 |
| 28.65 |
|
|
|
|
|
| 21.00 |
| 28.71 |
|
|
|
|
|
| 21.03 |
| 28.77 |
|
|
|
|
|
| 21.01 |
| 28.72 |
|
|
|
|
|
| 20.14 |
| 28.89 |
|
|
|
|
|
| 20.01 |
| 28.36 |
|
|
|
|
|
| 20.52 |
| 28.55 |
|
|
|
|
|
MHCC97H-miR-34a | 21.03 | 20.43 | 26.49 | 25.89 | 5.46 | −2.42 | 5.35 |
|
| 21.00 |
| 26.45 |
|
|
|
|
|
| 21.01 |
| 26.42 |
|
|
|
|
|
| 20.56 |
| 26.00 |
|
|
|
|
|
| 20.79 |
| 26.09 |
|
|
|
|
|
| 20.33 |
| 26.10 |
|
|
|
|
|
| 19.85 |
| 25.18 |
|
|
|
|
|
| 19.68 |
| 25.12 |
|
|
|
|
|
| 19.66 |
| 25.10 |
|
|
|
|
| MHCC97H+5-FU | 18.76 | 19.30 | 25.03 | 25.54 | 6.24 | −1.64 | 3.12 |
|
| 18.42 |
| 25.02 |
|
|
|
|
|
| 18.62 |
| 25.05 |
|
|
|
|
|
| 19.01 |
| 25.42 |
|
|
|
|
|
| 19.10 |
| 25.32 |
|
|
|
|
|
| 19.24 |
| 25.40 |
|
|
|
|
|
| 20.20 |
| 26.33 |
|
|
|
|
|
| 20.01 |
| 26.15 |
|
|
|
|
|
| 20.32 |
| 26.11 |
