Introduction
Colorectal cancer (CRC), usually starting as a
harmless polyp, is one of the most common gastrointestinal
malignancies. Globally, CRC is the third most common cancer in the
diagnosis of male diseases, ranking second in women. Approximately
1.4 million new CRC cases and 700,000 CRC deaths were evaluated in
2012. It has been found that the incidence of CRC is highest in
North America, Europe, and Australia/New Zealand, and other
countries with lower history rate are currently facing increased
risk (1). The risk factors of CRC
include environment (such as obesity, alcohol consumption and
tobacco smoking), heredity and gene-environment interactions
(2). Stool tests, sigmoidoscopy or
colonoscopy, and virtual colonoscopy are common clinical screening
methods for CRC (3). Although the
fecal occult blood testing and sigmoidoscopy can provide valuable
information for the guide therapy of CRC, they are not sufficient
to confirm clinical outcomes. Therefore, seeking new effective
diagnostic methods and exploring the canceration mechanism from
polyp becomes urgent. It has been reported that bioinformatics
analysis of gene expression profiles could clearly explain the
occurrence, development, metastasis and prognosis of tumors, and is
a hot spot in the current basic and clinical research (4–6).
Long non-coding RNA (lncRNA) is a novel class of
transcripts with 200 nucleotides that do not serve as a template
for protein (7), but they are
involved in a variety of biological processes, such as epigenetic
regulation, chromosome remodeling and gene expression regulation
(8). Studies have found that lncRNAs
are abnormally expressed in many tumors, including non-small cell
lung cancer, cervical cancer, hepatocellular cancer, and human
breast cancer (9–12). The expression level of other
transcripts as miRNA sponges could be regulated by these lncRNAs
through competitive endogenous RNAs (ceRNAs) network (13,14).
Furthermore, some canonical oncogenic pathways can be revealed by
ceRNA interaction network (15), and
Wang et al have reported that the newly identified ceRNA
network contributes to the exploration of the regulatory mechanisms
of ceRNA mediated by lncRNA in the nosogenesis of muscle-invasive
bladder cancer (16). However, it
remains a challenge to identify lncRNA biomarkers of CRC, and to
understand the functional roles of ceRNAs mediated by lncRNA in
CRC.
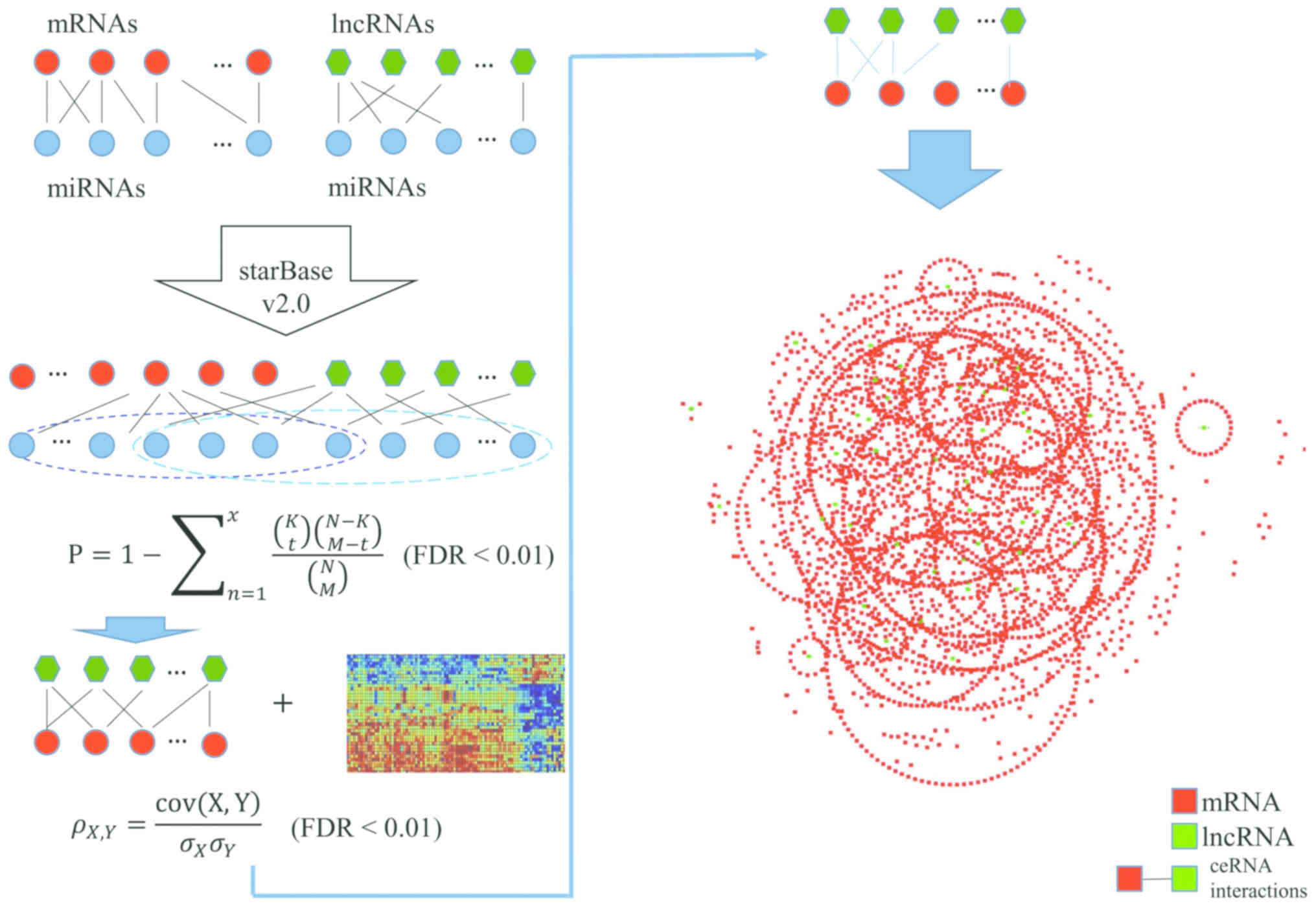
In the present study, a functional lncRNA-mediated
ceRNA network (LMCN) associated with CRC was constructed using a
multi-step computational method, and the relevant lncRNAs were
identified based on the constructed landscape map. Then we carried
out functional enrichment analyses for mRNAs which were
significantly associated with lncRNAs, and used the functions of
the mature mRNAs to forecast the lncRNA functions.
