Introduction
In-depth studies have demonstrated that tumor
tissues are composed of cells at different stages of
differentiation, including a subset of cells termed cancer stem
cells (CSCs) that have the potential for self-renewal and
differentiation. These tumor-initiating cells are involved in
tumorigenesis, development, metastasis, recurrence and drug
resistance. A determining role of CSCs in tumorigenesis has been
recognized in breast and hematological cancers, as well as in
nervous system tumors (1,2). The continuous development of the CSC
theory has brought new ideas to tumorigenesis research and novel
methods for diagnosing and treating cancer.
Hepatocellular carcinoma (HCC) is among the most
aggressive human diseases, and its pathogenesis remains unclear.
There is extensive evidence that HCC is a stem cell-derived
disease. Hepatic cancer stem cells (HCSCs) are the initiating cells
of HCC and are important in maintaining HCC growth, metastasis and
recurrence, whereas non-stem cell-like HCC cells do not have
self-renewal capacity and ultimately die after limited
proliferation (3). The novel CSC
theory could have a significant impact on the traditional HCC
treatment strategy. Currently, all HCC cells are targeted by
surgical treatment, radiotherapy and/or chemotherapy. Although
eliminating the vast majority of HCC cells with limited
proliferative capacity can promote tumor regression, it cannot
eradicate the tumor source (4).
Compared with non-stem cell-like HCC cells, HCSCs have high
proliferative capacity and tumorigenicity and can effectively
initiate the DNA SOS response and increase the expression of drug
resistance-related proteins that facilitate chemotherapy resistance
and allow tumor cells to survive treatment, which ultimately
results in HCC recurrence. Therefore, it is necessary to
effectively eradicate HCSCs to achieve therapeutic efficacy against
HCC. This may be possible if specific stem cell signals are
inhibited using gene therapy, while at the same time attacking
proliferating cells by conventional therapy. However, key genes and
related pathways in HCSC research are still preliminary (5,6).
With support from the National Natural Science
Foundation of China, in the present study we isolated an expressed
sequence tag (EST) fragment that was highly expressed in HCC tissue
using the suppression subtractive hybridization technique and then
obtained a full-length cDNA sequence (1476 bp) using the rapid
amplification of cDNA ends (RACE) method. A GeneBank search
revealed that it was identical to a recently reported novel gene
with unknown function (GeneBank accession number: BC047440). The
gene is located on 20q11.22. PROSITE analysis showed that this gene
might contain three protein kinase C phosphorylation sites, two
casein kinase II phosphorylation sites, two myristyl sites and one
asparagine-linked glycosylation site. The subcellular localization
of the protein is likely cytosolic. The BC047440-encoded protein is
predicted to be composed of 200 amino acids, with an isoelectric
point of 6.32 and a molecular weight of 22.6 kDa. Our previous
studies suggested that the novel HCC-related gene BC047440 is
associated with HCC development and progression. Furthermore,
evidence suggests that BC047440 plays a crucial role in mediating
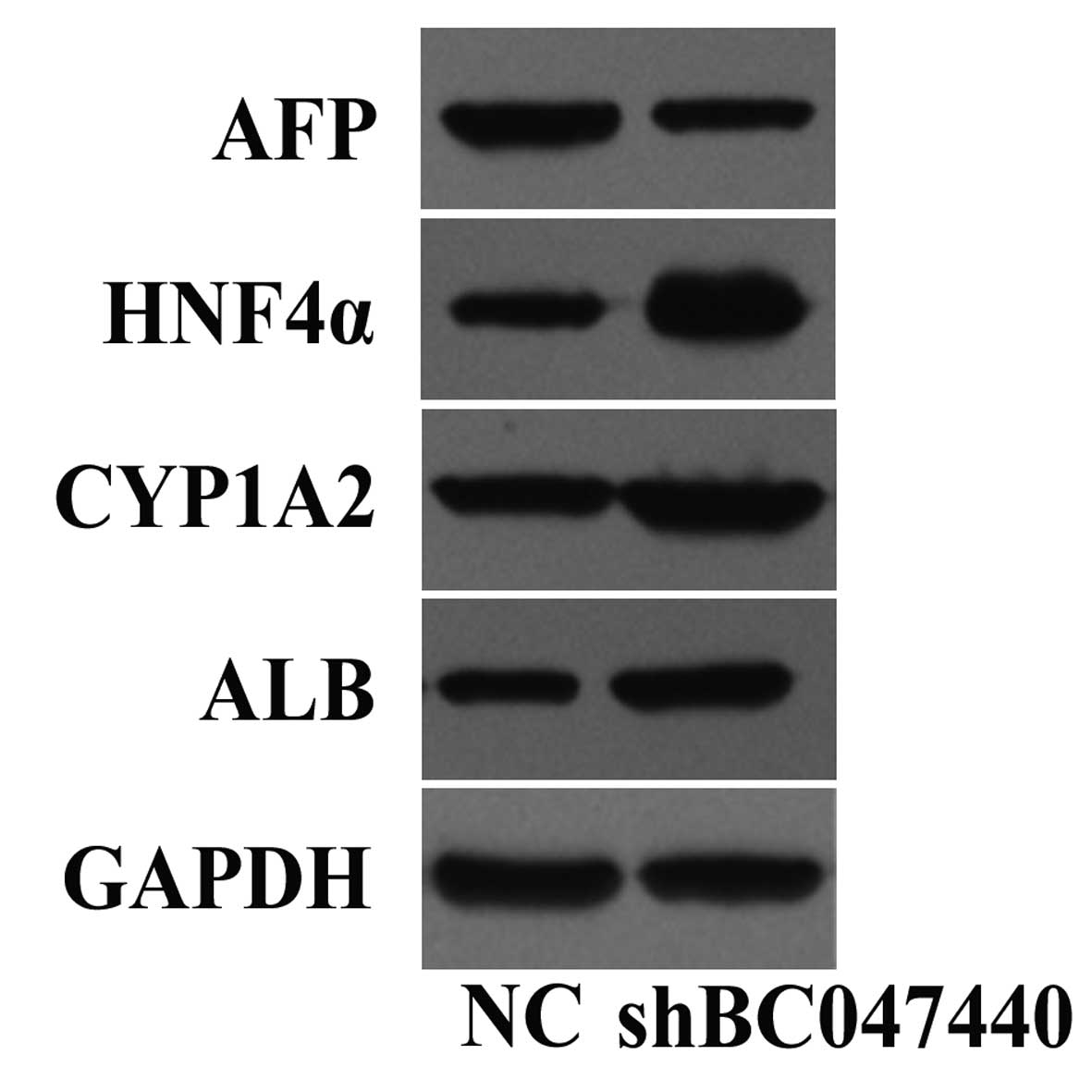
HCC proliferation and differentiation (Fig. 1) of HCC cells (7–9).
Nevertheless, few studies have assessed the role of BC047440 in
HCSCs. Based on our previous findings regarding the determining
role of HCSCs in tumorigenesis, we hypothesized that BC047440 might
be involved in maintaining HCSC malignant behavior (including
proliferation and differentiation) and might be useful for
gene-targeted therapies.
In order to further elucidate the specific mode of
BC047440 action, in the present study, we investigated the effect
of BC047440 inhibition on HCSC proliferation and differentiation.
We also elucidated the molecular mechanisms that underlie these
processes. To the best of our knowledge, this is the first report
describing the effects of BC047440 inhibition in HCSCs. Loss of
BC047440 gene expression results in the proliferation inhibition of
HCSCs with concomitant differentiation, eventually leading to
tumorigenicity suppression. The results of the present research
support a novel role of BC047440 in hepatic tumorigenesis and will
help revolutionize therapeutic strategies and provide unique
opportunities for HCC gene therapy.
Materials and methods
Animals
Fischer 344 rats (250–300 g body weight) were
purchased from the Shanghai Slack Experimental Animal Center,
China. Nude mice were purchased from the Laboratory Animal Center
of Beijing Xiehe Medical University, China. All rats were housed
under standardized conditions [room at constant temperature (25°C)
with alternating 12-h periods of light and darkness] and fed a
standard rat diet with free access to water. Food and water were
given ad libitum. All experimental procedures involving
animals were approved by the Local Animal Care and Use Committee of
the Third Military Medical University.
Isolation of hepatic normal stem cells
(HNSCs) and HCSCs and cell culture
HNSCs and HCSCs were respectively obtained by
procedures previously reported by our group (10,11).
