Introduction
Glioma is the most common primary malignant tumor of
the central nervous system in adults. Despite modern diagnosis and
treatment, prognosis of the disease remains poor, with the median
survival time ranging from 12–14 months (1). The local tissue destruction caused by
the invasion of glioma cells within the brain is often lethal. This
is the cause of high morbidity and mortality of malignant glioma
(2,3).
Identifying novel molecules that can repress the invasiveness and
metastasis of glioma cells will facilitate the development of novel
anti-glioma strategies.
Big mitogen-activated protein kinase 1 (BMK1) is a
newly identified member of the mitogen-activated protein (MAP)
kinase family (4). In our previous
research, it was demonstrated that BMK1 was upregulated and
associated with invasion in glioma (5). Furthermore, BMK1 is also overexpressed
in melanoma (6) and hepatocellular
carcinoma (7). However, the role of
BMK1 in the molecular pathogenesis of glioma remains largely
unknown.
MicroRNAs (miRNA/miRs) are a class of small,
noncoding single-stranded RNAs that modulate gene expression at the
post-transcriptional level (8).
Notably, previous studies have shown that miRNAs serve a similar
role to oncogenes or tumor suppressors by regulating the signaling
pathways involved by the target gene and by regulating the
formation and development of tumors (9,10). miRNAs
downregulate gene expression by binding to the 3′-untranslated
regions (UTRs) of the target mRNAs (8,11). A
single miRNA may have hundreds of target genes that regulate about
200 protein-coding genes. In terms of the number of confirmed
miRNAs, they regulate the expression of ~one-third of the
protein-coding genes (12).
Additionally, previous studies provide evidence that certain miRNAs
can function as potential biomarkers for cancer diagnosis,
progression, and response to treatment (13,14).
miR-143 is located on chromosome 5 position 33 in
the human genome. This miRNA is highly expressed in the colon and
is consistently reported as being downregulated in the colorectal
adenocarcinoma (15). Subsequent
studies have confirmed that miRNA-143 inhibits cell proliferation,
invasion and metastasis by regulating multiple target genes
(16–18). Of particular interest to us, similar
findings have been reported in pancreatic cancer, breast cancer,
and other solid tumors of epithelial origin (19,20).
However, the functions of miR-143 in the glioma that underlie their
presumed tumor suppressor activity have not been investigated.
The purpose of the present study was to explore the
association between BMK1 and miR-143. In the present study, it was
demonstrated that miR-143 acts as a novel BMK1 inhibitor that is a
valuable prognostic marker for patients with glioma. The in
vitro and in vivo results indicate that miR-143 is
significantly involved in glioma invasion and metastasis and the
expression of miR-143 is repressed by methylation of its promoter.
The present study may provide a therapeutic target molecule for
malignant glioma infiltration.
Materials and methods
Cell culture and reagents
The human cell lines U87 and HEB were obtained from
the American Type Culture Collection (Manassas, VA, USA) and were
cultured in Roswell Park Memorial Institute medium (RPMI)-1640
(HyClone; GE Healthcare, Chicago, IL, USA), supplemented with 10%
fetal bovine serum (FBS; HyClone; GE Healthcare) and 100 µg/ml
penicillin/streptomycin in a humidified atmosphere of 37°C and 5%
CO2. Notably, U87 cell line was recognized as
glioblastoma cell line of unknown origin. The human glioma cell
lines U251 (TCHu 58) and SHG44 (TCHu 48) was purchased from the
cell bank of the Chinese Academy of Sciences. All cell lines have
been authenticated by STR profiling.
Patients and tissue specimens
A total of 176 paraffin-embedded samples and 30
pairs of surgically resected fresh glioma tissues including their
corresponding adjacent non-cancerous tissues were collected from
the Affiliated Hospital of Weifang Medical University and the
Affiliated Yantai Yuhuangding Hospital of Qingdao University from
2008–2015. The detailed information of 176 patients with glioma is
listed in Table I. The inclusion
criteria for the patients is that they had gliomas and had agreed
to participate in the study. Samples were collected following a
protocol approved by the Institutional Review Board, and patients
gave their consent for the research of their tissue specimens in
the present study. The study protocol was reviewed and approved by
the Weifang Medical University Ethics Committee (approval no 99,
11-November-2016).
 | Table I.Clinicopathologic characteristics of
patients with glioma. |
Table I.
Clinicopathologic characteristics of
patients with glioma.
| Patient
characteristics | n (%) |
|---|
| Sex |
|
|
Male | 99
(56.2) |
|
Female | 77
(43.7) |
| Age (years) |
|
|
<40 | 81
(46.0) |
|
40-49 | 37
(21.0) |
|
50-59 | 27
(15.3) |
|
60-69 | 25
(14.2) |
|
70-79 | 6
(3.4) |
| WHO grading |
|
| Grade I
(pilocytic astrocytoma) | 24
(13.6) |
| Grade
II | 53
(30.1) |
| Grade
III | 56
(31.8) |
| Grade
IV | 43
(24.4) |
| Patient
survival |
|
|
Alive | 34
(19.3) |
|
Deceased | 142 (80.7) |
miRNA prediction
Two online miRNA prediction databases (www.microrna.org/microrna/home.do and
www.targetscan.org) were used. These were
searched for BMK1 and found that miR-143 was the most potential
regulator.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA extraction and RT-qPCR was performed as
previously described (21). Total
miRNA from cultured cells, surgically resected fresh tissues, and
paraffin-embedded glioma specimens was extracted using the mirVana
miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and RecoverALL Total Nucleic Acid Isolation kit
(Ambion; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Complementary DNA was synthesized with 5
ng of total RNA using the TaqMan miRNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc), and the
expression levels of miR-143 were measured using Hairpin-itTM
miRNAs qPCR Quantitation kit (Shanghai Genepharma, Co, Ltd.,
Shanghai, China). Reaction conditions: 95°C for 30 sec, 95°C for 3
sec, 60°C for 30 sec, a total of 40 cycles. miR-143 reverse
transcription primer sequences: Forward,
5′-TGTAGTTTCGGAGTTAGTGTCGCGC-3′; reverse,
5′-CCTACGATCGAAAACGACGCGAACG-3′. U6 primer sequences: Forward,
5′-GTTTTTTGTAGTTTTTGGAGTTAGTGTTGTGT-3′; reverse,
5′-CTCAACCTACAATCAAAAACAACACAAACA-3′. U6 was used as an internal
reference. Primers used for BMK1: Forward,
5′-CTGGCTGTCCAGATGTGAA-3′; reverse, 5′-ATGGCACCATCTTTCTTTGG-3′.
Analysis of relative gene expression data using RT-quantitative PCR
and the 2−ΔΔCq method (22).
Bisulfite sequencing and DNA
methylation
Genomic DNAs from NHA and glioma cell lines (the
cell density reached 90% confluency) were isolated using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and Genomic
DNAs from fresh glioma tissues were isolated using the DNeasy Blood
and Tissue kit (Qiagen, Valencia, CA, USA) according to the
manufacturer's protocol. Bisulfite sequencing was performed as
previously described (23). DNA
methylation analysis was performed using Epigenetek Methylflash
Methylated DNA quantification kit (cat. no. P1034±48) following the
manufacturer's protocol.
Plasmid, retroviral infection, and
transfection
miR-143 mimics (miR-143) oligonucleotides
(3′-CUCGAUGUCACGAAGUAGAGU-5′), miR-143 inhibitor (Ant-miR)
oligonucleotides (3′-UGGUCUCUACGUCGUGACGUGG-5′) and miR-143 mutant
oligonucleotides (3′-AUGUGUCUUAUGCAAGCUCGCA-5′) were synthesized by
GenePharma (Shanghai, China). They were transfected into glioma
cell lines by using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at a final concentration of 50 nM according to
the manufacturer's protocol. A DNA fragment containing the
hsa-miR-143 precursor with 300 bp flanking sequence of each side
was amplified into retroviral transfer plasmid pMSCV-puro (Shanghai
GenePharma). A blank plasmid was used as a negative control (NC).