Materials and methods
Data source
Expression profiles of lncRNA and mRNA
in CRC
In the present study, the gene expression profile
dataset no. GSE31737, deposited by Loo et al (17) in the Gene Expression Omnibus (GEO)
database (http://www.ncbi.nlm.nih.gov/geo/) based on the GPL5175
platform [HuEx-1_0-st; Affymetrix Human Exon 1.0 ST Array:
transcript (gene) version; Affymetrix; Thermo Fisher Scientific,
Inc., Waltham, MA, USA], was subjected to bioinformatics analysis.
A total of 80 chips were involved in the dataset, including 40
colon cancer tissues (tumor group) and 40 paired adjacent-normal
colon tissues (control group). After preprocessing and mapping
between genes and probes, we obtained 14,451 gene expression
profile data.
Identification of miRNA-target
interactions
Firstly, interactions between miRNA and mRNA, and
between miRNA and lncRNA were obtained from the starBase v2.0 which
provided the most comprehensive decoding of miRNA-ceRNA,
miRNA-ncRNA and protein-RNA interaction networks from large-scale
CLIP-Seq data (18). Then a new
expression profile was acquired through taking the intersection
between the genes in the above-mentioned expression profile data of
CRC and the mRNAs and lncRNAs, respectively, which were located in
the interactions of miRNA-mRNA and lncRNA-miRNA. Subsequently, the
interaction relationship of the new gene expression profile were
extracted from the downloaded interactions of miRNA-mRNA and
miRNA-lncRNA.
Construction of LMCN
The competitive lncRNA-mRNA interactions were
identified using a hypergeometric test that was able to effectively
assess the significance of the miRNAs shared by each lncRNA and
mRNA. There were a total of N miRNAs in the genome, where
K and M represented the number of miRNAs related to
the current lncRNA and mRNA, and x represented the number of
common miRNAs shared by lncRNA and mRNA. P-value was calculated in
order to evaluate the enrichment significance for that function
using the formula:
P=1-∑n=1x(kt)(N-KM-t)(NM)
False discovery rate (FDR) correction was performed
to adjust the P-values. FDR<0.01 was used as the threshold of
LMCN.
The lncRNA-mRNA interaction-pairs screened out above
were co-expression analyzed through calculating their Pearson's
correlation coefficient in the control and tumor group. The
calculation formula used was:
ρX,Y=cov(X,Y)σXσY
of which cov(X,Y) represents the covariance of
variables X and Y, and σX and σY represent
the standard deviations of X and Y, respectively. FDR<0.01 was
set as the threshold. The lncRNA-mRNA interaction-pairs whose
difference values of Pearson's correlation coefficient was >0.3
between control and tumor groups were taken as the significant
co-expression ceRNA interactions (19). The ceRNA network was constructed and
described using a Cytoscape software (v3.5.1; Cytoscape Consortium,
New York, NY, USA). A complete bipartite graph was involved in this
model in which an edge was achieved from each apex of the lncRNA
set to each vertex of the mRNA set.
We studied the relationship between the number of
genes and the degree of distribution, and the fitting coefficient
R2 of the power-law Y = aXb of the objective
networks was detected (20). The
built-in Network Analyzer tool (Agilent Technologies, Inc., Santa
Clara, CA, USA) was used to analyze the node degree and the
betweenness centrality (BC). BC represents a central metric for the
nodes in the network. It can be obtained by computing the number of
the shortest paths through the node, from each node to all others.
This property reflects the control ability or the probability to
exert the ‘gate-keeping’ effect of a node in the ceRNA network.
Generally, a higher degree implies that the node is a hub involved
in more ceRNA interactions. A higher BC indicates that the node is
a bottleneck that functions as a bridge to connect different
network modules.
Screening of hub lncRNAs
LMCN can provide an all-sided landscape of all
possible ceRNA interactions, which contributes in exploring the
regulatory roles of the lncRNAs. Moreover, more detailed view on
the synergy between lncRNAs and competing mRNAs can be confirmed by
analyzing the regional sub-networks. A high-competitive sub-network
was extracted from the LMCN based on a Pearson's correlation
coefficient threshold >0.5. lncRNAs with more degrees and BCs
were selected out as hub lncRNAs involved in CRC.
Function prediction of lncRNAs
Functions of the hub lncRNAs can be studied with a
‘guilt by association’ (GBA) approach (21,22). It
has been stated by GBA principle that genes that participate in the
same biological process are inclined to be correlated (or have
similar properties, such as similar expression patterns), allowing
statistically to infer previously unknown functions of a gene based
on some previous knowledge on other genes and correlative data
(23,24). In this study, the GBA meant that the
function of lncRNAs could be deduced though carrying out a
functional enrichment analysis for the mature mRNAs which were
significantly associated with lncRNAs.
Function and pathway enrichment analysis were
performed to evaluate the lncRNAs competitive mediated modules with
functionality obtained from the Gene Ontology (GO) database
(25) as well as pathways acquired
from the Reactome pathway database (26). The significance thresholds were set as
P<0.01 in functional analysis, and P<0.05 in pathway analysis
after Fisher's correction.