Cells were grown in Williams’ E medium (AppliChem GmbH, Darmstadt,
Germany) supplemented with 10% fetal bovine serum (FBS;
Gibco/Invitrogen, Grand Island, NY, USA), 100 IU/ml penicillin, 400
IU/l trypsin and 100 μg/ml streptomycin. The cells were plated in
75-cm2 flasks and cultured at 37°C with 5%
CO2 and 95% humidified air. The medium was changed every
2 days.
Analysis of the BC047440 gene expression
between HNSCs and HCSCs
HNSCs and HCSCs were processed for protein
extraction, and western blot analysis was performed according to
the published method. The primary antibodies used were:
anti-BC047440 (diluted 1:500, self-prepared) and
anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; diluted
1:400, mouse monoclonal C-2; Santa Cruz Biotechnology, Santa Cruz,
CA, USA).
short hairpin RNA (shRNA) lentiviral
infection of HCSCs
HCSCs were plated at 2×105 cells/well in
1 ml of Williams’ E supplemented with 10% FBS in a 12-multiwell
plate. Infection was performed at a cell confluency of ~50–70%
using commercial shRNA lentiviral particles (Shanghai GeneChem Co.,
Ltd., Shanghai, China) according to the manufacturer’s protocol. An
invalid RNAi sequence was used as a negative control (NC).
Puromycin was used to screen stably transfected clones. Established
stable cell lines, HCSCs (shBC047440) or HCSCs (NC), were then
cultured in medium containing 0.5 μg/ml puromycin. Knockdown of
BC047440 was verified by western blot analysis for successful
silencing.
The effects of BC047440 on HCSC
proliferation
Annexin V/propidium iodide (PI)
assay
To exclude apoptosis-related effects, Annexin V
assays were performed using an apoptosis detection kit [Annexin
V-fluorescein isothiocyanate (FITC)/PI Staining kit; Immunotech
Co., Marseille, France] as described in the manufacturer’s
instructions. Briefly, the different interfered cells were
collected, washed in cold phosphate-buffered saline (PBS),
incubated for 15 min with a fluorescein-conjugated Annexin V and PI
and analyzed using flow cytometry (Becton-Dickinson, San Jose, CA,
USA).
Cell counting
HCSCs (shBC047440) and HCSCs (NC) were digested with
0.25% trypsin and 0.01% ethylenediaminetetraacetic acid (EDTA),
adjusted to a density of 1×104 cells/ml and seeded in
12-well plates (1×104 cells/well). Each group of cells
included 3 parallel samples. Every day during a period of 7 days,
the cells of the 4 parallel samples in each group were trypsinized
and the cell number was counted under an inverted microscope
(Olympus, Tokyo, Japan).
Colony formation assay
Approximately 3×102 cells from the
different interfered cells were plated in six-well dishes. After 7
days, cells were fixed with 20% methanol and stained with 1%
crystal violet. Colonies consisting of >50 cells were counted
per well, and each experiment was performed in triplicate.
Analyses of the expression of the
cellular proliferation marker Ki-67
Flow cytometric analysis was performed for the Ki-67
studies. Briefly, the different interfered cells were harvested,
fixed in 70% (v/v) ethanol, washed and incubated for 20 min with 1%
bovine serum albumin (BSA). The cells were then incubated with
1:200 diluted anti-Ki-67 (FITC conjugated; Bioscience, Beijing,
China). The data were obtained and analyzed using flow
cytometry.
Cell cycle distribution analysis
Cell cycle analysis assays are used in flow
cytometry. HCSCs (shBC047440) and HCSCs (NC) were fixed in ice-cold
70% (v/v) ethanol for 48 h at 4°C. They were then washed once with
PBS and centrifuged at 200 × g for 10 min, followed by treatment
with RNase A at 1 mg/ml for 30 min at 37°C. After staining with 40
μl of 0.1 mg/l PI, histograms of DNA content were analyzed using
flow cytometry to determine cell cycle distribution (G0/G1, S, and
G2/M phase).
Western blot analysis for the effects of
BC047440 on the activation of nuclear nuclear factor-κB (NF-κB) in
HCSCs
The preparation of cytoplasmic and nuclear extracts
was performed using the Nuclear Extract kit (HyClone-Pierce, Logan,
UT, USA) according to the manufacturer’s instructions. To measure
nuclear NF-κB/p65 and cytoplasmic NF-κB/p65 expression, western
blot analysis was performed. Related fusion proteins were
identified using anti-NF-κB/p65 primary antibody (Santa Cruz
Biotechnology).
The effects of BC047440 on HCSC
differentiation
Detection of stem cell markers by flow
cytometric analysis
The stem-cell-associated markers of different
interfered cells were analyzed by flow cytometric analysis.
Briefly, at 7 days of HCSC (shBC047440) and HCSC (NC) growth in the
presence of hepatocyte growth factor (HGF), the cells were prepared
as single cell suspensions at a density of 1×106
cells/ml using Dulbecco’s modified Eagle’s medium (DMEM)
(containing 20% FBS) and incubated for 15–30 min at room
temperature to block non-specific sites. These cells were then
washed twice with PBS and re-suspended in 990 μl PBS. Subsequently,
10 μl of antibodies, including CD133 (PE-conjugated; BioLegend, San
Diego, CA, USA) and EpCAM (FITC-conjugated, BioLegend), were added
to each cell suspension. After 30 min of incubation at 4°C in the
dark, the cells were washed twice with PBS, fixed in 0.1%
formaldehyde and analyzed using flow cytometry.
Measurement of albumin (ALB),
α-fetoprotein (AFP) and urea levels
The conditioned media from HCSCs (shBC047440) and
HCSCs (NC) cultured with HGF were collected at day 7 and were used
for assaying AFP and ALB production with enzyme-linked
immunosorbent assay (ELISA) kits (Cusabio Biotech Co., Ltd., Wuhan,
China) according to the manufacturer’s protocol. The cells from
each condition were incubated with NH4Cl (5 mM/ml;
Sigma, St. Louis, MO, USA) for 24 h in 5% CO2 at 37°C on
day 7. Following incubation, the supernatants were collected and
the urea concentrations were measured using a colorimetric assay
kit (Gdb Corp., San Diego, CA, USA).
Transmission electron microscopy
(TEM)
Morphological changes in HCSCs (shBC047440) and
HCSCs (NC) treated with HGF were evaluated by TEM. After incubation
with HGF for 7 days, the cells were digested with trypsin and fixed
in 2.5% glutaraldehyde precooled at 4°C for 2 h. To make ultra-thin
sections for copper staining, cells were washed with PBS, fixed in
1% osmic acid for an additional hour, dehydrated in acetone and
embedded in epoxide resin. After the sections were stained with
uranyl acetate and lead citrate, the ultrastructural features of
cells were observed under an electron microscope (JEM-2000EX; JEOL
Ltd., Tokyo, Japan).