|
|
|
|
|
MHCC97H-miR-34a+5-FU | 19.97 | 19.78 | 25.00 | 24.92 | 5.14 | −2.74 | 6.68 |
|
| 19.83 |
| 25.02 |
|
|
|
|
|
| 19.70 |
| 25.03 |
|
|
|
|
|
| 21.02 |
| 26.15 |
|
|
|
|
|
| 21.01 |
| 26.09 |
|
|
|
|
|
| 21.11 |
| 26.10 |
|
|
|
|
|
| 18.15 |
| 23.62 |
|
|
|
|
|
| 18.57 |
| 23.63 |
|
|
|
|
|
| 18.63 |
| 23.61 |
|
|
|
|
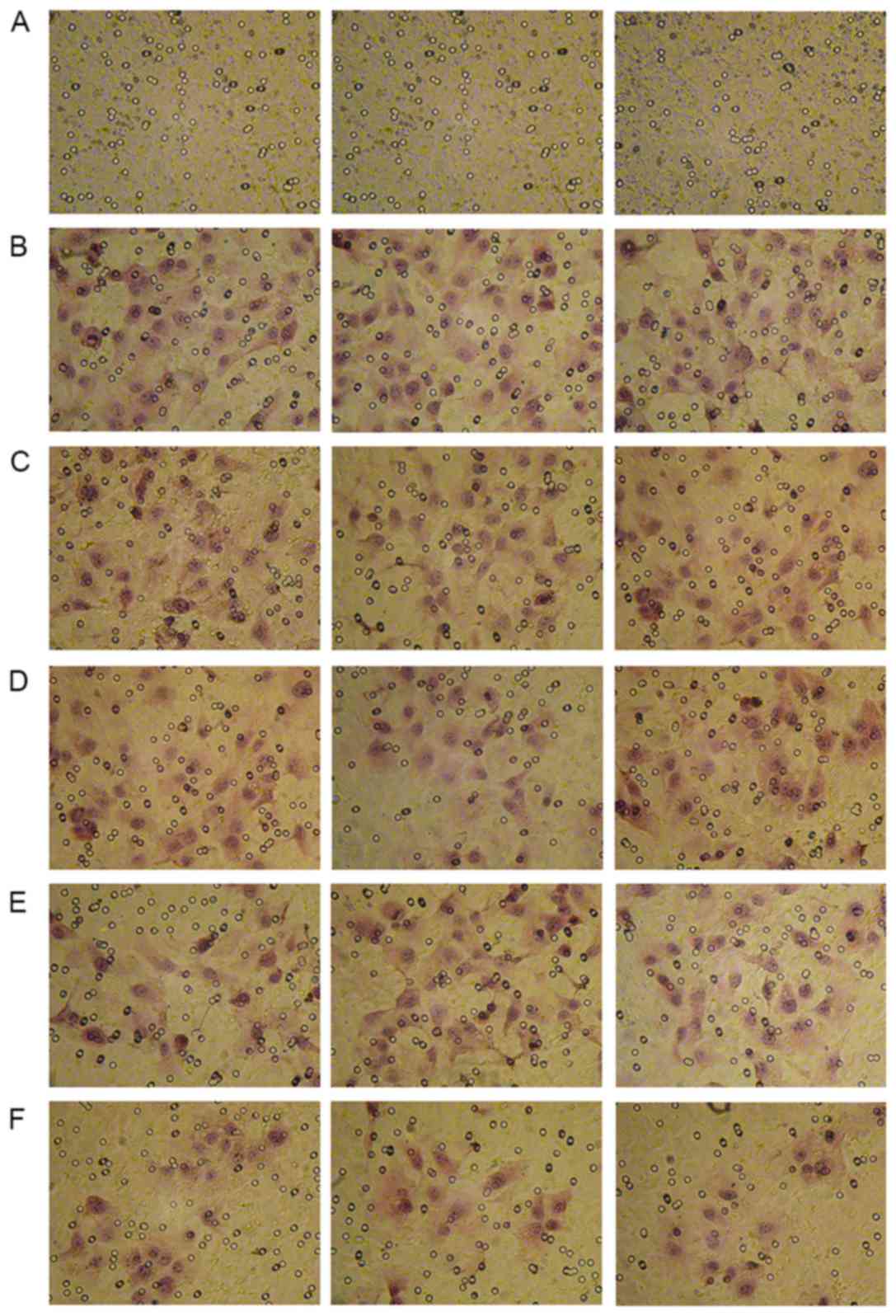
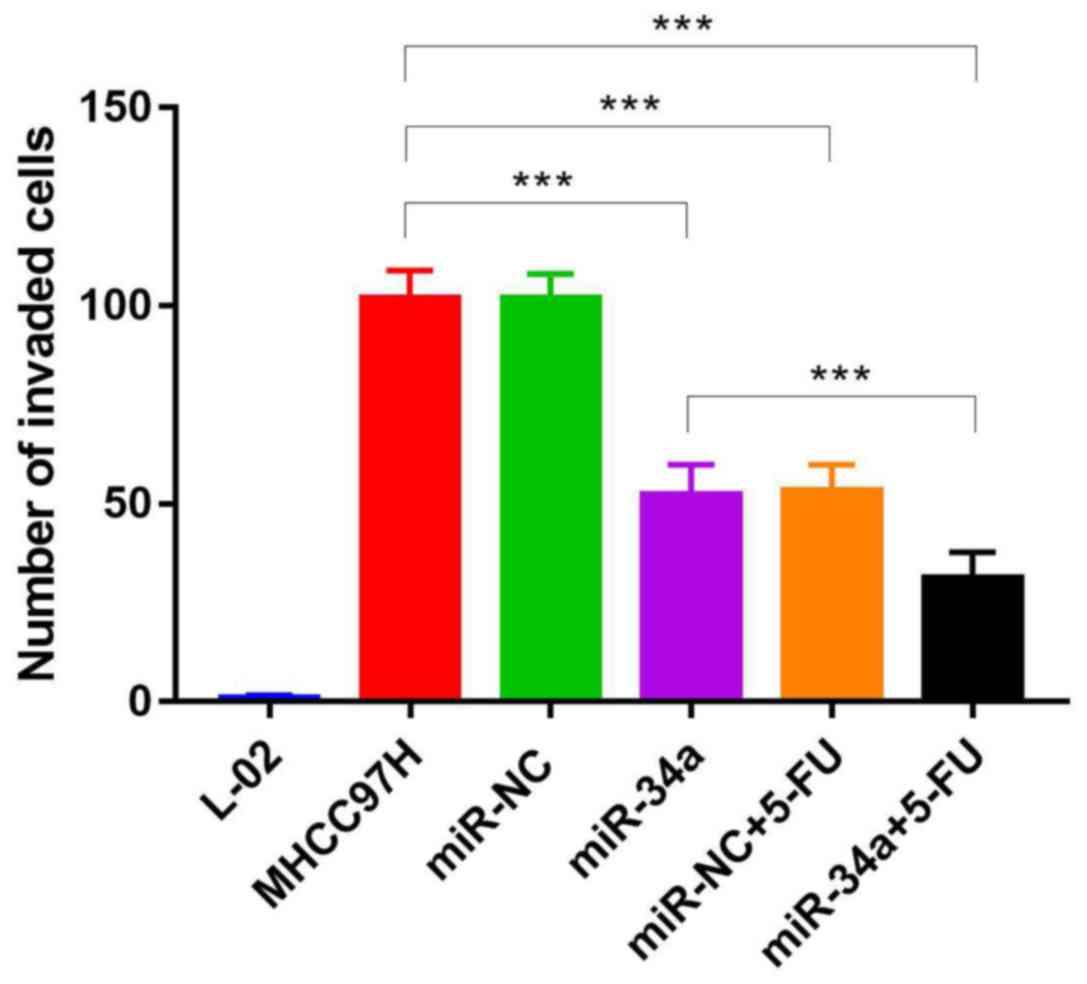
miR-34a inhibits cell invasion
To study the effect of miR-34a on metastasis,
invasion analysis of transfected and drug-treated cells was carried
out using Transwell assay. The overexpression of miR-34a had a
similar effect as 5-FU treatment as both treatments were able to
inhibit cell invasion. The mean number of invaded cells in five
random fields were 51.36±8.43 in the overexpressed miR-34a group.