The open reading frames of BMK1 genes generated by PCR were cloned
into retroviral vector pMSCV-neo (Clontech Laboratories, Inc.,
Mountainview, CA, USA). Retroviral production and infection were
performed as previously described (24). The 3′UTRs of BMK1 were amplified and
cloned into the downstream region of a luciferase gene in a
modified pGL3 control vector (Promega Corporation, Madison, WI,
USA), as described previously (25).
The U251/miR-143 cells were transfected with
pMSCV-neo-BMK1 plasmid or pMSCV-neo vector using
Lipofectamine® (Invitrogen, Carlsbad, CA, USA) following
the manufacturer's protocol. Briefly, the cells were plated at a
density of 5×105 cells/well in a 6-well plates to grow
overnight, and then they were transected with 4 µg plasmid using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the protocol. After transfection, cells
were cultured at 37°C, 5% CO2, overnight. Then cells
were trypsinised, diluted and reseeded into 10 cm culture dishes.
Single cell clones were isolated for clone expansion. Stable
transfected cell clones were named U251/miR-143+BMK1 and
U251/miR-143+CON cells.
Western blot analysis
Western blot analysis was carried out using standard
methods. After treatment, the cells were treated with lysis buffer
(Cell Signaling, Danvers, MA, USA) on ice for 20 min. Subsequently,
the cell lysates were centrifuged at 1,5000 × g at 4°C for 30 min,
the supernatant was collected as the total cellular protein
extract. The total protein extract was quantified by using the BCA
protein assay kit (Pierce, Rockford, IL, USA). Equal amounts of
protein (10 µg/lane) were separated on 10% SDS-polyacrylamide gel
and transferred onto nitrocellulose membranes. The membrane was
incubated for 2 h in PBS plus 0.1% Tween-20 and 5% non-fat skim
milk to block non-specific binding. The following antibodies were
used in the present study: BMK1 (cat. no. sc-81460, 1:1,000; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA), phosphorylated
(p)-cofilin (cat. no. sc-21867-R, 1:1,000; Santa Cruz
Biotechnology, Inc.), cofilin (cat. no. sc-53934, 1:1,000; Santa
Cruz Biotechnology, Inc.), β-actin (4970, 1:1,000; Cell Signaling
Technology, Inc.), and the secondary antibody: Horseradish
peroxidase-linked anti-mouse IgG antibody (cat. no. 58802, 1:2,000;
Cell Signaling Technology, Inc. Danvers, MA, USA). A reference
housekeeping protein (β-actin) was used to normalize the average
protein expression. All experiments were repeated at least three
times. After incubated with antibodies, the membranes were placed
into TBST (10 mM Tris-HCL, 150 mM Nacl and 0.05% Tween-20, pH=7.6),
and shaken for 5 min at room temperature for 3 times. Western blots
were visualized by using enhanced chemiluminescence reagents
(Pierce; Thermo Fisher Scientific, Inc.). The protein band
intensity was quantified using the ImageJ 1.44p software
(https://imagej.nih.gov/ij/download.html).
Cell viability assay
Cell viability was detected via MTT assays (Sigma;
Merck KGaA, Darmstadt, Germany). All experiment steps were
performed using 2×103 of cells seeded on 96-well plates.
After the cells were cultured for different time points, the
culture medium was removed. Subsequently, the cells were incubated
with MTT solution (50 µl of 2 mg/ml MTT powder in PBS; 100 µl
RPMI-1640 containing 10% FBS) at 37°C for 4 h. Subsequently, the
culture medium was removed and DMSO (150 µl; Sigma; Merck KGaA) was
added into each well for 30 min until all crystals were dissolved.
The absorbance was directly proportional to the number of viable
cells.
Chemotaxis assay
Chemotaxis assay was performed as described
previously (26). Briefy, IGF-1 was
loaded into the lower chemotaxis chamber and 5×105
cell/ml cells were added to the upper chambers. The polycarbonate
filter (Neuroprobe, Cabin John, MD, USA) was inserted between the
chambers. The number of migrating cells were counted. Chemotaxis
index=the migrating cell number in a chemo-attractant gradient/the
migrating cell number in a medium control.
Cellular F-Actin measurement
The F-actin content was done as described previously
(27). After reaching 70–80%
confluency, U87 cells were followed by the stimulation of 50 ng/ml
IGF-1 at 37°C at different time points. Then the cells were fixed,
permeabilized, and incubated with Oregon Alexa-Flour 568 phalloidin
at room temperature for 2 h. After washing 5 times, the labeled
phalloidin was extracted by using methanol at 4°C for 90 min. The
fluorescence was captured at Ex/Em 578/600 nm in each sample and
normalized against the total protein content as analyzed by a BCA
kit (Pierce; Thermo Fisher Scientific, Inc.).
Scratch assay
The control cells and cells transfected with miR-143
mimic (2×105/ml) were seeded in 6-well plates. 48 h
later, cross lines were made using a 200 µl sterile pipette tip. At
0, 6, 12, 18, 24 h, the cells were imaged by using a Olympus
inverted microscope (CH-BI45-T; Olympus). Experiments were repeated
three times.
Matrigel invasion assay
A Boyden chamber invasion assay was performed as
described previously (28). The
glioma cells in serum-free RPMI-1640 at a density of
2×105 cells/ml were loaded on the membrane in the upper
chamber. All assays were repeated at least three times
independently.
Luciferase reporter assay
To investigate whether miR-143 directly regulates
BMK1 expression, the sequence of the 3′-UTR of BMK1 was inserted
downstream of a Renilla luciferase open reading frame. A total of
100 ng pGL3-BMK1-3′UTR plasmid, 1ng pRL-TK renilla plasmid
(Promega, Madison, WI, USA) and miR-143 mimics or miR-NC (Thermo
Fisher Scientific, Inc.) were transfected into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). At 48 h after transfection, the activity of
firefly luciferase was measured by using the Dual-Luciferase
reporter assay kit (Promega, Madison, WI, USA). The data were
normalized with Renilla luciferase activity.
RNA immunoprecipitation (RIP)
Immunoprecipitation of miRNA ribonucleoprotein
(miRNP) with anti-Ago1 (1:3,000; cat. no. ab5070; Abcam, Cambridge,
UK) or IgG (1:2,000; cat. no. ab97051; Abcam) was performed as
previously described (29). The RNA
that was immunoprecipitated with anti-Ago1 or IgG antibodies was
extracted using TRIzol LS (Invitrogen; Thermo Fisher Scientific,
Inc.) as described previously (30).
Intracranial brain tumor xenografts
and H&E staining
Adult male Sprague-Dawley rats (n=16) weighing
between 200–250 g were purchased from Wei Tong Li Hua experimental
animal Co. (Beijing, China). Intracranial brain tumor xenografts
were established with U87/NC (5×105) and U87/miR-143
(5×105) stereotactically implanted into the brain of
four-week-old male severe immunodeficient mice with eight mice per
group. The study protocol was reviewed and approved by Weifang
Medical University Ethics Committee (approval no 129,
11-November-2016). Using a microliter syringe connected to the
manipulating arm of the stereotactic apparatus, glioma cells were
injected into the caudate nucleus at a depth of 4.0-4.5 mm from the
dura. The glioma-enduring mice were euthanized 30 days after tumor
cell injection, and the whole brains were removed. The maximum
diameter of the tumor was about 1cm. Subsequently,
paraffin-embedded tissues were sectioned into 5 µm slices and
subjected to H&E staining. H&E staining was performed as
described previously (31). The
tissue slices were stained in hematoxylin for 5 min and in eosin
for 10 sec. All the procedures were performed at room temperature.