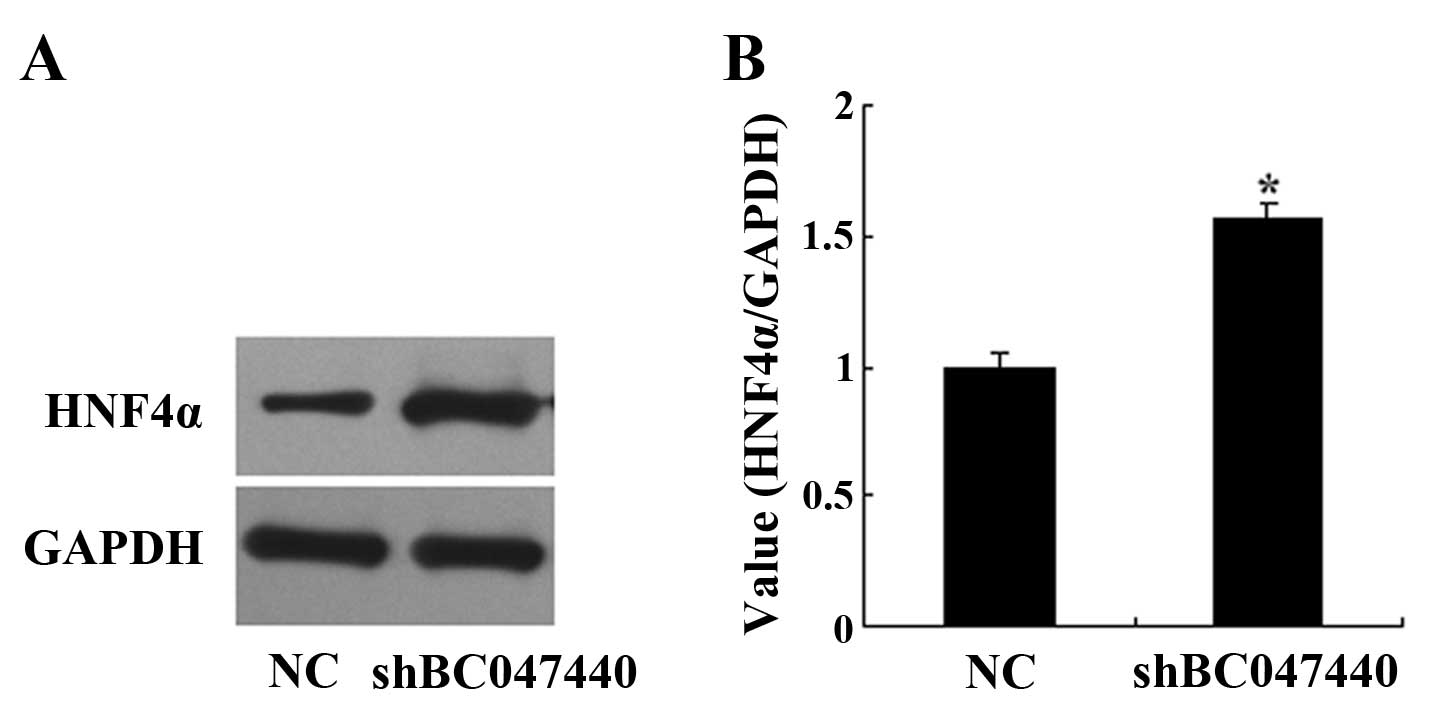
Western blot analysis for the effects of
BC047440 on hepatocyte nuclear factor 4α (HNF4α) expression
HNF4α expression was measured by western blot
analysis. Western blot analysis was performed using techniques
previously described. Briefly, HNF4α fusion protein was identified
using anti-HNF4α primary antibody (diluted 1:500, rabbit
polyclonal; Santa Cruz Biotechnology) and the horseradish
peroxidase (HRP)-conjugated anti-rabbit IgG (diluted 1:2,000) as
secondary antibody.
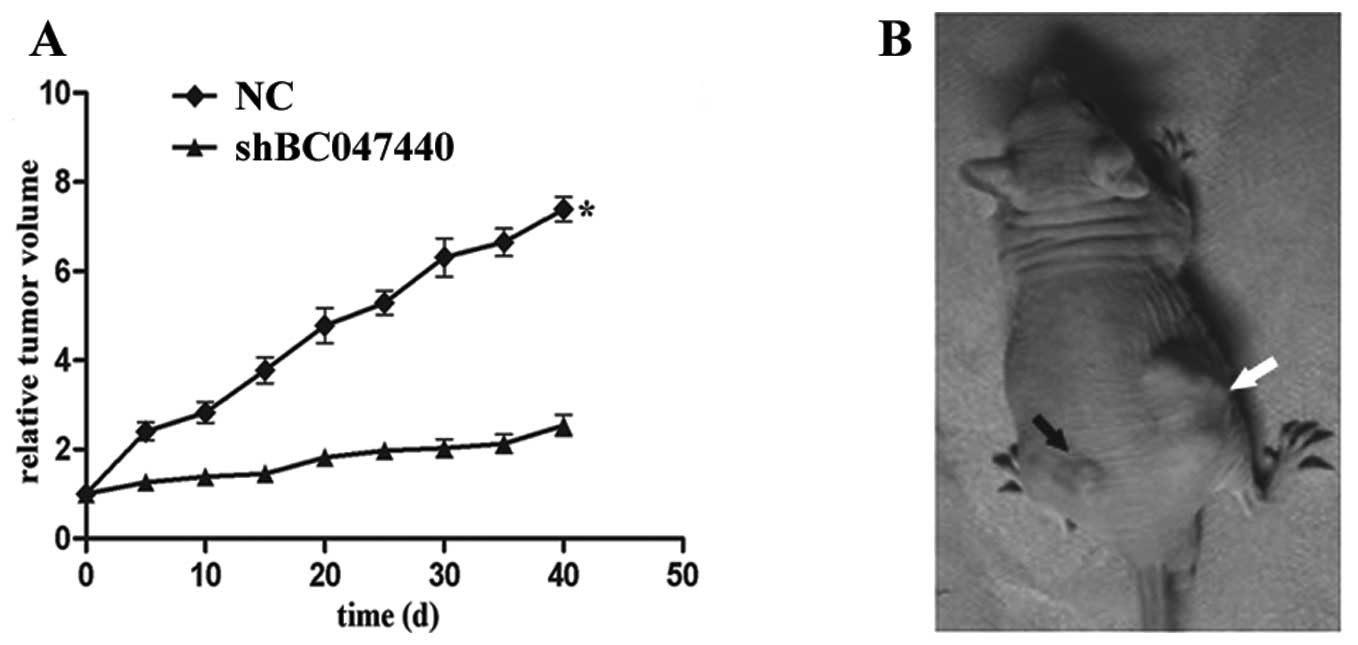
Tumorigenesis assay
To determine the effects of BC047440 knockdown on
the tumorigenic potential of HCSCs in vivo, we compared the
growth of tumors produced by injection of 2×106 HCSCs
(shBC047440) on the left side of the back with tumors produced by
injection of 2×106 HCSCs (NC) on the right side of the
same animal’s back. Tumor size was measured twice weekly. Tumor
volume (V) was calculated using the following formula: V = (a × b ×
c)/2, where a and b are the shorter and longer diameters of each
tumor, respectively, and c is the thickness. The animals were
sacrificed after 40 days.
Statistical analysis
The significance of differences between the groups
was determined with one-way ANOVA (SPSS 10.0 statistical software).
The results with a P-value ≤0.05 were considered statistically
significant. Data are expressed as the mean ± the standard error of
the mean of separate experiments (n≥3, where n represents the
number of independent experiments).
Results
Isolation of HNSCs and HCSCs
According to the special density characteristic of
stem cells, we have enriched special density HNSCs and HCSCs via a
density gradient centrifugation-centered method (10,11).
Our previous data revealed that the derived cells exhibit stem
cell-like characteristics, including high tumorigenicity.
Increased BC047440 expression in
HCSCs
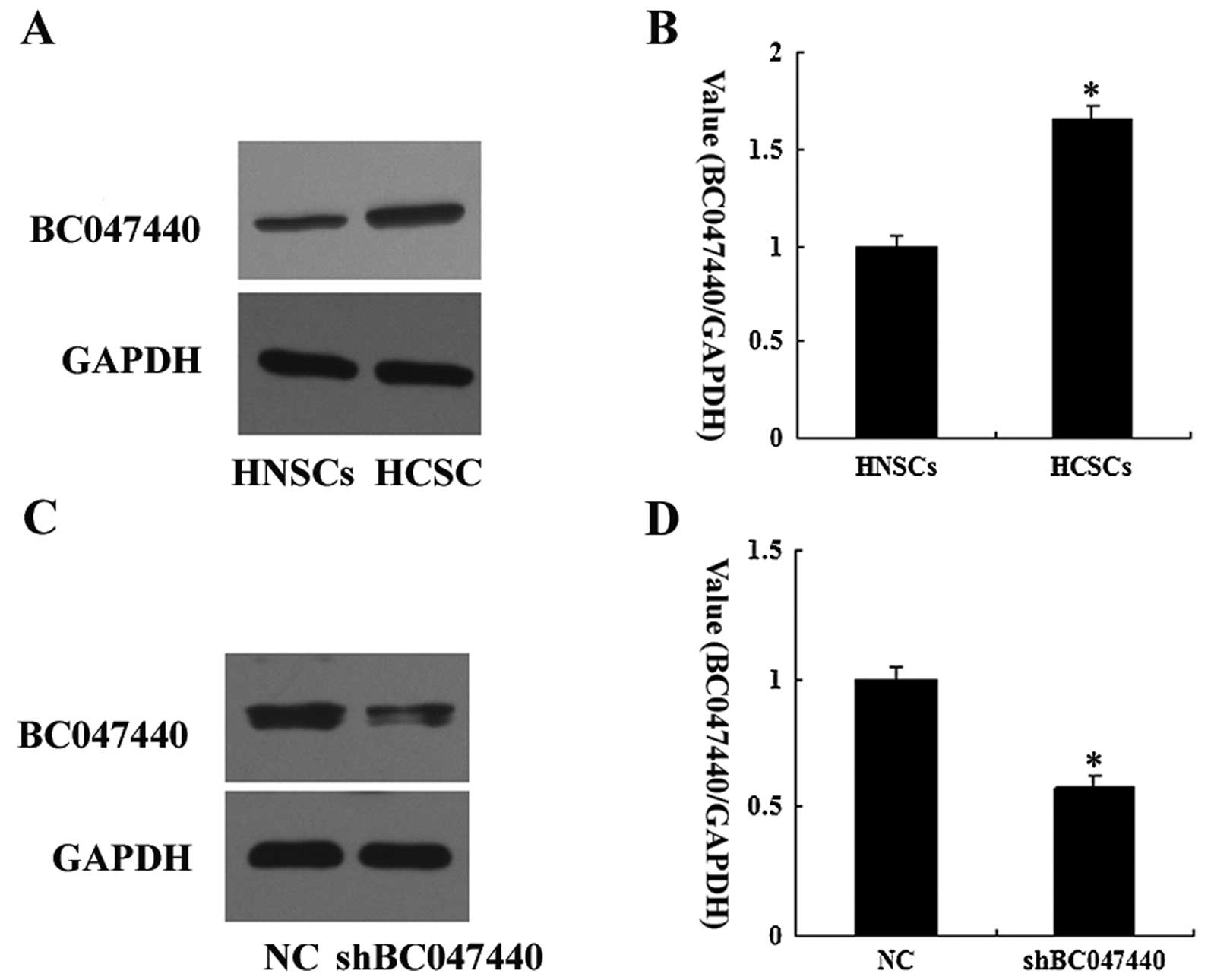
BC047440 expression was compared between HCSCs and
HNSCs by western blot analysis. The levels of BC047440 expression
in HCSCs were higher than those for HNSCs (Fig. 2A and B).