Results
The LMCN reveals specific topological
properties
The LMCN was built with a multi-step method to
assess the lncRNA-mediated ceRNA interactions. For miRNA-lncRNA
interactions and miRNA-mRNA interactions in CRC, we collected and
integrated full-scale datasets from the starBase v2.0 which
provided experimentally supported high confidence miRNA-target
interactions. A total of 265,728 pairs of miRNA-mRNA interactions
as well as 598 pairs of lncRNA-miRNA interactions were acquired
from a new expression profile covering a total of 8,522 genes
consisting of 8,471 mRNAs and 51 lncRNAs. Based on the studies that
lncRNA transcripts competed with endogenous mRNAs by binding to
miRNAs that could be predicted with current miRNA target prediction
approach (27,28). The significance of common miRNAs
shared by each lncRNA-mRNA pair was examined with a hypergeometric
test. Then 51 lncRNAs, 8,125 mRNAs and 3,586 lncRNA-mRNA pairs were
obtained. Pearson's correlation coefficient of the identified
candidate ceRNA pair was calculated to confirm active ceRNA pairs
in CRC. Eventually, we constructed and graphically simulated the
LMCNs using the significantly co-expressed lncRNA-mRNA ceRNA pairs
(Fig. 1). There were 51 lncRNAs,
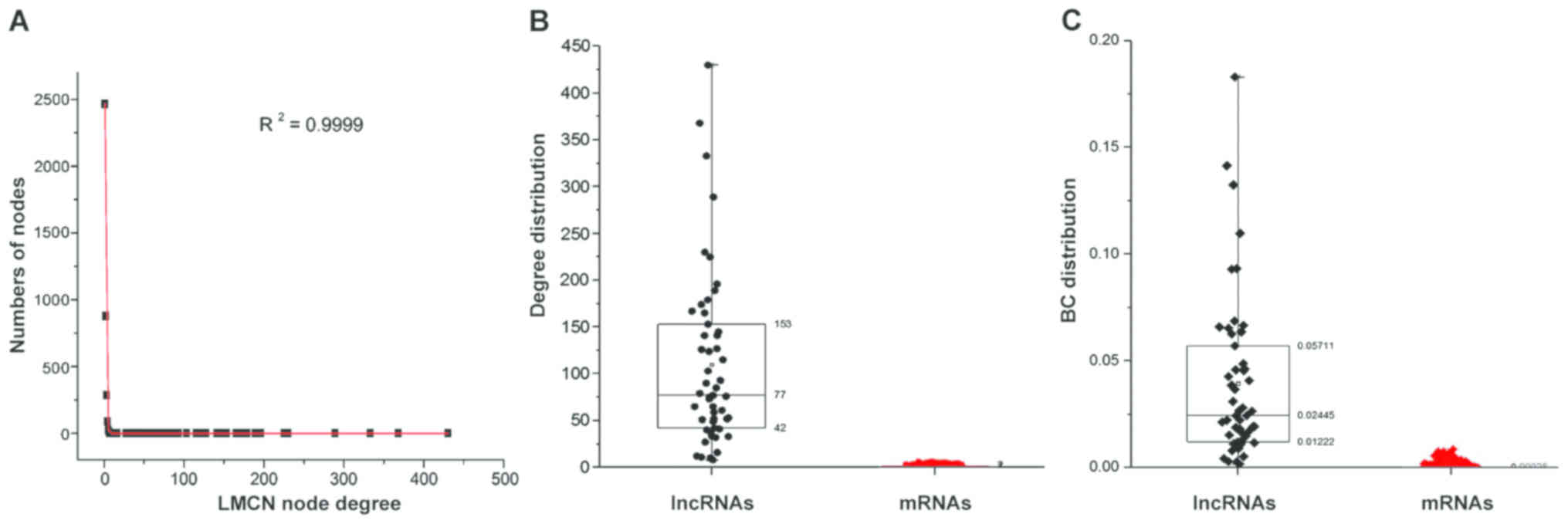
3,752 mRNAs, and 5,617 ceRNA interactions in the LMCN. The
statistics of nodes degree (R2=0.9999) showed power law
distributions (Fig. 2A), indicating
that the LMCN related to CRC is a scale-free network. Moreover, the
lncRNA nodes exhibited more specific topological properties with
more degrees and BCs in comparison with mRNA nodes (Fig. 2B and C).