The images were captured using a light microscope system (Olympus
Corporation, Tokyo, Japan).
5-AZA-2-deoxycytidine treatment
Glioma cells were seeded in 10 cm dishes
(1×106 cells per dish) one day before drug treatment.
The cells were treated with 1uM 5-AZA-2-deoxycytidine (5-AZA-dC;
Sigma; Merck KGaA) every 24 h for 3 days.
Statistical analysis
All statistical analyses were performed using SPSS
13.0 statistical software package (SPSS, Inc., Chicago, IL, USA).
The χ2 test was used to analyze the association between
miR-143 expression and clinicopathologic characteristics. Survival
curve was evaluated using the Kaplan-Meier method and differences
were assessed using the log-rank test. Statistical significance for
comparisons between groups was determined using two-tailed unpaired
Student's t-test or analysis of variance. Multiple comparison
between the groups was performed using S-N-K method, following
ANOVA. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-143 is predicted to target
BMK1
To investigate the potential miRNA regulators of
BMK1 which was known to be overexpressed in glioma (5), two online miRNA target prediction
databases were used (miRNA.org; www.microrna.org and Targetscan; www.targetscan.org), and miR-143 was selected as the
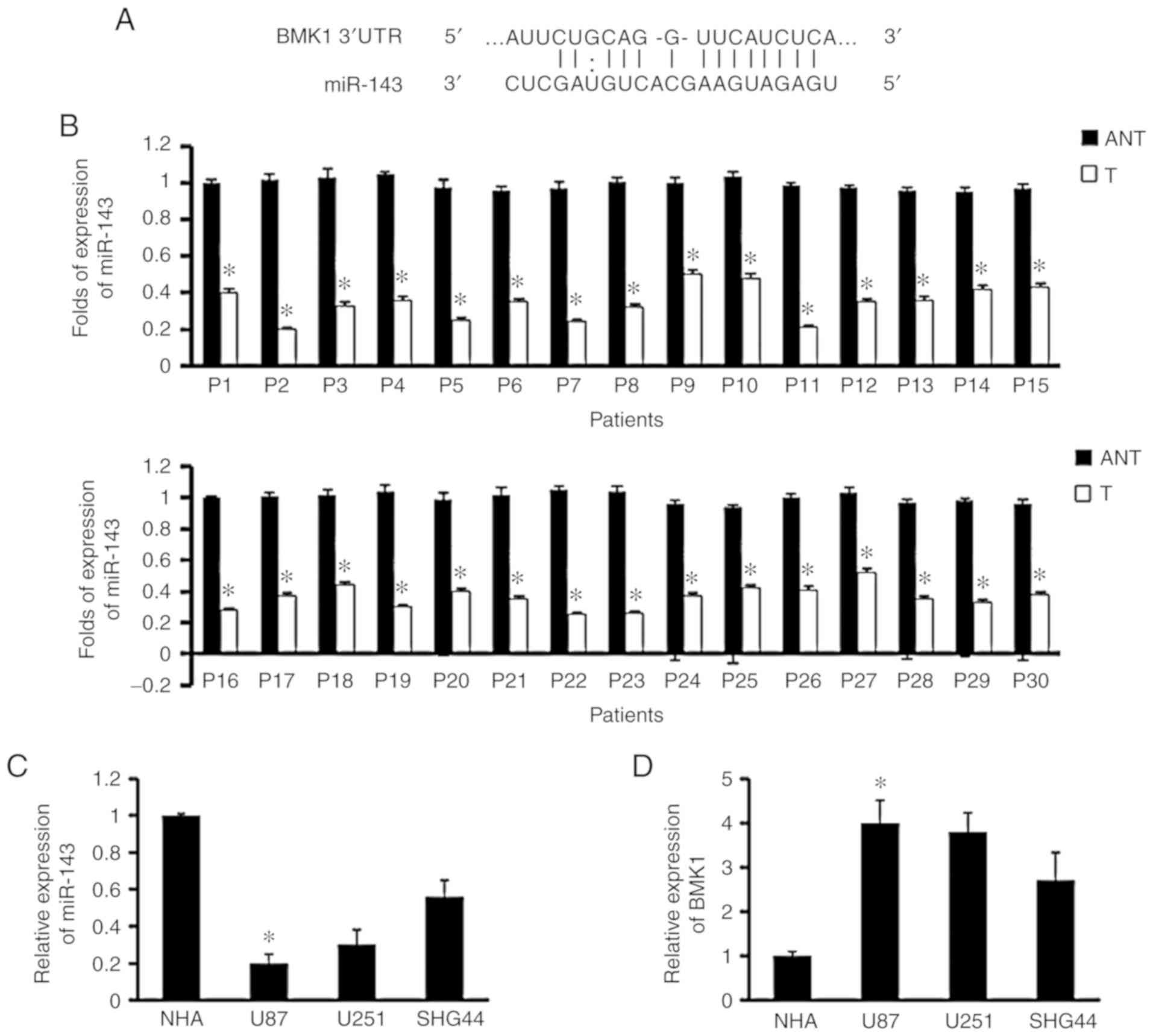
most potential regulator (Fig. 1A).
Paired glioma (T) and adjacent non-tumor tissues (ANT) were
comparatively evaluated by RT-qPCR analysis, with each pair
obtained from the same patient. The results showed miR-143
expression was downregulated in 30 pairs of glioma tumor tissues
compared with their ANT (Fig. 1B).
Furthermore, similar results were found in glioma cell lines and
normal brain glial cell line (NHA; Fig.
1C). In contrast, the expression of BMK1 in glioma cell lines
was higher than that in NHA (Fig.
1D). These data suggested that the expression of miR-143 in
glioma was significantly reduced.
Decreased expression of miR-143 is
associated with the clinicopathological features of glioma
In order to further understand the association
between miR-143 expression and clinicopathological features of
glioma, the expression levels of miR-143 in 176 paraffin-embedded
glioma samples was examined by RT-qPCR. The detailed information of
176 glioma patients is listed in Table
I. It was found that miR-143 expression was strongly associated
with World Health Organization (WHO) grade (32) and the survival status of glioma
patients, but not with age and sex (Table II).
 | Table II.Association between clinicopathologic
features and expression of miR-143 in patients with glioma. |
Table II.
Association between clinicopathologic
features and expression of miR-143 in patients with glioma.
|
| miR-143
expression |
|
|---|
|
|
|
|
|---|
| Patient
characteristics | Low/none | High | P-value |
|---|
| Sex |
|
| 0.651 |
|
Male | 61 | 38 |
|
|
Female | 50 | 27 |
|
| Age(years) |
|
| 0.377 |
|
≤45 | 66 | 43 |
|
|
>45 | 45 | 22 |
|
| WHO grade |
|
| <0.001 |
| I and
II | 33 | 44 |
|
| III and
IV | 78 | 21 |
|
| Survival |
|
| 0.031 |
|
Alive | 16 | 18 |
|
|
Deceased | 95 | 47 |
|
In addition, Kaplan-Meier analysis using log-rank
test was performed to assess the effect of miR-143 expression on
the survival of patients. Patients with tumors exhibiting low
miR-143 expression had shorter overall survival than those with
high expression of it (P<0.05, Fig.
1E). The median survival time of patients with low miR-143
expression (14±1.497 months, 95% confidence interval:
11.065-16.935) was significantly shorter than those with high
miR-143 expression (35±3.527 months, 95% confidence interval:
28.087-41.913). Furthermore, similar results were found in either
WHO grade I+II subgroup (P<0.05, Fig.