shRNA-mediated downregulation of BC047440
expression
To confirm the silencing of BC047440 expression in
HCSCs, western blot analysis was performed. Western blot analysis
after shRNA treatment showed a marked decrease in BC047440 protein
in HCSCs (Fig. 2C and D). These
data show that the lentiviral-mediated shRNA used in the present
study was effective in silencing BC047440 gene expression in
HCSCs.
Effects of BC047440 on HCSC
proliferation
To determine whether BC047440 effects HCSC
proliferation, we first examined the effect of BC047440 depletion
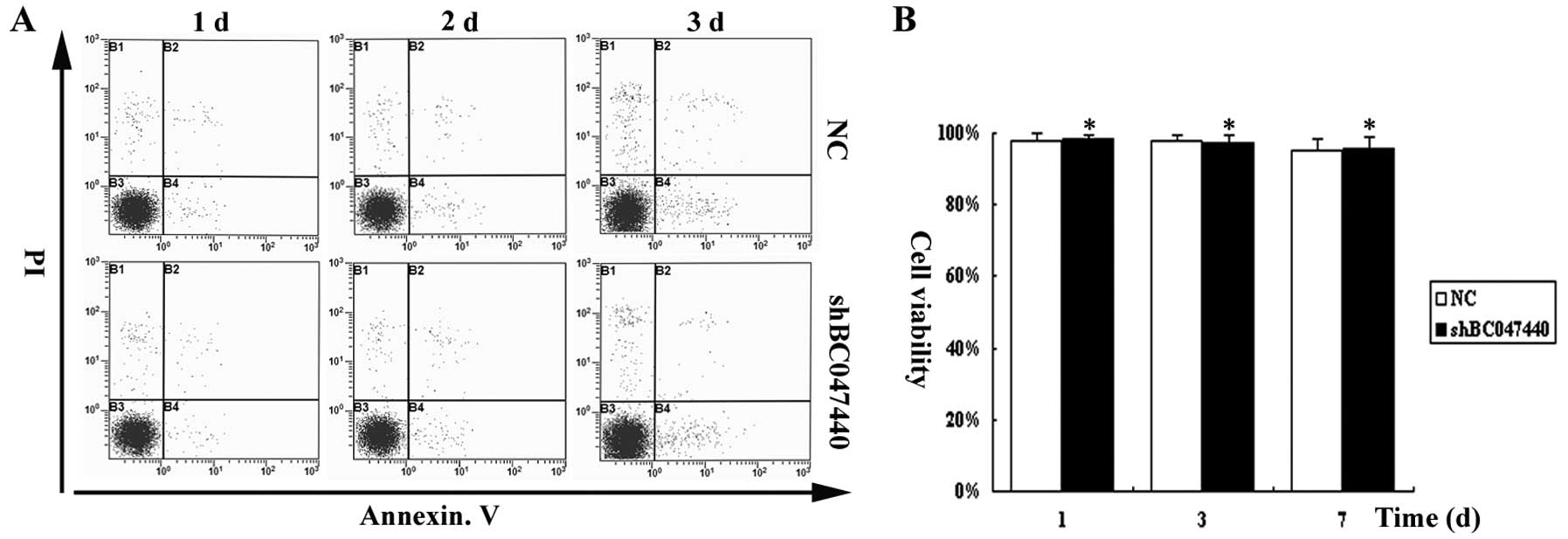
on cell viability using Annexin V assays. As shown in Fig. 3, over the time course of the
experiment, BC047440 depletion did not affect the viability of
HCSCs and apoptosis had no effect on cell proliferation. Cell
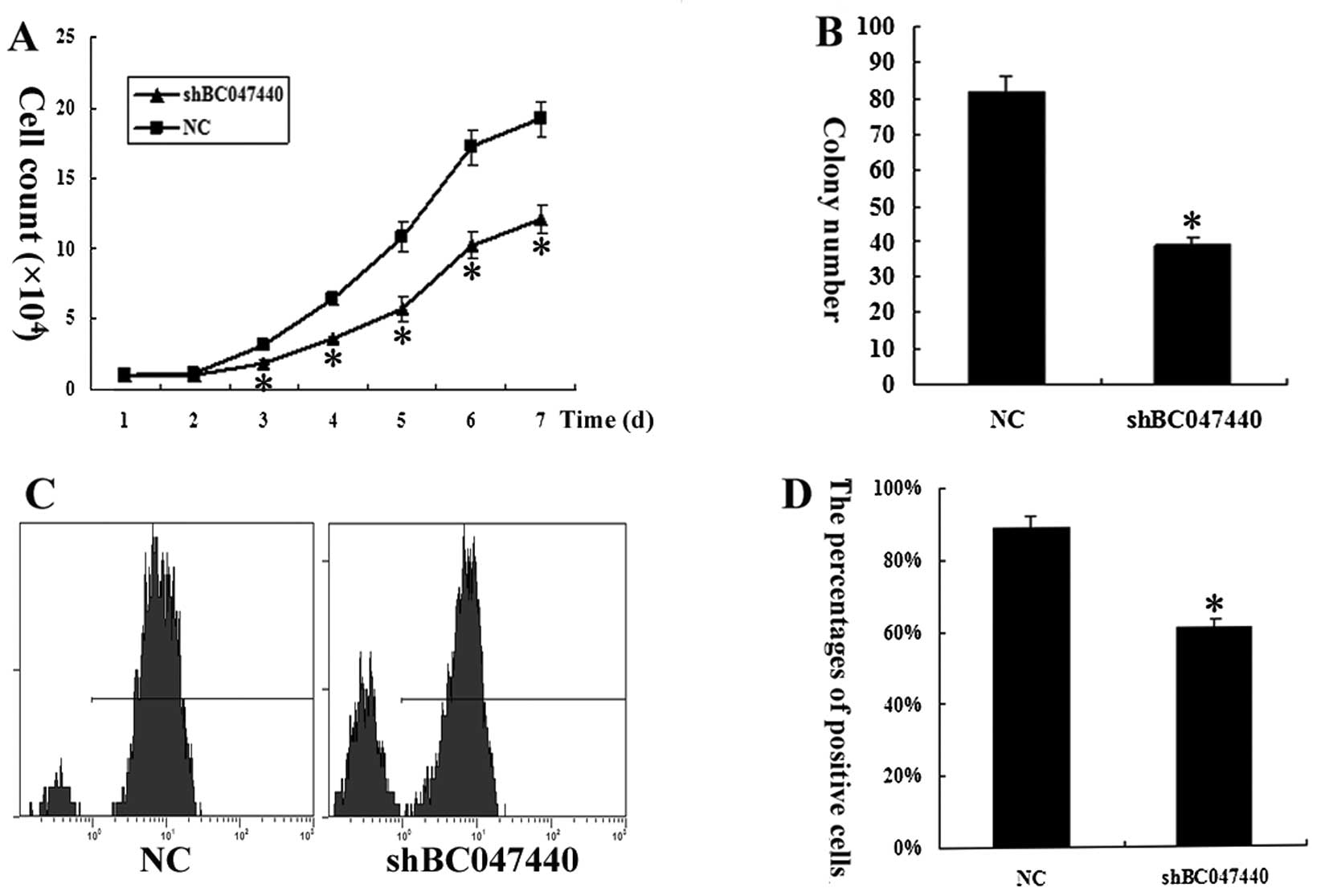
counting showed that BC047440 depletion significantly suppressed
the proliferation of HCSCs (Fig.