The mean number of invaded cells in five random fields were
52.11±9.42 in the drug-treated MHCC97H-miR-NC group. While the mean
number of invaded cells in five random fields were 30.24±7.85 in
the drug-treated MHCC97H-miR-34a group. Furthermore, overexpressing
miR-34a was able to increase the effect of 5-FU, leading to reduced
cell invasion (P<0.05) (Figs. 3
and 4; Table II).
 | Table II.Effect of miR-34a or 5-FU on MHCC97H
cells invasion. |
Table II.
Effect of miR-34a or 5-FU on MHCC97H
cells invasion.
| Group | Mean number of
invaded cells (mean ± SD) | P-value (compared
with MHCC97H) |
|---|
| L-02 | 0 | – |
| MHCC97H | 105.14+15.87 | – |
| miR-NC | 103.26±20.59 | – |
| miR-34a | 51.36±8.43 | <0.05 |
| miR-NC+5-FU | 52.11+9.42 | <0.05 |
| miR-34a+5-FU | 30.24±7.85 | <0.05 |
Discussion
In recent years, miR-34a has been increasingly
studied. Previous studies have revealed miR-34a to be decreased in
tumors, compared with normal tissue (25). miR-34a is a member of the miR-34
family, and has been demonstrated to modulate critical gene
transcripts involved in tumorigenesis, but its role in
tumorigenesis remains unknown. miR-34a may be activated by p53 to
induce apoptosis and inhibit tumor growth (13–16);
however, it frequently causes dysfunctions or mutations in tumors
(17–21). Luo et al (22) demonstrated that miR-34a was able to
suppress the migration of breast cancer cells via targeting Fra-1.
Wang et al (23) observed an
inverse association between PD-L1 and miR-34a expression in a
number of AML samples. miR-34a, as a putative binder of the
PD-L1-3′UTR, overexpression in HL-60 and Kasumi-1 cells could block
PD-L1 expression (23). There is hope
to utilize miR-34a for diagnosis and therapy. Gallardo et al
(26) reported that miR-34 was able
to act as a prognostic marker of non-small-cell lung cancer, and
Fang et al (27) indicated
that miR-34a was able to be used to detect diffuse large B-cell
lymphoma. Furthermore, Wiggins et al (28) reintroduced chemically synthesized
miR-34a to cancer cells to drive a therapeutic response. The
results of the present study also suggested that overexpressing
miR-34a, which was depleted in cancer cells, was able to inhibit
cell invasion. According to its importance in tumorigenesis,
chemically synthesized miRNA mimic may be used to simulate
endogenous miRNA to target genes and inhibit tumor development.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WW and HT contributed in the design of the study,
developed the methodology, collected the data, performed the
experiments, analysis and wrote the manuscript; LT contributed to
the design of the study, critically revised the manuscript and
approved the final version to be published. All authors agreed to
be accountable for all aspects of the study.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Berezikov E, Cuppen E and Plasterk RHA:
Approaches to microRNA discovery. Nat Genet. 38 (Suppl):S2–S7.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Neely LA, Patel S, Garver J, Gallo M,
Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S and Rooke J:
A single-molecule method for the quantitation of microRNA gene
expression. Nat Methods. 3:41–46. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shyu AB, Wilkinson MF and van Hoof A:
Messenger RNA regulation: To translate or to degrade. EMBO J.
27:471–481. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Callis TE, Chen JF and Wang DZ: MicroRNAs
in skeletal and cardiac muscle development. DNA Cell Biol.