1F) or grade III+IV subgroup (P<0.05, Fig. 1G). Taken together, these results
demonstrated that miR-143 could be a valuable prognostic marker for
glioma patients at all disease stages, but this needs to be
confirmed with larger sample size.
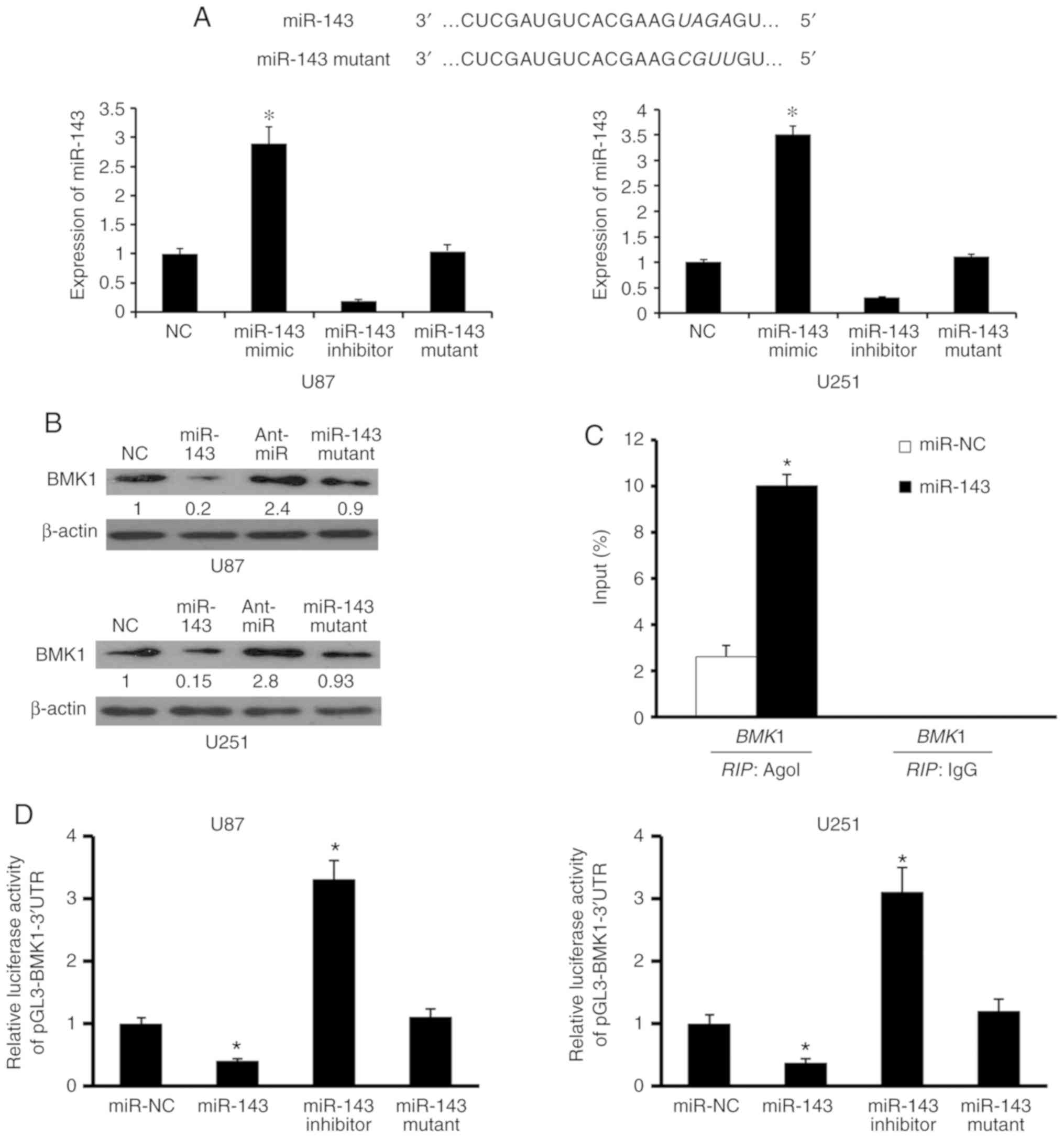
miR-143 directly targets BMK1
To verify our assumption that miR-143 could regulate
the expression of BMK1, firstly miR-143 expression was analyzed by
RT-qPCR after being transfected with negative control (NC), miR-143
mimic (miR-143), miR-143 inhibitor (Ant-miR), and miR-143 mutant in
the U87 and U251 cell lines respectively, and obtained the stable
transfected cell lines (Fig. 2A).
BMK1 protein levels were significantly reduced in miR-143 mimic
transfected cells but elevated in miR-143 inhibitor transfected
cells compared with those in the corresponding control cells
(Fig. 2B). Furthermore, RIP analysis
following miR-143 transfection indicated that mRNAs of BMK1 could
be specifically recruited to the miRNP complex isolated by
anti-Ago1 antibody (Fig. 2C). Based
on these results, it may be hypothesized that miR-143 targets BMK1
in glioma, in vitro.
To further investigate whether this regulation is
due to the fact that miR-143 binds to the 3′UTR of BMK1, the 3′UTR
of BMK1 was cloned downstream of a luciferase reporter gene
(wt-BMK1) and wt-BMK1 vector and negative control, miR-143, miR-143
inhibitor, or miR-143 mutant were co-transfected into U87 and U251
cells. miR-143 regulated BMK1 expression through a significant
reduction or addition of luciferase activity in cells transfected
with miR-143 mimic or miR-143 inhibitor, respectively, compared
with control cells (Fig. 2D).
However, it was observed that relative luciferase activity was
normal in cells co-transfected with BMK1 3′UTR and miR-143 mutant.
Taken together, these results indicated that miR-143 can regulate
directly BMK1 expression in glioma cells.