4A). Colony formation analysis showed that BC047440-depleted
HCSCs had a markedly reduced capacity to form colonies compared
with the control cells (Fig. 4B).
Moreover, cell proliferation was assessed by Ki-67 expression
analysis (Fig. 4C and D). The
expression of Ki-67 in HCSCs (shBC047440) decreased significantly
(n=3, P<0.05) compared to HCSCs (NC). The results suggest that
the silencing of the BC047440 gene resulted in the inhibition of
HCSC proliferation.
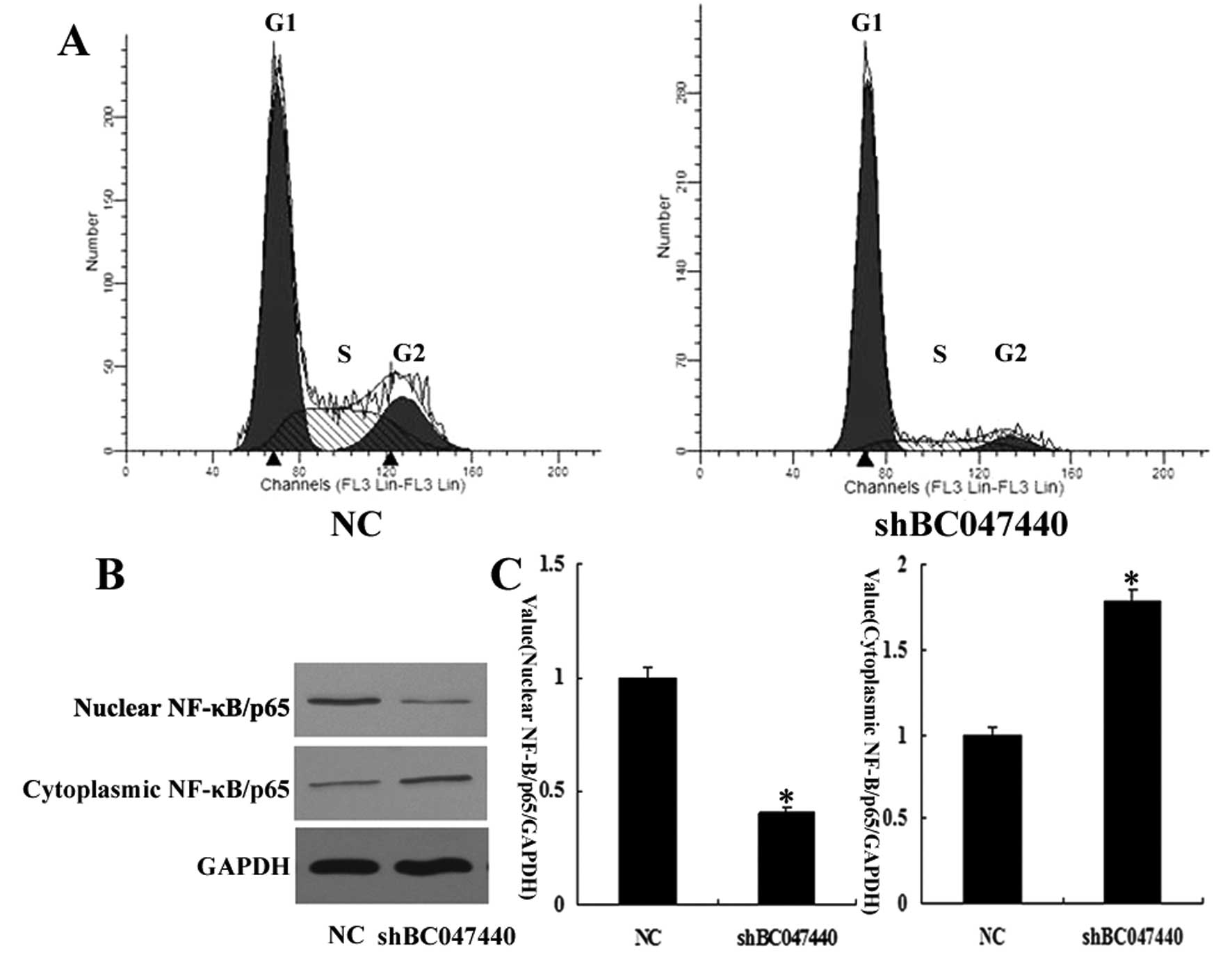
To further investigate the effects of BC047440 on
HCSC proliferation, we used flow cytometry to determine the cell
cycle distribution of HCSCs (shBC047440) and HCSCs (NC). As shown
in Fig. 5A, there was a higher
proportion of G0–G1 phase cells (81.47%) in HCSCs (shBC047440)
compared to HCSCs (NC) (58.09%). A compensatory decrease in the S
(12.48%) and G2/M phase (6.05%) proportions was also detected as
compared with the control in S (26.29%) and G2/M phases (15.62%).
The data indicate that the downregulation of BC047440 expression in
HCSCs may have a significant effect on cell proliferation.
Since NF-κB activation controls the expression of a
number of genes involved in cell growth and survival through direct
and indirect mechanisms, we investigated whether BC047440 knockdown
has an impact on NF-κB activation in HCSCs. We revealed that
BC047440 knockdown caused an increase in NF-κB levels in the
cytoplasmic fraction of HCSCs with a simultaneous decrease in the
nuclear fraction (Fig. 5B and C).
The results indicate that BC047440 knockdown suppresses
constitutive activation of NF-κB in HCSCs.
Effects of BC047440 on HCSC
differentiation
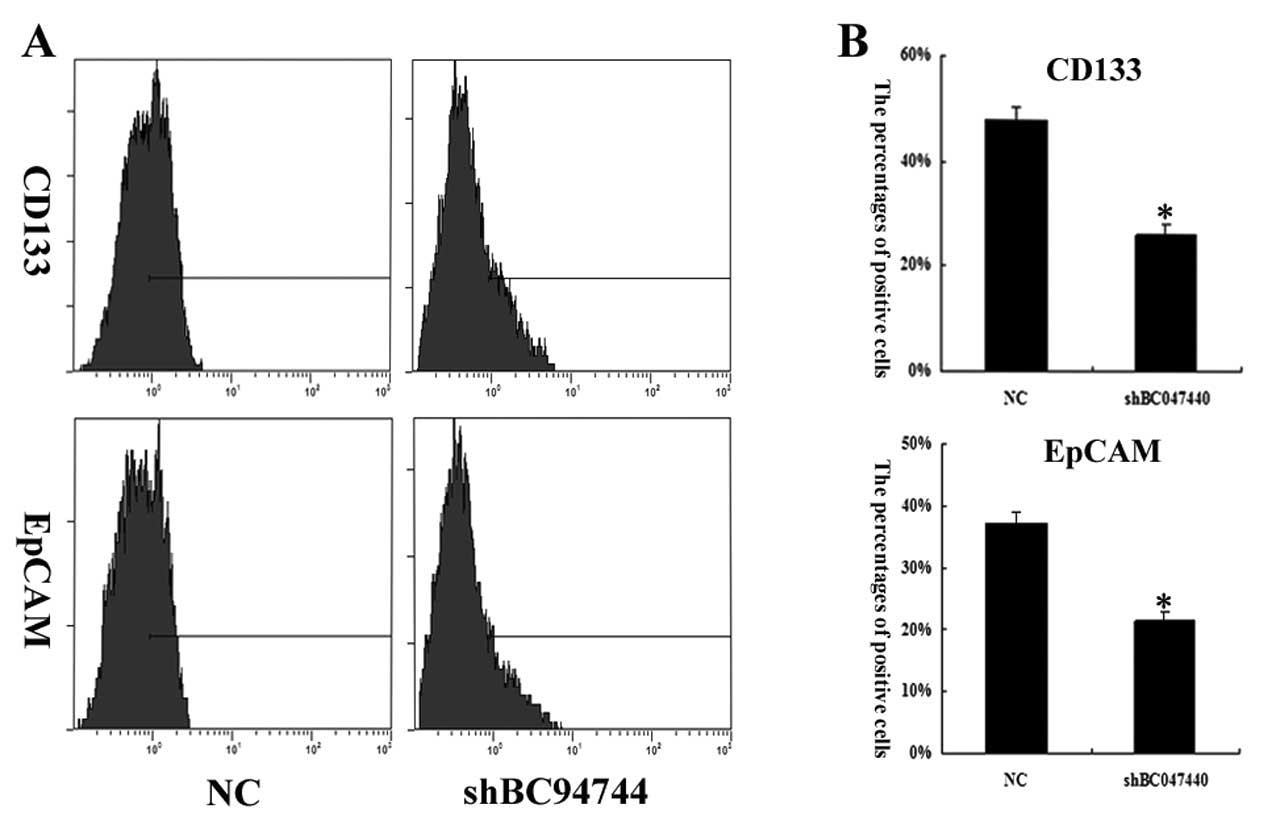
To determine the stem properties of cells in
different interfered cells, CD133 and EpCAM were used as a probe to
quantitatively analyze the profile of stem-like cells in HCSCs
(shBC047440) and HCSCs (NC). On day 7 of cell growth in the
presence of HGF, CD133 percentages in HCSCs (NC) and HCSCs
(shBC047440) were 47.9±2.5 and 26.1±2.1%, respectively. EpCAM
percentages in HCSCs (NC) and HCSCs (shBC047440) were 37.1±1.9 and
21.4±1.3%, respectively (Fig. 6).