HCP5, EPB41L4A-AS1, SNHG12, and
LINC00649 are the hub lncRNAs in CRC
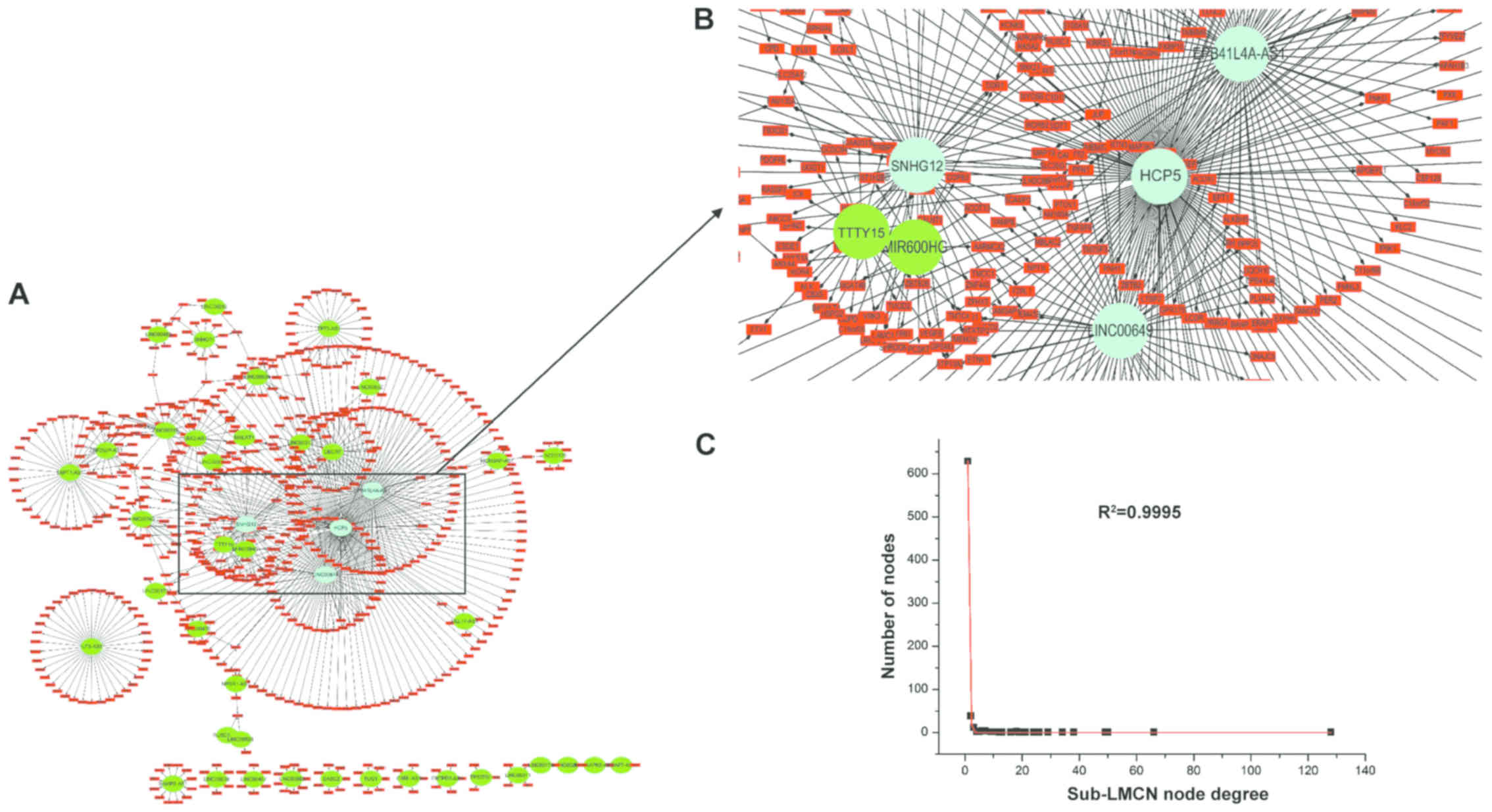
To determine the hub lncRNAs with highly specific
properties in CRC, a sub-LMCN covering lncRNA-mRNA co-expression
interactions with Pearson's correlation coefficient deference
>0.5 between tumor and control groups was constructed (29). Results showed that the sub-LMCN also
had a power-law distribution scaleless structure (Fig. 3C; R2=0.9995). The landscape
map contained 42 lncRNAs, 681 mRNAs, and 730 ceRNA interactions
(Fig. 3A). We found that HCP5,
EPB41L4A-AS1, SNHG12, and LINC00649 had more node degrees and BCs
in the sub-LMCN than others (Fig.
3B), indicating that they may be the prognostic markers in
CRC.
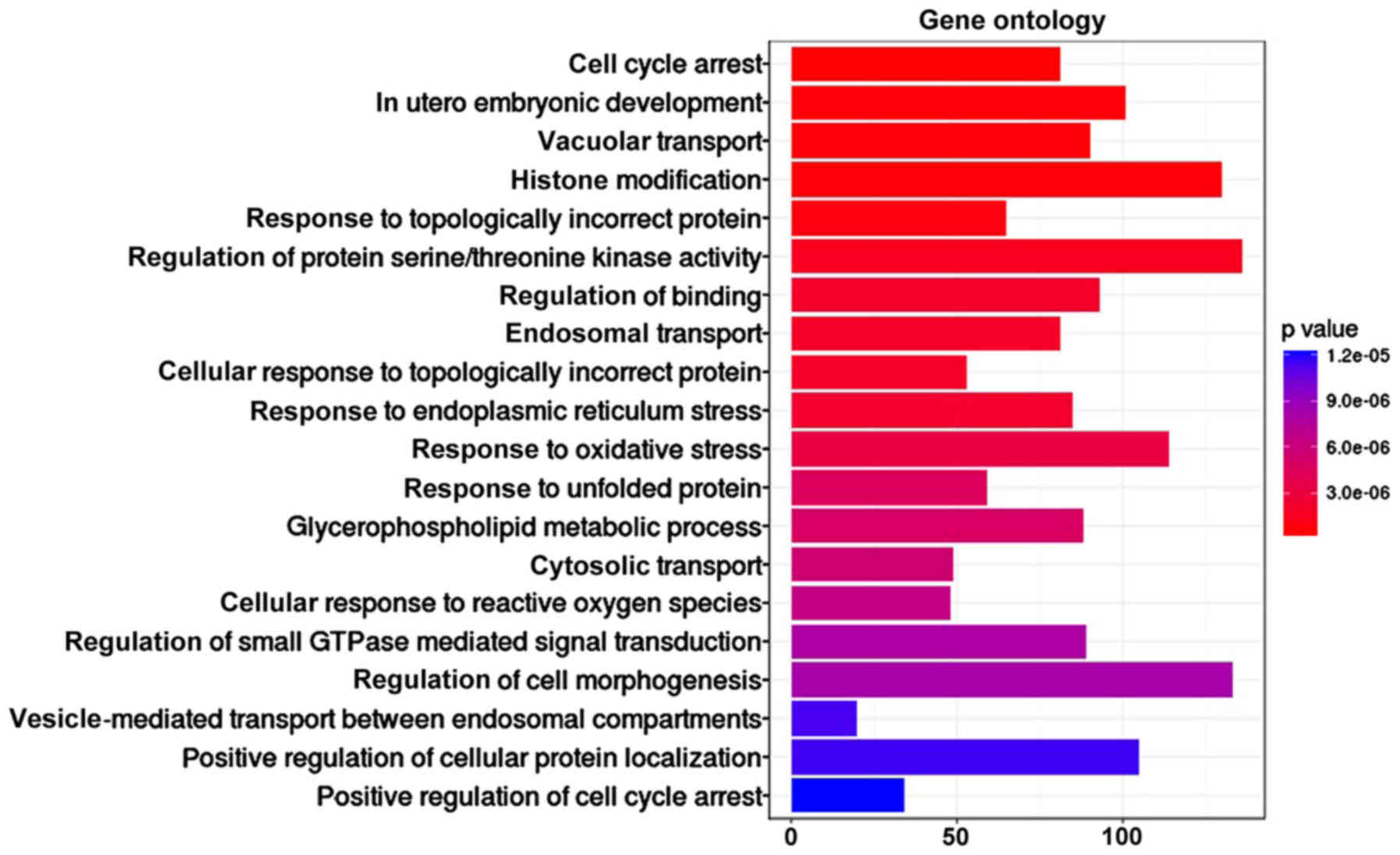
Sixty-three functions and 66 pathways
enriched from GO and Reactome pathway database
To further validate the potential functional
implication of lncRNAs, function and pathway enrichment analysis of
mRNAs in the sub-LMCN were conducted based on GO and Reactome
pathway database. The mRNAs associated with hub lncRNAs were
obviously enriched in 63 GO terms, especially involved in cell
cycle arrest, vacuolar transport, histone modification, and
regulation of protein serine/threonine kinase activity (Fig. 4), and 66 Reactome pathways, including
GPCR pathway, signaling by Rho GTPases, axon guidance and
developmental biology (Table I).
 | Table I.The top 20 enriched Reactome pathways
of mRNAs associated with lncRNAs. |
Table I.
The top 20 enriched Reactome pathways
of mRNAs associated with lncRNAs.
| No. | Reactome
pathway | P-value (FDR) | No. of genes |
|---|
| 1 | GPCR downstream
signaling | 2.64E-29 | 76 |
| 2 | Signaling by
GPCR | 4.09E-25 | 93 |
| 3 | GPCR ligand
binding | 6.93E-13 | 33 |
| 4 | Class A/1
(Rhodopsin-like receptors) | 1.31E-09 | 22 |
| 5 | Signaling by Rho
GTPases | 1.01E-06 | 125 |
| 6 | Peptide
ligand-binding receptors | 1.05E-06 | 11 |
| 7 | Complement
cascade | 7.60E-06 | 1 |
| 8 | Axon guidance | 2.07E-05 | 110 |
| 9 | Developmental
Biology | 5.78E-05 | 158 |
| 10 | G α (i) signalling
events | 5.78E-05 | 21 |
| 11 | Initial triggering
of complement | 3.70E-04 | 1 |
| 12 | Membrane
trafficking | 3.70E-04 | 70 |
| 13 | RHO GTPase