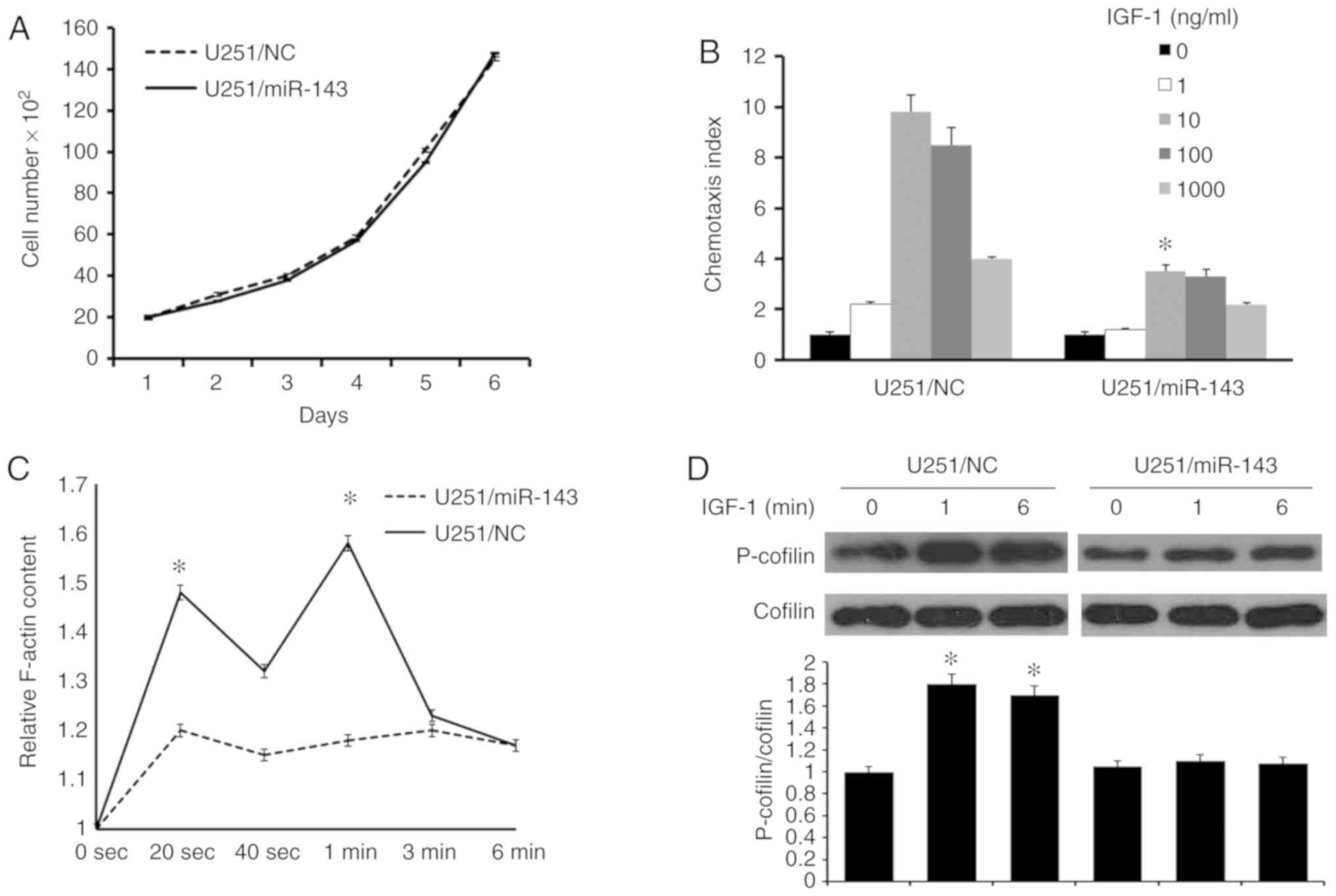
Restoration of miR-143 in glioma cells
inhibits cellular migration
To identify the effects of miR-143 on glioma cell
migration, firstly, the cell viability rate in U251/miR-143 and
U251/NC cells was investigated, in vitro. The same number of
U251/miR-143 and U251/NC cells were plated at the same time and
cell numbers were counted at the following days. The result showed
that increased miR-143 expression did not cause significant changes
in cell viability (Fig. 3A).
Following this, cell chemotaxis assay which was induced by IGF-1,
was performed. The result suggested decreased chemotaxis in
U251/miR-143 cells compared with U251/NC cells (Fig. 3B). The key to chemotaxis is
ligand-induced cytoskeleton rearrangement (33). Quantitative F-actin polymerization
assay revealed that IGF-1 induced transient actin polymerization at
20 and 60 sec in U251/NC cells. Whereas in U251/miR-143 cells,
F-actin polymerization was significantly reduced in spite of IGF-1
stimulation (Fig. 3C). These results
indicated that miR-143 serves an important role in the migration of
glioma cells.
Previous studies have proven that F-actin dynamics
is regulated by phosphorylating cofilin at Ser3, which polymerizes
actin, generates protrusions and determines the direction of cell
migration (34). Thus, the
phosphorylation level of cofilin in U251 cells was investigated. As
shown in Fig. 3D, cofilin was rapidly
activated by the IGF-1 at 1 and 6 min. However, in the U251/miR-143
cells, the IGF-1-induced phosphorylation of cofilin was impaired.
These results indicate that miR-143 is a key factor in the
IGF-1-induced cofilin recycling.
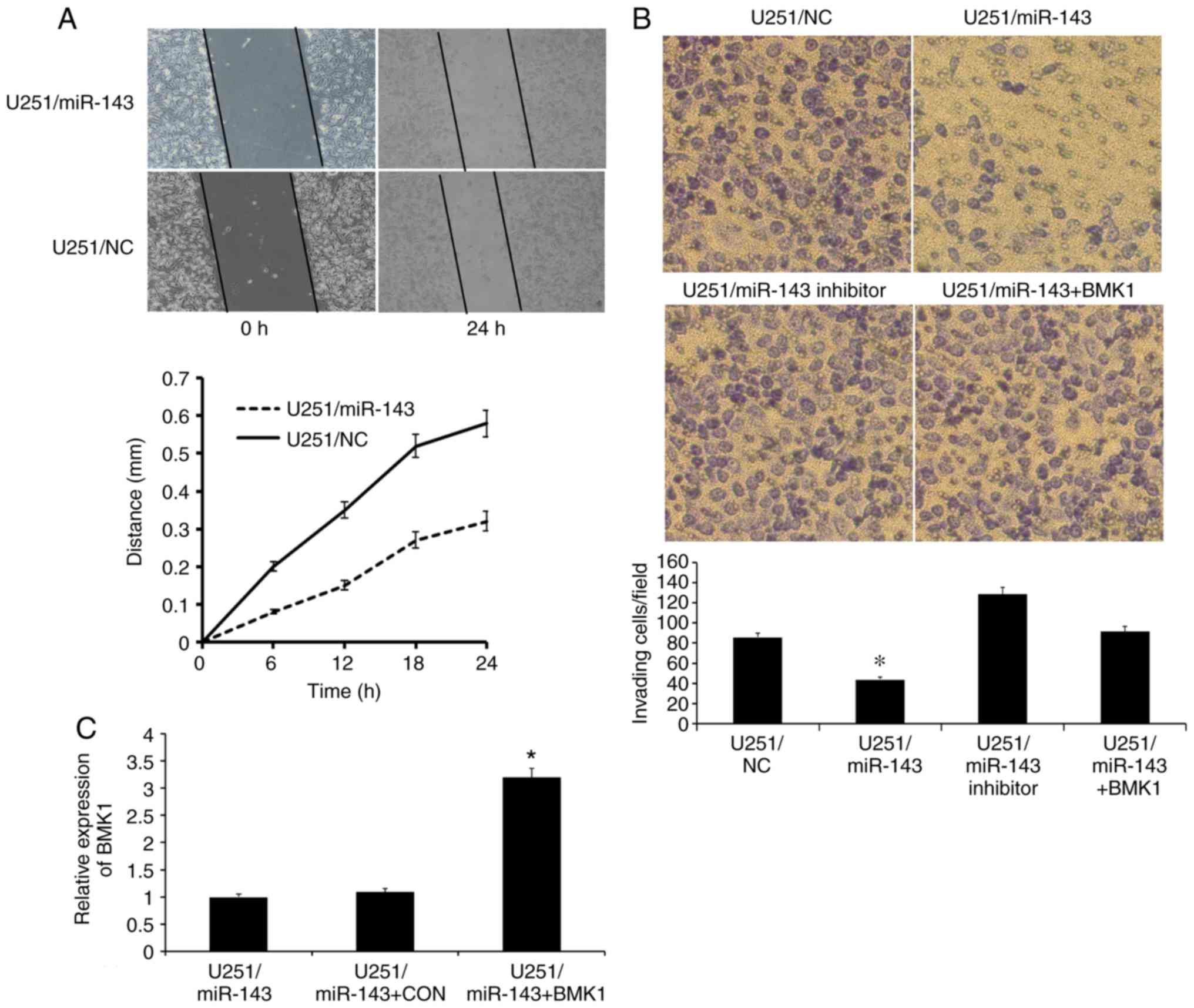
Increased expression of miR-143
inhibits glioma cell invasion and migration
To verify our hypothesis that miR-143 serves an
important role in glioma invasion through BMK1, scratch and
Matrigel invasion assay were performed. The scratch assay is one of
the few cell migration assays, which can be estimated at fixed time
points (35). Several hours after
wounding, it took U251/miR-143 cells a longer time to fill the gap,
further supporting a defect in migration (Fig. 4A). Subsequently, Matrigel assay was
performed and significant reductions of invasion by 48% in
U251/miR-143 compared with the control cells was observed. However,
there was no significant change in the invasion ability of
U251/miR-143 inhibitor compared with U251/NC. For further
confirmation, U251 cells were co-transfected with miR-143 and BMK1.