These data indicated that BC047440 can modulate the stemness
properties of HCSCs.
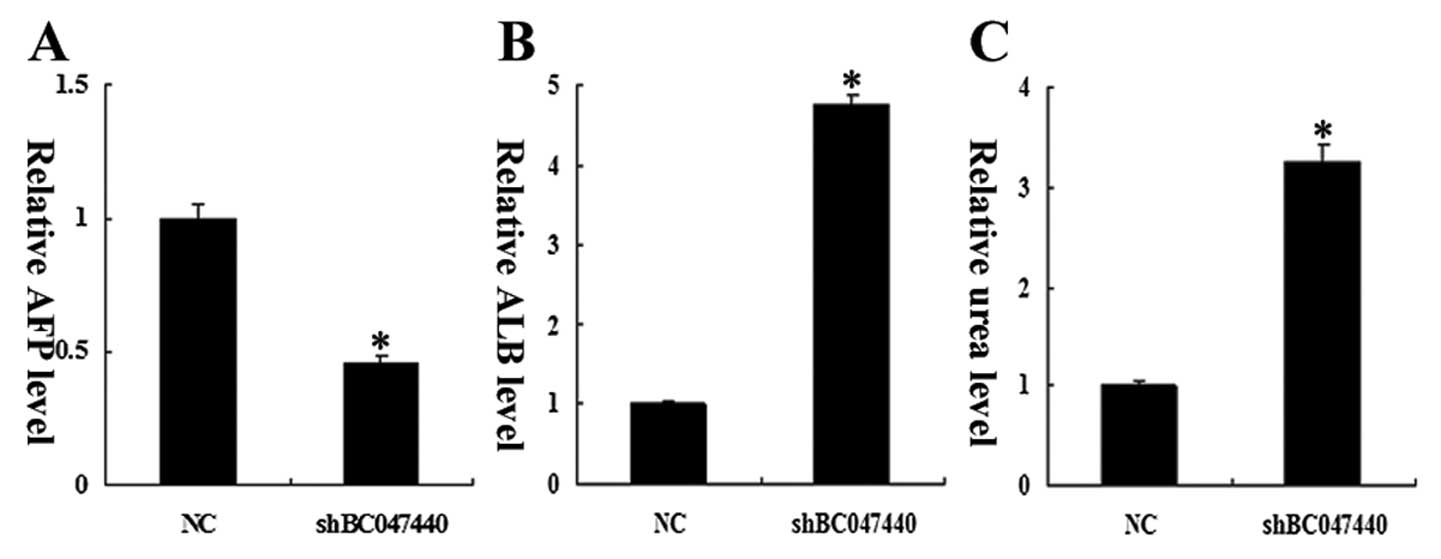
Stemness properties were also evaluated at the
functional level. On day 7 of cells treated with HGF, the
production of ALB in HCSCs (shBC047440) was significantly higher
than in HCSCs (NC), and an expected decrease in AFP production was
observed on day 7 in HCSCs (shBC047440) compared to HCSCs (NC)
(Fig. 7A and B). In addition, upon
treatment with HGF, HCSCs (shBC047440) produced significantly
higher levels of urea compared to HCSCs (NC) (Fig. 7C).
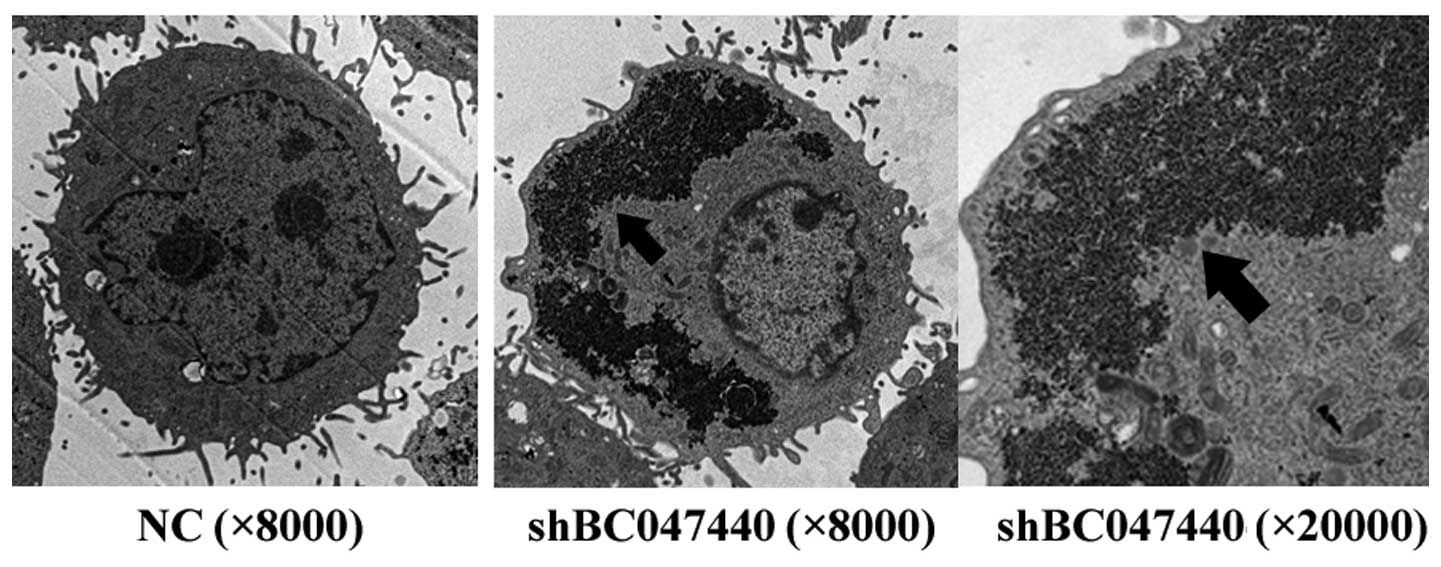
To evaluate the fine structural characteristics of
HCSCs (shBC047440) and HCSCs (NC), the cells cultured in the
presence of HGF on day 7 were examined using TEM (Fig. 8). HCSCs (shBC047440) exhibited
morphological features of early differentiation similar to those of
hepatocytes. Numerous mitochondria, lysosomes, prominent nucleoli,
well-developed Golgi apparatuses, rough and smooth endoplasmic
reticulum and especially glycogensomes (markers of mature
hepatocytes) were observed in the cytoplasm of HCSCs (shBC047440).
The nuclei displayed a low nuclear to cytoplasmic ratio and the
cells were polarized, as shown by the presence of bile canaliculi.
In contrast, the ultrastructural features of HCSCs (NC) were with
large and regular nuclei, abundant ribosomes and mitochondria,
poorly developed Golgi apparatus and endoplasmic reticulum, seldom
lysosomes and high nuclear-cytoplasmic ratio. The images suggest
that BC047440 can modulate HCSC differentiation and that
suppression of BC047440 results in inducing HCSC differentiation
into hepatocytes.