26:219–225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Giannakakis A, Coukos G, Hatzigeorgiou A,
Sandaltzopoulos R and Zhang L: miRNA genetic alterations in human
cancers. Exp Opin Biol Ther. 7:1375–1386. 2007. View Article : Google Scholar
|
|
8
|
Hatfield S and Ruohola-Baker H: microRNA
and stem cell function. Cell Tissue Res. 331:57–66. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahashi K, Yan I, Wen HJ and Patel T:
microRNAs in liver disease: From diagnostics to therapeutics. Clin
Biochem. 46:946–952. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomimaru Y, Eguchi H, Nagano H, Wada H,
Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M,
Umeshita K, et al: MicroRNA-21 induces resistance to the
anti-tumour effect of interferon-α/5-fluorouracil in hepatocellular
carcinoma cells. Br J Cancer. 103:1617–1626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Karakatsanis A, Papaconstantinou I,
Gazouli M, Lyberopoulou A, Polymeneas G and Voros D: Expression of
microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c,
miR-221, miR-222, and miR-223 in patients with hepatocellular
carcinoma or intrahepatic cholangiocarcinoma and its prognostic
significance. Mol Carcinog. 52:297–303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bommer GT, Gerin I, Feng Y, Kaczorowski
AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al:
p53-mediated activation of miRNA34 candidate tumor-suppressor
genes. Curr Biol. 17:1298–1307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Raver-Shapira N, Marciano E, Meiri E,
Spector Y, Rosenfeld N, Moskovits N, Bentwich Z and Oren M:
Transcriptional activation of miR-34a contributes to p53-mediated
apoptosis. Mol Cell. 26:731–743. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tarasov V, Jung P, Verdoodt B, Lodygin D,
Epanchintsev A, Menssen A, Meister G and Hermeking H: Differential
regulation of microRNAs by p53 revealed by massively parallel
sequencing: miR-34a is a p53 target that induces apoptosis and
G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ichimura A, Ruike Y, Terasawa K, Shimizu K
and Tsujimoto G: MicroRNA-34a inhibits cell proliferation by
repressing mitogen-activated protein kinase kinase 1 during
megakaryocytic differentiation of K562 cells. Mol Pharmacol.
77:1016–1024. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Di Martino MT, Leone E, Amodio N, Foresta
U, Lionetti M, Pitari MR, Cantafio ME, Gullà A, Conforti F, Morelli
E, et al: Synthetic miR-34a mimics as a novel therapeutic agent for
multiple myeloma: In vitro and in vivo evidence. Clin Cancer Res.
18:6260–6270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li L, Xie X, Luo J, Liu M, Xi S, Guo J,
Kong Y, Wu M, Gao J, Xie Z, et al: Targeted expression of miR-34a
using the T-VISA system suppresses breast cancer cell growth and
invasion. Mol Ther. 20:2326–2334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pang RT, Leung CO, Lee CL, Lam KK, Ye TM,
Chiu PC and Yeung WS: MicroRNA-34a is a tumor suppressor in
choriocarcinoma via regulation of Delta-like1. BMC Cancer.
13:252013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guessous F, Zhang Y, Kofman A, Catania A,
Li Y, Schiff D, Purow B and Abounader R: microRNA-34a is tumor
suppressive in brain tumors and glioma stem cells. Cell Cycle.
9:1031–1036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo YP, Zhou H, Krueger J, Kaplan C, Liao
D, Markowitz D, Liu C, Chen T, Chuang TH, Xiang R and Reisfeld RA:
The role of proto-oncogene Fra-1 in remodeling the tumor
microenvironment in support of breast tumor cell invasion and
progression. Oncogene. 29:662–673. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Li J, Dong K, Lin F, Long M,
Ouyang Y, Wei J, Chen X, Weng Y, He T and Zhang H: Tumor suppressor
miR-34a targets PD-L1 and functions as a potential
immunotherapeutic target in acute myeloid leukemia. Cell Signal.
27:443–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thor T, Künkele A, Pajtler KW, Wefers AK,
Stephan H, Mestdagh P, Heukamp L, Hartmann W, Vandesompele J,
Sadowski N, et al: miR-34a deficiency accelerates medulloblastoma
formation in vivo. Int J Cancer. 136:2293–2303. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gallardo E, Navarro A, Viñolas N, Marrades
RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez
J and Monzo M: miR-34a as a prognostic marker of relapse in
surgically resected non-small-cell lung cancer. Carcinogenesis.
30:1903–1909. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH,
Liu L, Fan L, Miao KR, Liu P, Xu W and Li JY: Serum microRNAs are
promising novel biomarkers for diffuse large B cell lymphoma. Ann
Hematol. 91:553–559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wiggins JF, Ruffino L, Kelnar K, Omotola
M, Patrawala L, Brown D and Bader AG: Development of a lung cancer
therapeutic based on the tumor suppressor microRNA-34. Cancer Res.
70:5923–5930. 2010. View Article : Google Scholar : PubMed/NCBI
|