As shown in Fig. 4B, the invasion
ability of U251/miR-143+BMK1 recovered compared with U251/miR-143
(Fig. 4B). RT-qPCR results confirmed
that the BMK1 expression was indeed increased in the
U251/miR-143+BMK1 cells compared with the U251/miR-143 cells and
U251/miR-143+CON cells (Fig. 4C).
These results suggested that restoration of miR-143 inhibits the
invasion and migration of glioma cells through BMK1.
miR-143 inhibits glioma invasion by
downregulating BMK1 in vivo
To assess the biological functions of miR-143 and
BMK1 in glioma in vivo, U87/NC and U87/miR-143 cells were
implanted stereotactically into the brains of glioma-bearing mice
(n=8). The number of satellite tumors (tumor foci not connected
with the main tumor) was regarded as a semi-quantitative
measurement of tumor invasion (36).
The number of satellite tumors which have migrated away from the
main tumor mass or were projections from the main tumors was
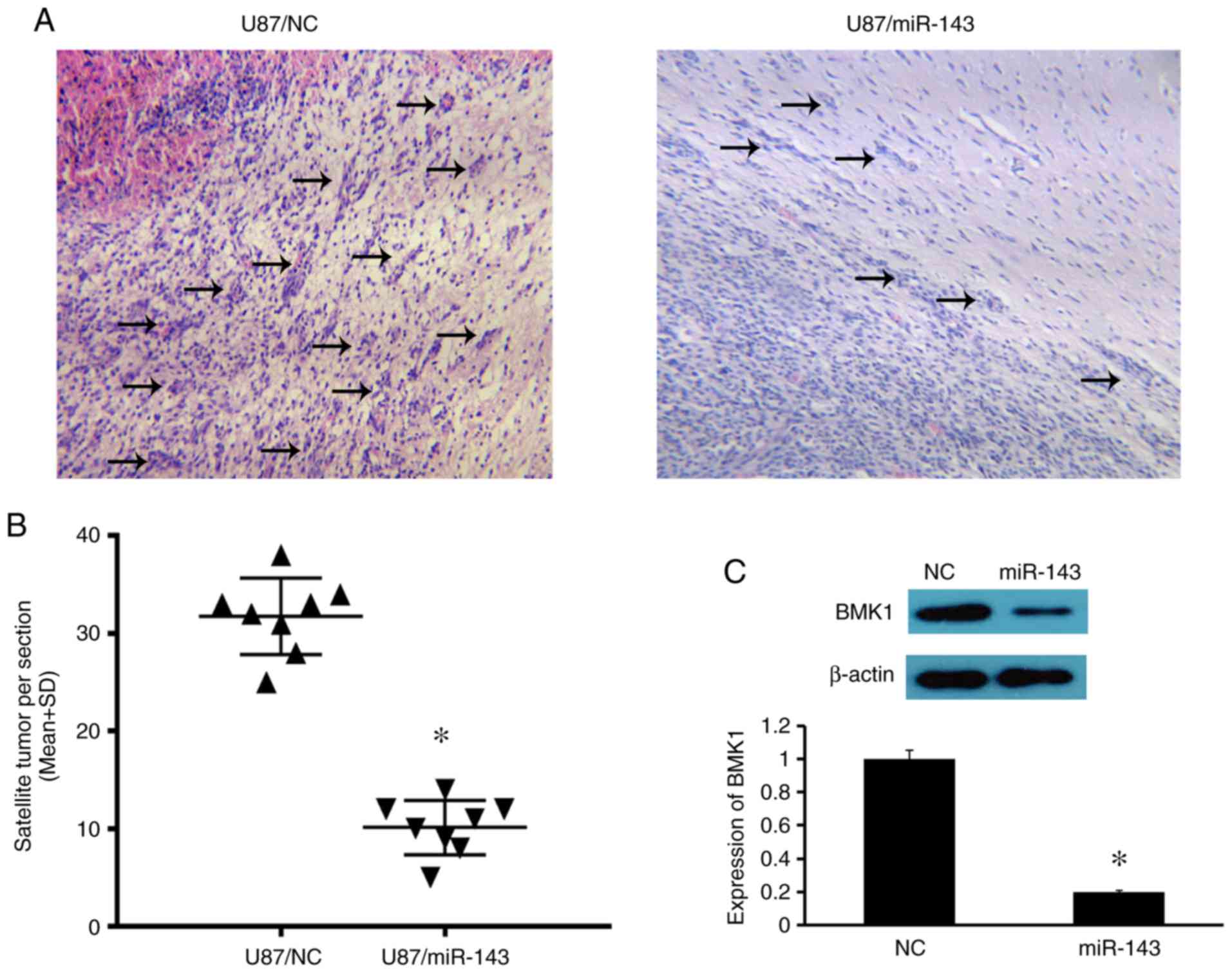
counted 30 days after tumor cell injection. As shown in Fig. 5A and B, the number of satellite tumors
was less in the brains of mice injected with U87/miR-143 cells than
that in the mice injected with U87/NC cells (10.1 vs. 31.7
satellite tumors per section, P<0.05; Fig. 5B). The expression of BMK1 in the tumor
xenograft was also investigated. As expected, BMK1 expression was
lower in the mice injected with U87/miR-143 cells than in the mice
injected with U87/NC cells (Fig. 5C).
Taken together, these results indicate that miR-143 can
significantly inhibit the expression of BMK1 and suppress the
invasion of glioma cells, in vivo.