effectors | 3.70E-04 | 85 |
| 14 | Asparagine
N-linked glycosylation | 1.12E-03 | 46 |
| 15 | UPR | 1.12E-03 | 37 |
| 16 | Sema4D in
semaphorin signaling | 1.66E-03 | 16 |
| 17 | Biological
oxidations | 2.43E-03 | 16 |
| 18 | Scavenging heme
from plasma | 2.43E-03 | 1 |
| 19 | Immunoregulatory
interactions between a lymphoid and a non-lymphoid cell | 2.47E-03 | 9 |
| 20 | Translocation of
GLUT4 to the plasma membrane | 2.85E-03 | 27 |
Discussion
Considering the characteristics with hidden
tumorigenesis, apt to local invasion and metastasis of CRC, what we
primarily do for good prognosis is to ascertain pathogenesis, make
early diagnosis and targeted individualized treatment.
Bioinformatics is one of the interdisciplinaries involved in many
fields, and one of its core elements is to study the internal
expression regulatory mechanisms of genes and reveal essential laws
of human diseases. lncRNA is an important type of gene expression
regulatory factor in bioinformatics, and is involved in many human
diseases, such as tumors (30),
cardiovascular disease (31),
inflammation (32) and autoimmune
disease (33). The interconnections
of ceRNAs play a crucial role in the development and physiology of
tumors (34,35). In the present study, we constructed a
functional LMCN to explore the intermodulation relationship between
lncRNAs and mRNAs, and predicted the functions and pathways
involved in lncRNAs across 80 CRC samples. Firstly, the LMCN was
constructed through integrating the expression profiles of
genome-wide lncRNA/mRNA and comprehensive interactions of
miRNA-target. The LMCN presented a scale-free network conforming to
power-law distributions, and the lncRNA nodes exhibited more
specific topological properties with more degrees and BCs than mRNA
nodes. Thus, these results indicated that LMCN has similar
properties with many biological networks, and can be greatly
organized into a structured rather than random network through a
set of core lncRNA-mRNA competing principles (36,37). Then
a sub-LMCN was established to identify the hub and bottleneck
lncRNAs with a threshold of Pearson's correlation coefficient
deference >0.5. We found that HCP5, EPB41L4A-AS1, SNHG12, and
LINC00649 have more node degrees and BCs in the landscape map.