HNF4α is important for hepatocyte development,
differentiation and function. We investigated whether BC047440
knockdown alters the expression of HNF4α. As shown in Fig. 9, BC047440 knockdown induced an
increase in HNF4α protein levels.
BC047440 knockdown inhibits the
tumorigenic capacity of HCSCs in vivo
To examine the effect of BC047440 depletion on cell
tumorigenic capacity in vivo, xenograft tumor growth assays
were performed in nude mice. Compared with HCSCs, injection of
BC047440-silent HCSCs led to markedly decreased tumor volume
(P<0.05; Fig. 10). Taken
together, these data show that decreased expression of BC047440 in
HCSCs effectively suppressed the tumorigenic capacity of HCSCs
in vivo.
Discussion
HCSC-specific phenotypes and mechanisms that relate
to functions in tumorigenicity, HCC progression, and therapeutic
resistance have been identified. These results indicate that HCSCs
may contribute to the failure of existing therapies to consistently
eradicate malignant tumors (12–15).
Therefore, HCSCs represent novel and translationally relevant
targets for clinical cancer therapy. Notably, proof-of-principle
experiments have strengthened the rationale for developing
HCSC-targeted therapeutic modalities that might complement more
conventional HCC therapies (16).
The malignant biological behavior of CSCs is mainly
due to increased proliferation and immature state. Recent research
showed that malignant behavior was most likely maintained and
regulated by various genes and related pathways. For example, the
Wnt pathway is closely related to CSC self-renewal and
proliferation. Studies have shown that Wnt pathway activation plays
an important role in maintaining the self-renewal and proliferation
of leukemia stem cells and epithelium-derived CSCs (17). The activation of the Hedgehog
pathway has been found in multiple myeloma and chronic myelogenous
leukemia CSCs (18). In normal
cells, p21 is an important regulator of the cell cycle. Studies of
leukemia stem cells indicated that the expression of proto-oncogene
PMLRAR upregulated p21 expression, which was closely related to
leukemia stem cell proliferation (19). The proto-oncogene Bmil can promote
self-renewal and proliferation in normal cells, and recent studies
reported that Bmil was overexpressed in the brain and in
colon-derived CSCs (20,21). In addition, other factors such as
Notch and human telomerase also play important roles in maintaining
CSC function (22). The malignant
behavior of CSCs and the complicated gene activation and related
pathways that maintain CSC behavior can lead to drug resistance and
the failure of traditional treatments. Therefore, we speculate that
novel therapeutic approaches that target the genes and pathways
responsible for maintaining CSC malignant behavior could improve
cancer treatment. Fan et al (23) treated medulloblastoma cells with
γ-secretase inhibitor to block Notch signaling and found that the
proliferative ability of CD133+ cells was significantly
reduced; however, the proliferative ability of CD133−
cells was not affected. Verma et al (24) downregulated the expression of
β-catenin, the key protein in the Wnt pathway in colon cancer
cells, and found that the proliferative capacity of CSCs was
effectively inhibited. Piccirillo et al (25) applied exogenous bone morphogenetic
protein to induce glioblastoma stem cell differentiation and
observed a significant reduction in the tumorigenic ability of the
entire tumor cell population. The above studies suggested that
manipulating the genes and related pathways responsible for
maintaining CSC function could reduce proliferative capacity and
induce CSC differentiation, improving the likelihood of achieving
better treatment outcomes. Targeting CSCs is a novel and effective
therapeutic strategy to inhibit tumor occurrence and growth.
However, compared with the research on CSCs in solid tumors, HCSC
research is still preliminary. There are relatively few studies
regarding the effects of key genes and related pathways on HCSC
malignant behavior. In addition, CSCs and normal stem cells share
various pathways, such as Notch, Wnt and Bmi, and applying specific
gene therapies targeting those pathways might raise safety
concerns. Therefore, conducting in-depth research on the specific
genes and pathways responsible for maintaining HCSC functions might
lay a foundation for investigating the molecular mechanism of CSC
pathogenesis, which will provide new CSC intervention targets.
The current studies regarding the roles of BC047440,
a new HCC-related gene, in HCC were all completed by our research
group, and the results have been widely recognized (7–9). In a
previous investigation, we found that downregulated BC047440
expression can inhibit HepG2 cell proliferation, decreasing
cellular AFP expression and significantly increasing functional
gene expression in normal hepatocytes, such as ALB and cytochrome
P450 1A2 (CYP1A2) (Fig. 1). The
above results suggested that BC047440 might be an important
molecule to mediate HCC proliferation and differentiation of HCC
cells and might be useful for gene-targeted therapies. Recent
studies confirmed that the existence of HCSCs was the main reason
why traditional therapies designed to reduce tumor volume failed.
How to inhibit HCSC functions in tumorigenesis and maintain HCC to
achieve better treatment effects has become a hot topic. A growing
body of evidence suggests that compared with traditional
therapeutic approaches, HCSC-targeting therapy that aims to inhibit
HCSC proliferation and promote tumor cell differentiation is a more
effective way to eradicate HCC and reduce recurrence and metastasis
rates. The development of HCSC-targeting therapy has always been an
important research topic in our laboratory. However, it still faces
many challenges, such as identifying the specific molecules
involved in maintaining HCSC malignant behavior. At present, few
such studies have been reported. Due to its important role in HCC,
we considered whether BC047440 could be one of the specific
molecules involved in maintaining HCSC malignant behavior. To test
the above hypothesis, we employed gene chips to screen for genes
that are differentially expressed between HCSCs and HNSCs and found
that BC047440 expression was significantly upregulated in HCSCs
(data not shown). This result was subsequently confirmed with
western blot analysis. Furthermore, we also applied an RNAi
technique to interfere with BC047440 expression in HCSCs. Since the
proliferative capacity of HCSCs and differentiation degree are
closely related to their malignant behavior and given the important
role of BC047440 in HCC proliferation and differentiation, we
hypothesized that the absence of BC047440 might inhibit HCSC
proliferation and promote HCSC differentiation. As we speculated,
knockdown of BC047440 by shRNA in HCSCs leads to proliferation
inhibition and differentiation induction in vitro and alters
the expression of proliferation and differentiation-associated
molecules. The in vivo experiment results indicated that
inhibiting BC047440 expression significantly decreased HCSC
tumorigenicity. Based on our study results, we described that
BC047440 plays an important role in the proliferation and
differentiation of HCSCs. However, further studies should be
conducted to investigate this putative process in more detail.
By conducting studies on BC047440, we found that it
might affect multiple proliferation- and differentiation-related
proteins: i) NF-κB is a pleiotropic and multifunctional nuclear
transcription factor involved in mediating tumor growth,
proliferation, escaping apoptosis, invasion and metastasis
(26). Studies have shown that
NF-κB is activated in a variety of CSCs, and inhibiting NF-κB
activation can inhibit CSC proliferation and induce apoptosis
(27). We previously found that
BC047440 can promote HepG2 proliferation by activating NF-κB
signaling. After inhibiting BC047440 expression in HCSCs, the NF-κB
level was increased in the cytosol but decreased in the nucleus.
Given the close relationship between NF-κB activation and CSC
proliferation described above, as well as results from our studies,
we considered NF-κB might play an important role in
BC047440-regulated HCSC proliferation. However, NF-κB activation
might not be the initiating factor of the proliferation signal
pathway in HCSCs. Rather, it might be a key factor in the signaling
pathway to promote cell proliferation, and other genetic products
might be involved. For example, the essential upstream molecules of
NF-κB, such as TRAF2, TRAF5, TRAF6, TRAF1, RIP, TLR, Myd88, IRAK1,
IRAK2, TAK1 and TAK2, can interact with NF-κB to maintain its
sustained activity and promote HCSC proliferation. However, which
gene products play roles in the NF-κB signaling pathway are
unknown, as are the specific mechanisms of those gene products. At
present, our understanding regarding the regulation of
BC047440/NF-κB signaling in HCSC proliferation remains preliminary.
ii) HNF4α is a member of the nuclear receptor superfamily and is
highly expressed in mature hepatocytes. It is an important
transcription factor that regulates hepatocyte development,
differentiation and function. It plays a critical role in liver
development, as well as gene transcription and regulation in adult
hepatocytes. In addition, HNF4α overexpression can facilitate tumor
cell transformation into lower degrees of malignancy and reverse
the de-differentiated stage of HCC cells (28). We examined the expression level of
HNF4α in preliminary experiments and found that the absence of
BC047440 expression was accompanied by significantly elevated HNF4α
expression in HepG2 cells, suggesting that BC047440 can promote
HepG2 differentiation through HNF4α signaling (Fig. 1). Inhibiting BC047440 expression in
HCSCs leads to significantly elevated HNF4α expression. In-depth
studies on whether BC047440 can regulate HCSC differentiation
through HNF4α signaling will help to elucidate the mechanism of
tumorigenesis in HCC and provide important experimental evidence
for the application of gene-targeted therapy in HCC treatment. iii)
BC047440 dysregulation can lead to transcriptional activation of
downstream proteins that play important roles in HCSC proliferation
and differentiation. However, the specific functions of induced
gene expression are not fully understood. The identification of
specific downstream target molecules in the BC047440 signaling
pathway requires further research.