Downregulation of miR-143 in glioma
cell lines and tissues is due to the DNA methylation
In mammals, DNA methylation serves a crucial role in
the regulation of gene expression and chromatin structure. In many
tumors, decreased miRNA expression is due to the hypermethylation
of the promoter region (37). To
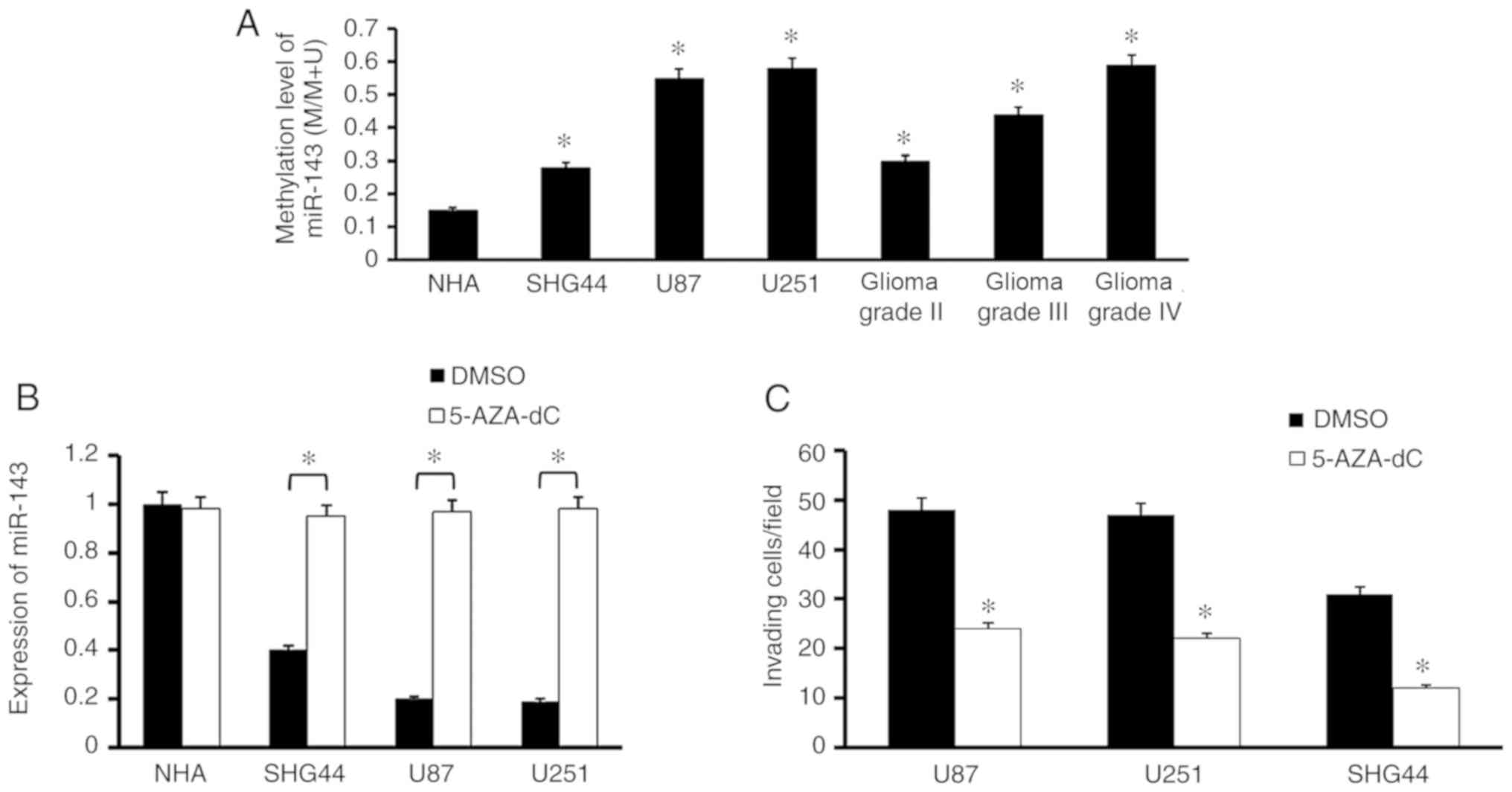
verify this hypothesis, we explored the methylation level of
miR-143 in both glioma cell lines and tissues. The results showed
that the methylation level of miR-143 in genomic DNA obtained from
glioma cell lines and clinical tissue samples was higher than that
from NHA cell line. Notably, it also showed a tendency associated
with WHO grading, strongly demonstrated that promoter methylation
serves an important role in miR-143 downregulation (Fig. 6A).
To further explore the effect of DNA methylation,
the NHA cell line and glioma cell lines were treated with a DNA
methylation inhibitor 5-AZA-dC and found that it promoted the
expression of miR-143 (Fig. 6B).
Additionally, the inhibition of DNA methylation significantly
inhibited the invasive ability of U87, U251 and SHG44 cells
(Fig. 6C). In summary, these findings
strongly suggested that the expression of miR-143 was regulated by
DNA methylation in glioma cells and tissues.
Discussion
Recent reports suggest that miRNA-143 inhibits cell
proliferation, invasion and metastasis by regulating multiple
target genes (16). The present study
provided the first evidence that miR-143 expression was
significantly low in glioma tissues and cell lines. In particular,
it was demonstrated that the expression of miR-143 had a strong
association with the WHO grade and survival rates in patients with
glioma.
In a previous study, BMK1 was highly expressed in
glioma tissues and cell lines. It promoted the invasion and
migration of glioma cells acting as a predictor of poor prognosis
for patients with glioma (5). In the
present study, miR-143 was predicted to act as a novel BMK1
suppressor. By transduction of miR-143 into glioma cells, it was
demonstrated that miR-143 could repress BMK1 expression.
Furthermore, 3′UTR luciferase assay and RIP analysis confirmed that
miR-143 inhibited BMK1 via direct binding to the 3′UTR of BMK1.
Polarized cell migration, which is tightly regulated
during tissue development, chemotaxis and wound healing, is highly
associated with the tumors infiltration and invasion (38). In the present study, the restoration
of miR-143 in the chemotaxis, wound healing and matrigel invasion
assay severely impaired the migration and invasive ability of
glioma cell lines. This result suggested that miR-143 may be an
essential factor in glioma cell migration and invasion. The rescue
assays drew the conclusion that miR-143 inhibited glioma invasion
by downregulating BMK1. Actin polymerization and the subsequent
formation of membrane protrusions are necessary for polarized cell
migration. Actin regulates the motility of cancer cell through
cytoskeletal rearrangement (39).
Previous studies have proven that F-actin dynamics are regulated by
phosphorylating cofilin at Ser3 (34). The present results demonstrated that
miR-143 participated in the IGF-1-induced F-actin polymerization to
mediate cytoskeletal rearrangement, which is vital in glioma cell
migration and invasion. Furthermore, miR-143 is a key factor in the
IGF-1-induced cofilin recycling. Therefore, it may be speculated
that overexpression of miR-143 downregulated the expression of
BMK1, which in turn regulated the phosphorylation of cofilin and
F-actin polymerization.
A previous study showed that miR-143 could inhibit
viability of A172 cells (40). In the
present study, increased miR-143 expression did not cause
significant changes in cell viability. The potential reasons of
this distinction include different glioma cell lines, different
cell density and different observed time. In addition,
downregulation of miR-143 expression did not significantly enhance
the invasion ability of glioma cell lines. The reason may be that
the cell lines used were highly invasive glioblastoma cell lines,
which is hard to further increase the invasion.
Epigenetic modifications have been shown to display
a tight association with carcinogenesis and to serve an important
role in miRNA expression (41,42). The
present data suggested that decreased miR-143 expression is due to
the hypermethylation of the upstream promoter region in glioma cell
lines and tissues. This conclusion was confirmed through treatment
with 5-aza-dC, a DNA methyltransferase inhibitor, which restored
miR-143 expression in glioma cell lines and reduced the invasion of
cancer cells. Based on these findings, the methylation status of
miR-143 may serve as a potential biomarker for the prognosis of
glioma, but this needs to be further verified.
In summary, miR-143 expression levels were lower in
glioma cells and that miR-143 inhibited glioma cell invasion and
migration. The molecular mechanisms of miR-143 and BMK1 mediating
in cells migration and invasion was also investigated. Future
studies could examine the therapeutic potential of miR-143 and
identify additional genome-wide targets of this microRNA.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Scientific Foundation of China (grant nos. 81872163,
81672631, 81072068, 81472365, and 81501185), The Young and
Middle-Aged Scientists Research Awards Foundation of Shandong
Province (grant no. 2010BSB14050), Foundation of Shandong
Educational Committee (grant no. J14LK13), Scientific Foundation of
Shandong Province (grant nos. ZR2017PH079, ZR2014HM003, and
ZR2015HM028), Shandong Provincial Key Research & Development
Project (grant no. 2017GSF218043). Shandong Province outstanding
youth scientist foundation plan (grant no. BS2013YY020).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
BZ conceived and designed the present study. BZ,
WYC, ZQL, CR and PY performed the experiments and analyzed data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Samples were collected following a protocol approved
by the Institutional Review Board, and patients gave their consent
for the research of their tissue specimens in the present study.
The study protocol was reviewed and approved by the Weifang Medical
University Ethics Committee (approval no 99, 11-November-2016). The
study protocol for the xerograph study in mice was reviewed and
approved by Weifang Medical University Ethics Committee (approval
no 129, 11-November-2016).
Patient consent for publication
Written informed consent for publication was
obtained from all patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rousseau A, Mokhtari K and Duyckaerts C:
The 2007 WHO classification of tumors of the central nervous
system-what has changed? Curr Opin Neurol. 21:720–727. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohgaki H and Kleihues P: Genetic pathways
to primary and secondary glioblastoma. Am J Pathol. 170:1445–1453.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Demuth T and Berens ME: Molecular
mechanisms of glioma cell migration and invasion. J Neurooncol.