Finally, function enrichment analyses of mature mRNAs from GO and
Reactome pathway databases were carried out to predict the
functions of the obviously relative lncRNAs based on the GBA
principle. As a result, the lncRNAs were found to be involved in 63
GO terms and 66 Reactome pathways, including the biological process
of cell cycle arrest, vacuolar transport, histone modification,
regulation of protein serine/threonine kinase activity, and the
pathways of signaling by Rho GTPases, GPCR pathway, axon guidance,
developmental biology had lower P-values and covered more
mRNAs.
lncRNAs regulate the expression level of
protein-coding genes in different ways, such as basic regulation
process of genes and post-transcriptional regulation (38–40). In
this study, we found that HCP5, EPB41L4A-AS1, SNHG12, and LINC00649
present stronger centrality and bridge-joint, meaning that they are
more important than other lncRNAs in the onset and progress of CRC.
Some investigations have shown that SNHG12 could increase cell
cycle-related protein expression and suppress caspase-3 expression
in CRC and human osteosarcoma cells. Moreover, silencing SNHG12
expression can inhibit triple-negative breast cancer cell
proliferation, and it has been found that SNHG12, as an endogenous
sponge of miR-199a/b-5p, regulates MLK3 expression in
hepatocellular carcinoma and affects the activation of the NF-κB
pathway (41–44). Therefore, SNHG12 may be considered as
a potential biomarker and a promising therapeutic target for CRC
and other cancers. It has been reported that (HCP5) is highly
expressed in glioma tissues and U87 and U251 cells, which can
promote the malignant biological behavior of glioma cells by
enhancing proliferation, migration as well as invasion, and
inhibiting apoptosis (45).
Furthermore, a single nucleotide polymorphism in HCP5 (rs2244546)
has been confirmed as a strong predictor of hepatitis C
virus-related hepatocellular carcinoma in the Swiss Hepatitis C
Cohort Study (46). EPB41L4A-AS1 was
found a risky lncRNA in ovarian cancer (47). HCP5 and EPB41L4A-AS1 could be boldly
inferred as potential lncRNA biomarkers for the diagnosis,
evaluation and gene-targeted therapy of CRC, but further studies
needs to be carried out to verify this inference.
GO analysis suggested that the target genes of hub
lncRNAs are significantly associated with cell cycle and histone
modification. Cell cycle regulation is of great significance in
maintaining homeostasis and preventing cancer, and its abnormity
has been long regarded as an important intermediate link for the
tumor development (48–50). The basic function of histone
modification is to regulate gene expression, and phosphorylation of
histones is not only an important intermediate step of certain
signal transduction pathways, but also affects cell division and
cycle through combining with other types of modification (51,52).
Therefore, the result in this study was consistent with previous
reports, and the occurrence and development of CRC are closely
related to cell cycle and histones modification. The reactome
pathway analysis showed that the lncRNAs might participate in the
Rho GTPases and axon guidance pathways. Rho GTPases family is an
intracellular signal transducer that connects cell surface signals
to multiple intracellular reactions. They playe a critical role in
various cellular processes, including cell morphology, gene
transcription, cell cycle progression, and cell adhesion (53,54).
Furthermore, it has been reported that oncogene-specific cell
migration and invasion pathways are mediated by Rho GTPases in
colon cancer cells (55). Semaphorins
primarily known as ligands for plexins and neuropilins are key
members of axon guidance molecules (56). Many studies have found that semaphorin
proteins have certain modulatory effects on tumorous occurrence and
development (57–59), for example, the axon guidance molecule
semaphorin 3F was found in normal neuroendocrine cells, while it
was lost in most human primary tumors and all metastases. Axon
guidance molecule semaphorin 3F plays a negative regulator role in
the development of tumor in ileal neuroendocrine tumors (60). Additionally, the inhibition of
migration, invasion and epithelial to mesenchymal transition has
been revealed because of the overexpression of SEMA3A by
suppressing the expression of NF-κB and SNAI2 in head and neck
squamous cell carcinoma (HNSCC); whereas, a shorter overall
survival and more important independent prognostic significance
could be found in HNSCC patients with lower SEMA3A expression
(61). Therefore, axon guidance cues
could be promising targets for personalized anticancer therapies.
In short, the results of the present study indicated that
cancer-related LMCN contributes in improving the identification of
biomarkers and promotes the development of CRC therapy.
In conclusion, we have provided the LMCN landscape
across 80 CRC samples with a new method. The specific topological
properties and synergistic competition effects of lncRNAs may
reveal the regulatory interactions of coding mRNAs in CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MH analyzed the data and was also a major
contributor in writing the manuscript. YL and YX made substantial
contribution to the conception and design of the study. MH, YL
performed statistical analysis. YX was involved in the drafting of
the manuscript and gave the final approval for publication. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marley AR and Nan H: Epidemiology of
colorectal cancer. Int J Mol Epidemiol Genet. 7:105–114.
2016.PubMed/NCBI
|
|
3
|
Bretthauer M: Colorectal cancer screening.
J Intern Med. 270:87–98. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Itzel T, Scholz P, Maass T, Krupp M,
Marquardt JU, Strand S, Becker D, Staib F, Binder H, Roessler S, et
al: Translating bioinformatics in oncology: Guilt-by-profiling
analysis and identification of KIF18B and CDCA3 as novel driver
genes in carcinogenesis. Bioinformatics. 31:216–224. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fortney K and Jurisica I: Integrative
computational biology for cancer research. Hum Genet. 130:465–481.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hao L, Leng J, Xiao R, Kingsley T, Li X,
Tu Z, Yang X, Deng X, Xiong M, Xiong J, et al: Bioinformatics
analysis of the prognostic value of Tripartite Motif 28 in breast
cancer. Oncol Lett. 13:2670–2678. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fritah S, Niclou SP and Azuaje F:
Databases for lncRNAs: A comparative evaluation of emerging tools.