We preliminarily confirmed the novel HCC-related
gene BC047440 as an important molecule that induces malignant
behavior in HCSCs. The absence of BC047440 may be associated with
reduced HCSC proliferation and increased differentiation. However,
elucidating the specific mode of action requires further studies.
Furthermore, the precise molecular mechanism of BC04744’s
involvement in maintaining HCSC malignant behavior remains unclear.
The answers to these questions will provide a foundation to
elucidate the function and molecular mechanism of BC047440 during
HCC onset and development, which can provide theoretical and
practical evidence for developing a novel gene target for HCC
therapy.
Acknowledgements
The authors thank Juan Li for her excellent
technical assistance. The present study was funded by the Chinese
National Natural Science Foundation (grant no. 81372561, 81302168)
and the Natural Science Foundation of Chongqing (grant no.
cstc2012jjA10079).
Abbreviations:
|
AFP
|
α-fetoprotein
|
|
ALB
|
albumin
|
|
BSA
|
bovine serum albumin
|
|
CSCs
|
cancer stem cells
|
|
CYP1A2
|
cytochrome P450 1A2
|
|
DMEM
|
Dulbecco’s modified Eagle’s medium
|
|
EDTA
|
ethylenediaminetetraacetic acid
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
EST
|
expressed sequence tag
|
|
FBS
|
fetal bovine serum
|
|
FITC
|
fluorescein isothiocyanate
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
HCC
|
hepatocellular carcinoma
|
|
HCSCs
|
hepatic cancer stem cells
|
|
HGF
|
hepatocyte growth factor
|
|
HNF4α
|
hepatocyte nuclear factor 4α
|
|
HNSCs
|
hepatic normal stem cells
|
|
HRP
|
horseradish peroxidase
|
|
LSD
|
least significant difference
|
|
NC
|
negative control
|
|
NF-κB
|
nuclear factor-κB
|
|
PBS
|
phosphate-buffered saline
|
|
RACE
|
rapid amplification of cDNA ends
|
|
shRNA
|
short hairpin RNA
|
|
TEM
|
transmission electron microscopy
|
|
V
|
volume
|
References
|
1
|
Shiozawa Y, Nie B, Pienta KJ, Morgan TM
and Taichman RS: Cancer stem cells and their role in metastasis.
Pharmacol Ther. 138:285–293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng X and O’Neill HC: Oncogenesis and
cancer stem cells: current opinions and future directions. J Cell
Mol Med. 13:4377–4384. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamashita T and Wang XW: Cancer stem cells
in the development of liver cancer. J Clin Invest. 123:1911–1918.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Padhya KT, Marrero JA and Singal AG:
Recent advances in the treatment of hepatocellular carcinoma. Curr
Opin Gastroenterol. 29:285–292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wilson GS, Hu Z, Duan W, et al: Efficacy
of using cancer stem cell markers in isolating and characterizing
liver cancer stem cells. Stem Cells Dev. 22:2655–2664. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee H, Kim JB, Park SY, Kim SS and Kim H:
Combination effect of paclitaxel and hyaluronic acid on cancer
stem-like side population cells. J Biomed Nanotechnol. 9:299–302.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng L, Liang P, Li J, et al:
ShRNA-targeted COMMD7 suppresses hepatocellular carcinoma growth.
PLoS One. 7:e454122012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng L, Huang X, Liang P, et al: BC047440
overexpression is a risk factor for tumor invasion and poor
prognosis in hepatocellular carcinoma. Hepatogastroenterology.
57:919–925. 2010.PubMed/NCBI
|
|
9
|
Zheng L, Liang P, Zhou J, Huang X, Wen Y,
Wang Z and Li J: BC047440 antisense eukaryotic expression vectors
inhibited HepG2 cell proliferation and suppressed xenograft
tumorigenicity. Braz J Med Biol Res. 45:97–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu WH, Li R and Dou KF: Convenient and
efficient enrichment of the CD133+ liver cells from rat
fetal liver cells as a source of liver stem/progenitor cells. Stem
Cell Rev. 7:94–102. 2011.PubMed/NCBI
|
|
11
|
Liu WH, Wang X, You N, et al: Efficient
enrichment of hepatic cancer stem-like cells from a primary rat HCC
model via a density gradient centrifugation-centered method. PLoS
One. 7:e357202012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li CH, Wang YJ, Dong W, et al: Hepatic
oval cell lines generate hepatocellular carcinoma following
transfection with HBx gene and treatment with aflatoxin B1 in vivo.
Cancer Lett. 311:1–10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chiba T, Kita K, Zheng YW, et al: Side
population purified from hepatocellular carcinoma cells harbors
cancer stem cell-like properties. Hepatology. 44:240–245. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J and
Li J: Cancer stem/progenitor cells are highly enriched in
CD133+CD44+ population in hepatocellular
carcinoma. Int J Cancer. 126:2067–2078. 2010.PubMed/NCBI
|
|
15
|
Espandiari P, Robertson LW, Srinivasan C
and Glauert HP: Comparison of different initiation protocols in the
resistant hepatocyte model. Toxicology. 206:373–381. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamashita T, Honda M, Nakamoto Y, et al:
Discrete nature of EpCAM+ and CD90+ cancer
stem cells in human hepatocellular carcinoma. Hepatology.
57:1484–1497. 2013.PubMed/NCBI
|
|
17
|
Malanchi I, Peinado H, Kassen D, et al:
Cutaneous cancer stem cell maintenance is dependent on β-catenin
signalling. Nature. 452:650–653. 2008.
|
|
18
|
Zhao C, Chen A, Jamieson CH, et al:
Hedgehog signalling is essential for maintenance of cancer stem
cells in myeloid leukaemia. Nature. 458:776–779. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Viale A, De Franco F, Orleth A, et al:
Cell-cycle restriction limits DNA damage and maintains self-renewal
of leukaemia stem cells. Nature. 457:51–56. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui H, Hu B, Li T, Ma J, Alam G, Gunning
WT and Ding HF: Bmi-1 is essential for the tumorigenicity of
neuroblastoma cells. Am J Pathol. 170:1370–1378. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim JH, Yoon SY, Kim CN, et al: The Bmi-1
oncoprotein is overexpressed in human colorectal cancer and
correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett.
203:217–224. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song K, Wu J and Jiang C: Dysregulation of
signaling pathways and putative biomarkers in liver cancer stem
cells (Review). Oncol Rep. 29:3–12. 2013.PubMed/NCBI
|
|
23
|
Fan X, Matsui W, Khaki L, Stearns D, Chun
J, Li YM and Eberhart CG: Notch pathway inhibition depletes
stem-like cells and blocks engraftment in embryonal brain tumors.
Cancer Res. 66:7445–7452. 2006. View Article : Google Scholar
|
|
24
|
Verma UN, Surabhi RM, Schmaltieg A,
Becerra C and Gaynor RB: Small interfering RNAs directed against
β-catenin inhibit the in vitro and in vivo growth of
colon cancer cells. Clin Cancer Res. 9:1291–1300. 2003.
|
|
25
|
Piccirillo SG, Reynolds BA, Zanetti N, et
al: Bone morphogenetic proteins inhibit the tumorigenic potential
of human brain tumour-initiating cells. Nature. 444:761–765. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
DiDonato JA, Mercurio F and Karin M: NF-κB
and the link between inflammation and cancer. Immunol Rev.
246:379–400. 2012.
|
|
27
|
Prud’homme GJ: Cancer stem cells and novel
targets for antitumor strategies. Curr Pharm Des. 18:2838–2849.
2012.PubMed/NCBI
|
|
28
|
Ning BF, Ding J, Yin C, et al: Hepatocyte
nuclear factor 4α suppresses the development of hepatocellular
carcinoma. Cancer Res. 70:7640–7651. 2010.
|