70:217–228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shukla A, Miller JM, Cason C, Sayan M,
MacPherson MB, Beuschel SL, Hillegass J, Vacek PM, Pass HI and
Mossman BT: Extracellular signal-regulated kinase 5: A potential
therapeutic target for malignant mesotheliomas. Clin Cancer Res.
19:2071–2083. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen W, Zhang B, Guo W, Gao L, Shi L, Li
H, Lu S, Liu Y and Li X: miR-429 inhibits glioma invasion through
BMK1 suppression. J Neurooncol. 125:43–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song C, Wang L, Xu Q, Wang K, Xie D, Yu Z,
Jiang K, Liao L, Yates JR, Lee JD and Yang Q: Targeting BMK1
impairs the drug resistance to combined inhibition of BRAF and
MEK1/2 in melanoma. Sci Rep. 7:462442017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rovida E, Di Maira G, Tusa I, Cannito S,
Paternostro C, Navari N, Vivoli E, Deng X, Gray NS, Esparís-Ogando
A, et al: The mitogen-activated protein kinase ERK5 regulates the
development and growth of hepatocellular carcinoma. Gut.
64:1454–1465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao L, Sun Y, Hou Y, Peng Q, Wang L, Luo
H, Tang X, Zeng Z and Liu M: miRNA expression analysis of
cancer-associated fibroblasts and normal fibroblasts in breast
cancer. Int J Biochem Cell Biol. 44:2051–2059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H,
Li W, Hu B, Cheng SY and Li M: Loss of miR-204 expression enhances
glioma migration and stem cell-like phenotype. Cancer Res.
73:990–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Krek A, Grun D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
16
|
Motoyama K, Inoue H, Takatsuno Y, Tanaka
F, Mimori K, Uetake H, Sugihara K and Mori M: Over- and
under-expressed microRNAs in human colorectal cancer. Int J Oncol.
34:1069–1075. 2009.PubMed/NCBI
|
|
17
|
Bandres E, Cubedo E, Agirre X, Malumbres
R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M and
García-Foncillas J: Identification by Real-time PCR of 13 mature
microRNAs differentially expressed in colorectal cancer and
non-tumoral tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schepeler T, Reinert JT, Ostenfeld MS,
Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ,
Kruhøffer M, Laurberg S, et al: Diagnostic and prognostic microRNAs
in stage II colon cancer. Cancer Res. 68:6416–6424. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Papaconstantinou IG, Manta A, Gazouli M,
Lyberopoulou A, Lykoudis PM, Polymeneas G and Voros D: Expression
of microRNAs in patients with pancreatic cancer and its prognostic
significance. Pancreas. 42:67–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
−145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun L, Zhang B, Liu Y, Shi L, Li H and Lu
S: miR125a-5p acting as a novel Gab2 suppressor inhibits invasion
of glioma. Mol Carcinog. 55:40–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ufkin ML, Peterson S, Yang X, Driscoll H,
Duarte C and Sathyanarayana P: miR-125a regulates cell cycle,
proliferation, and apoptosis by targeting the ErbB pathway in acute
myeloid leukemia. Leuk Res. 38:402–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang L, Lin C, Song L, Wu J, Chen B, Ying
Z, Fang L, Yan X, He M, Li J and Li M: MicroRNA-30e* promotes human
glioma cell invasiveness in an orthotopic xenotransplantation model
by disrupting the NF-κB/IκBα negative feedback loop. J Clin Invest.
122:33–47. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Yin C, Zhang B, Sun Y, Shi L, Liu N,
Liang S, Lu S, Liu Y, Zhang J, et al: PTTG1 promotes migration and
invasion of human non-small cell lung cancer cells and is modulated
by miR-186. Carcinogenesis. 34:2145–2155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Tu Y, Wen J, Yao F, Wei W and Sun S:
Role for ezrin in breast cancer cell chemotaxis to CCL5. Oncol Rep.
24:965–971. 2010.PubMed/NCBI
|
|
27
|
Wang LH, Xiang J, Yan M, Zhang Y, Zhao Y,
Yue CF, Xu J, Zheng FM, Chen JN, Kang Z, et al: The mitotic kinase
Aurora-A induces mammary cell migration and breast cancer
metastasis by activating the Cofilin-F-actin pathway. Cancer Res.
70:9118–9128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hall EH, Gurel V, Dahlberg AE, McMichael J
and Brautigan DL: Inhibition of human breast cancer Matrigel
invasion by Streptolysin O activation of the EGF receptor ErbB1.
Cell Signal. 23:1972–1977. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan LP, Seinen E, Duns G, de Jong D, Sibon
OC, Poppema S, Kroesen BJ, Kok K and van den Berg A: A high
throughput experimental approach to identify miRNA targets in human
cells. Nucleic Acids Res. 37:e1372009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng GZ, Zhang W and Wang LH: Regulation
of cancer cell survival, migration, and invasion by Twist: AKT2
comes to interplay. Cancer Res. 68:957–960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi L, Sun X, Zhang J, Zhao C, Li H, Liu
Z, Fang C, Wang X, Zhao C, Zhang X, et al: Gab2 expression in
glioma and its implications for tumor invasion. Acta Oncol.
52:1739–1750. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Villa C, Miquel C, Mosses D, Bernier M and
Di Stefano AL: The 2016 World Health Organization classification of
tumours of the central nervous system. Presse Med. 47:e187–e200.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He Y, Li D, Cook SL, Yoon MS, Kapoor A,
Rao CV, Kenis PJ, Chen J and Wang F: Mammalian target of rapamycin
and Rictor control neutrophil chemotaxis by regulating Rac/Cdc42
activity and the actin cytoskeleton. Mol Biol Cell. 24:3369–3380.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ghosh M, Song X, Mouneimne G, Sidani M,
Lawrence DS and Condeelis JS: Cofilin promotes actin polymerization
and defines the direction of cell motility. Science. 304:743–746.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Etienne-Manneville S and Hall A:
Integrin-mediated activation of Cdc42 controls cell polarity in
migrating astrocytes through PKCzeta. Cell. 106:489–498. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tran TT, Uhl M, Ma JY, Janssen L, Sriram
V, Aulwurm S, Kerr I, Lam A, Webb HK, Kapoun AM, et al: Inhibiting
TGF-beta signaling restores immune surveillance in the SMA-560
glioma model. Neuro Oncol. 9:259–270. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yim RL, Wong KY, Kwong YL, Loong F, Leung
CY, Chu R, Lam WW, Hui PK, Lai R and Chim CS: Methylation of
miR-155-3p in mantle cell lymphoma and other non-Hodgkin's
lymphomas. Oncotarget. 5:9770–9782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Noritake J, Watanabe T, Sato K, Wang S and
Kaibuchi K: IQGAP1: A key regulator of adhesion and migration. J
Cell Sci. 118:2085–2092. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang B, Gu F, She C, Guo H, Li W, Niu R,
Fu L, Zhang N and Ma Y: Reduction of Akt2 inhibits migration and
invasion of glioma cells. Int J Cancer. 125:585–595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu TG, Wang L, Li W, Li JZ and Li J:
miR-143 inhibits oncogenic traits by degrading NUAK2 in
glioblastoma. Int J Mol Med. 37:1627–1635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wynter CV: The dialectics of cancer: A
theory of the initiation and development of cancer through errors
in RNAi. Med Hypotheses. 66:612–635. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ren X, McHale CM, Skibola CF, Smith AH,
Smith MT and Zhang L: An emerging role for epigenetic dysregulation
in arsenic toxicity and carcinogenesis. Environ Health Perspect.
119:11–19. 2011. View Article : Google Scholar : PubMed/NCBI
|