RNA. 20:1655–1665. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dong Y, Huo X, Sun R, Liu Z, Huang M and
Yang S: LncRNA Gm15290 promotes cell proliferation and invasion in
non-small cell lung cancer through directly interacting with and
suppressing the tumor suppressor miR-615-5p. Oncol Res. May
5–2017.(Epub ahead of print). View Article : Google Scholar
|
|
10
|
Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N,
Yin Q, Li J and Sheng X: Long noncoding RNA GAS5, which acts as a
tumor suppressor via microRNA 21, regulates cisplatin resistance
expression in cervical cancer. Int J Gynecol Cancer. 27:1096–1108.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou Z, Xu X, Fu X, Tao S, Zhou J, Liu S
and Tan D: HBx-related long non-coding RNA MALAT1 promotes cell
metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am
J Cancer Res. 7:845–856. 2017.PubMed/NCBI
|
|
12
|
Huan J, Xing L, Lin Q, Xui H and Qin X:
Long noncoding RNA CRNDE activates Wnt/β-catenin signaling pathway
through acting as a molecular sponge of microRNA-136 in human
breast cancer. Am J Transl Res. 9:1977–1989. 2017.PubMed/NCBI
|
|
13
|
Ebert MS and Sharp PA: Emerging roles for
natural microRNA sponges. Curr Biol. 20:R858–R861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer
A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J,
et al: An extensive microRNA-mediated network of RNA-RNA
interactions regulates established oncogenic pathways in
glioblastoma. Cell. 147:370–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Niu L, Jiang S, Zhai J, Wang P,
Kong F and Jin X: Comprehensive analysis of aberrantly expressed
profiles of lncRNAs and miRNAs with associated ceRNA network in
muscle-invasive bladder cancer. Oncotarget. 7:86174–86185. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Loo LW, Cheng I, Tiirikainen M, Lum-Jones
A, Seifried A, Dunklee LM, Church JM, Gryfe R, Weisenberger DJ,
Haile RW, et al: cis-Expression QTL analysis of established
colorectal cancer risk variants in colon tumors and adjacent normal
tissue. PLoS One. 7:e304772012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((D1)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mukaka MM: Statistics corner: A guide to
appropriate use of correlation coefficient in medical research.
Malawi Med J. 24:69–71. 2012.PubMed/NCBI
|
|
20
|
Ravasz E, Somera AL, Mongru DA, Oltvai ZN
and Barabási AL: Hierarchical organization of modularity in
metabolic networks. Science. 297:1551–1555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X: Prediction of optimal gene
functions for osteosarcoma using network-based-guilt by association
method based on gene oncology and microarray profile. J Bone Oncol.
7:18–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peña-Castillo L, Tasan M, Myers CL, Lee H,
Joshi T, Zhang C, Guan Y, Leone M, Pagnani A, Kim WK, et al: A
critical assessment of Mus musculus gene function prediction using
integrated genomic evidence. Genome Biol. 9 Suppl 1:S22008.
View Article : Google Scholar
|
|
23
|
Pavlidis P and Gillis J: Progress and
challenges in the computational prediction of gene function using
networks: 2012–2013 update. F1000Res. 2:2302013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kominakis A, Hager-Theodorides AL, Zoidis
E, Saridaki A, Antonakos G and Tsiamis G: Combined GWAS and ‘guilt
by association’-based prioritization analysis identifies functional
candidate genes for body size in sheep. Genet Sel Evol. 49:412017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huntley RP, Sawford T, Mutowo-Meullenet P,
Shypitsyna A, Bonilla C, Martin MJ and O'Donovan C: The GOA
database: Gene Ontology annotation updates for 2015. Nucleic Acids
Res. 43((D1)): D1057–D1063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Haw R and Stein L: Using the reactome
database. Curr Protoc Bioinform. 38:1–23. 2012.
|
|
27
|
Zou T, Jaladanki SK, Liu L, Xiao L, Chung
HK and Wang JY, Xu Y, Gorospe M and Wang JY: H19 long noncoding RNA
regulates intestinal epithelial barrier function via microRNA 675
by interacting with RNA-binding protein HuR. Mol Cell Biol.
36:1332–1341. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dey BK, Pfeifer K and Dutta A: The H19
long noncoding RNA gives rise to microRNAs miR-675-3p and
miR-675-5p to promote skeletal muscle differentiation and
regeneration. Genes Dev. 28:491–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang P, Ning S, Zhang Y, Li R, Ye J, Zhao
Z, Zhi H, Wang T, Guo Z and Li X: Identification of
lncRNA-associated competing triplets reveals global patterns and
prognostic markers for cancer. Nucleic Acids Res. 43:3478–3489.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Misawa A, Takayama K, Urano T and Inoue S:
Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes
cell growth and inhibits apoptosis in prostate cancer cells. J Biol
Chem. 291:17861–17880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi Q and Yang X: Circulating microRNA and
long noncoding RNA as biomarkers of cardiovascular diseases. J Cell
Physiol. 231:751–755. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Su S, Liu J, He K, Zhang M, Feng C, Peng
F, Li B and Xia X: Overexpression of the long noncoding RNA TUG1
protects against cold-induced injury of mouse livers by inhibiting
apoptosis and inflammation. FEBS J. 283:1261–1274. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li
XM and Ye DQ: Emerging role of long noncoding RNAs in autoimmune
diseases. Autoimmun Rev. 14:798–805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Y, Xu Y, Feng L, Li F, Sun Z, Wu T,
Shi X, Li J and Li X: Comprehensive characterization of lncRNA-mRNA
related ceRNA network across 12 major cancers. Oncotarget.
7:64148–64167. 2016.PubMed/NCBI
|
|
35
|
Chiu YC, Wang LJ, Lu TP, Hsiao TH, Chuang
EY and Chen Y: Differential correlation analysis of glioblastoma
reveals immune ceRNA interactions predictive of patient survival.
BMC Bioinformatics. 18:1322017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rider AK, Milenković T, Siwo GH, Pinapati
RS, Emrich SJ, Ferdig MT and Chawla NV: Networks' characteristics
matter for systems biology. Netw Sci (Camb Univ Press). 2:139–161.
2014.PubMed/NCBI
|
|
37
|
Wang P, Lü J and Yu X: Identification of
important nodes in directed biological networks: A network motif
approach. PLoS One. 9:e1061322014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roberts TC, Morris KV and Weinberg MS:
Perspectives on the mechanism of transcriptional regulation by long
non-coding RNAs. Epigenetics. 9:13–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi X, Sun M, Wu Y, Yao Y, Liu H, Wu G,
Yuan D and Song Y: Post-transcriptional regulation of long
noncoding RNAs in cancer. Tumour Biol. 36:503–513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang JZ, Xu CL, Wu H and Shen SJ: LncRNA
SNHG12 promotes cell growth and inhibits cell apoptosis in
colorectal cancer cells. Braz J Med Biol Res. 50:e60792017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C,
Wang J, Tan Q, Cheng Y, Xia E, et al: C-MYC-induced upregulation of
lncRNA SNHG12 regulates cell proliferation, apoptosis and migration
in triple-negative breast cancer. Am J Transl Res. 9:533–545.
2017.PubMed/NCBI
|
|
43
|
Lan T, Ma W, Hong Z, Wu L, Chen X and Yuan
Y: Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in
hepatocellular carcinoma. J Exp Clin Cancer Res. 36:112017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ruan W, Wang P, Feng S, Xue Y and Li Y:
Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes cell proliferation and migration by upregulating
angiomotin gene expression in human osteosarcoma cells. Tumour
Biol. 37:4065–4073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H,
Xi Z, Li Z and Liu Y: Role of HCP5-miR-139-RUNX1 feedback loop in
regulating malignant behavior of glioma cells. Mol Ther.
24:1806–1822. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lange CM, Bibert S, Dufour JF, Cellerai C,
Cerny A, Heim MH, Kaiser L, Malinverni R, Müllhaupt B, Negro F, et
al: Swiss Hepatitis C Cohort Study Group: Comparative genetic
analyses point to HCP5 as susceptibility locus for HCV-associated
hepatocellular carcinoma. J Hepatol. 59:504–509. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
48
|
Xiao H, Xiao W, Cao J, Li H, Guan W, Guo
X, Chen K, Zheng T, Ye Z, Wang J, et al: miR-206 functions as a
novel cell cycle regulator and tumor suppressor in clear-cell renal
cell carcinoma. Cancer Lett. 374:107–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chukkapalli S, Amessou M, Dekhil H, Dilly
AK, Liu Q, Bandyopadhyay S, Thomas RD, Bejna A, Batist G and
Kandouz M: Ehd3, a regulator of vesicular trafficking, is silenced
in gliomas and functions as a tumor suppressor by controlling cell
cycle arrest and apoptosis. Carcinogenesis. 35:877–885. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ouyang Q, Xu L, Cui H, Xu M and Yi L:
MicroRNAs and cell cycle of malignant glioma. Int J Neurosci.
126:1–9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Banerjee T and Chakravarti D: A peek into
the complex realm of histone phosphorylation. Mol Cell Biol.
31:4858–4873. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kelly AE, Ghenoiu C, Xue JZ, Zierhut C,
Kimura H and Funabiki H: Survivin reads phosphorylated histone H3
threonine 3 to activate the mitotic kinase Aurora B. Science.
330:235–239. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zegers MM and Friedl P: Rho GTPases in
collective cell migration. Small GTPases. 5:e289972014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lin Y and Zheng Y: Approaches of targeting
Rho GTPases in cancer drug discovery. Expert Opin Drug Discov.
10:991–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Makrodouli E, Oikonomou E, Koc M, Andera
L, Sasazuki T, Shirasawa S and Pintzas A: BRAF and RAS oncogenes
regulate Rho GTPase pathways to mediate migration and invasion
properties in human colon cancer cells: A comparative study. Mol
Cancer. 10:1182011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Andermatt I, Wilson NH, Bergmann T, Mauti
O, Gesemann M, Sockanathan S and Stoeckli ET: Semaphorin 6B acts as
a receptor in post-crossing commissural axon guidance. Development.
141:3709–3720. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ge C, Li Q, Wang L and Xu X: The role of
axon guidance factor semaphorin 6B in the invasion and metastasis
of gastric cancer. J Int Med Res. 41:284–292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mumblat Y, Kessler O, Ilan N and Neufeld
G: Full-length semaphorin-3C is an inhibitor of tumor
lymphangiogenesis and metastasis. Cancer Res. 75:2177–2186. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gu C and Giraudo E: The role of
semaphorins and their receptors in vascular development and cancer.
Exp Cell Res. 319:1306–1316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bollard J, Massoma P, Vercherat C, Blanc
M, Lepinasse F, Gadot N, Couderc C, Poncet G, Walter T, Joly MO, et
al: The axon guidance molecule semaphorin 3F is a negative
regulator of tumor progression and proliferation in ileal
neuroendocrine tumors. Oncotarget. 6:36731–36745. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang Z, Chen J, Zhang W, Zheng Y, Wang Z,
Liu L, Wu H, Ye J, Zhang W, Qi B, et al: Axon guidance molecule
semaphorin 3A is a novel tumor suppressor in head and neck squamous
cell carcinoma. Oncotarget. 7:6048–6062. 2016.PubMed/NCBI
|