Introduction
Lung cancer, which remains the leading cause of
cancer-related mortality worldwide (1), is divided into small cell lung cancer
(SCLC) and non-SCLC (NSCLC). NSCLC accounts for 80–85% of all lung
cancer cases (2,3). Although the emergence of novel
therapeutic methods has significantly improved the treatment of
NSCLC, the prognosis remains poor, with a 5-year survival rate of
only 19.3% (4). Thus, there is an
urgent need for new therapies.
Autophagy is a highly conserved process in which
cellular components are captured and delivered to double-membrane
vesicles called autophagosomes, which are subsequently degraded by
the lysosomal system (5).
Autophagy plays an influential role in tumor development (6). It has been reported that the deletion
of Atg7 within tumor cells induces the inhibition of intracellular
autophagy, which has been shown in multiple models to impair their
growth (7). Chemical or genetic
autophagy inhibition delivered systemically blunted tumor growth
and invasion (8). Moreover,
strategies to inhibit autophagy to enhance reactive oxygen
species-induced oxidative damage for synergistic cancer therapy, as
well as new autophagy inhibitors, are emerging in clinical trials
for antitumor therapy (9,10). Unfortunately, almost all autophagy
inhibitors are highly toxic, which limits clinical application
(11–13). Therefore, it is particularly urgent
to develop new autophagy inhibitors with a low or no toxicity.
Pigment epithelial-derived factor (PEDF) is a member of the serine
protease superfamily and can regulate proteolytic cascades related
to key biological processes, such as blood coagulation,
inflammation and angiogenesis (14,15).
Studies have shown that PEDF is an effective tumor angiogenesis
inhibitor, which could inhibit cancer cell invasion and metastasis
to prevent cancer progression (16–18).
Zhang et al (19)
demonstrated that PEDF expression was reduced in NSCLC and was
correlated with clinical outcomes. Chen et al (20) suggested that the molecular impact
of PEDF on lung cancer cells and its clinical implications are
significant. PEDF is highly expressed in multiple tissues and is
essential for maintaining homeostasis; it is also a key regulator
of autophagy and energy metabolism (21,22).
Previous studies by the authors revealed that PEDF was involved in
the regulation of mitophagy levels and carbohydrate uptake and
metabolism in ischemic cardiomyocytes (23–25).
However, the effect of PEDF on intracellular autophagy in NSCLC
remains unclear.
Therefore, the present study evaluated the effect of
PEDF on autophagy status and the related mechanism in lung cancer
cells. It was demonstrated that PEDF significantly inhibited lung
cancer cell proliferation and viability by suppressing autophagy
through the downregulation of the adenosine
5′-monophosphate-activated protein kinase (AMPK)/Unc-51 like
autophagy-activated kinase 1 (ULK1) signaling pathway in NSCLC
cells.
Materials and methods
Materials
Anti-protein kinase C α (PKCα; cat. no. 2056),
anti-ULK1 (cat. no. 8054), anti-microtubule-associated protein
light chain 3-I (LC3-I; cat. no. 4108), anti-LC3-II (cat. no.
2775), anti-AMPKα (cat. no. 5832), anti-phosphorylated (p)-AMPKα
(Thr172, cat. no. 2535) and anti-p-ULK1 (cat. no. 5869) antibodies
were purchased from Cell Signaling Technology, Inc. Anti-p-PKCα
(cat. no. 07-790) antibody was obtained from MilliporeSigma.
Anti-β-actin (cat. no. 66009-1-lg) antibody was purchased from
ProteinTech Group, Inc. Bafilomycin A1 (BAF1; cat. no. S1413) was
purchased from Selleck Chemicals. Rapamycin (cat. no. HY-10219) was
purchased from MedChemExpress.
Cell culture and reagents
H460 and A549 human NSCLC cell lines were donated by
Dr Jingjun Han (Eighth Affiliated Hospital of Sun Yat-sen
University, Shenzhen, China) and cell line H1299 (cat. no.
SCSP-589) was purchased from the Cell Bank of the Chinese Academy
of Sciences (http://www.cellbank.org.cn), HBE135-E6E7 (referred to
as HBE hereafter; cat. no. CRL-2741) and hTERT lung fibroblasts
(referred to as fibroblasts hereafter; cat. no. CRL-4058) were
purchased from ATCC. All the cells were cultured in RPMI-1640
medium with 10% FBS (Cytiva), 100 µg/ml penicillin and 0.1 mg/ml
streptomycin. All cells were maintained in a cell culture incubator
at 37°C in a humidified atmosphere with 5% CO2. All
experiments were conducted in the exponential phase of the
cells.
Cell viability and (lactate
dehydrogenase) LDH release assay
Cell viability was assessed using Cell Counting
Kit-8 (CCK-8) (cat. no. C0038; Beyotime Institute of Biotechnology)
assay, according to the manufacturer's instructions. Briefly, NSCLC
cell lines, H1299, A549 and H460 were seeded in 96-well plates at a
density of 5×103 cells/well for 24 h. The mixture was
then treated with CCK-8 reagent and incubated at 37°C for an
additional 0.5-3 h. Cell viability was determined by measuring the
absorbance at 450 nm using a microplate reader. Each experiment was
repeated three times. LDH activity in cell supernatants was
detected with LDH Cytotoxicity Assay Kit (cat. no. 4744926001;
Roche Diagnostics), according to the manufacturer's
instructions.
Western blotting (WB)
Collected cells were mixed with lysis buffer (500
µl; Shanghai Aladdin Biochemical Technology Co., Ltd.) and placed
on ice to lyse for 25 min, followed by centrifugation at 12,000 × g
for 15 min at 4°C. A BCA protein Concentration Determination Kit
(cat. no. P0012; Beyotime Institute of Biotechnology) was used to
determine protein concentration in the supernatant. After mixing
the protein sample (5 µl) with 5X sodium dodecyl sulfate loading
buffer, the mixture was denaturated by boiling in a water bath for
10 min. The samples (20 µg per lane) were then electrophoresed on a
10% SDS-PAGE (100 V) and transferred to a PVDF membrane on ice (250
mA, 60 min). The PVDF membrane was then sealed with 50 g/l skimmed
milk at room temperature for 90 min. Subsequently, PVDF membranes
were incubated with primary antibodies overnight at 4°C. The
primary antibodies were as follows: Rabbit anti-human ULK1 [cat.
no. 8054; 1:1,000; Cell Signaling Technology, Inc. (CST)], LC3-I
(cat. no. 4108; 1:500; CST), LC3-II (cat. no. 2775; 1:500; CST),
AMPKα (cat. no. 5832; 1:1,000; CST), p-AMPKα (cat. no. 2535; 1:500;
CST), PI3K (cat. no. 4249; 1:1,000; CST), MAPK (cat. no. 4695;
1:1,000; CST), PEDF (cat. no. DF6547; 1:1,000; Affinity
Biosciences), extracellular signal-regulated protein kinase (ERK;
cat. no. 4348; 1:1,000; CST), p-ERK (cat. no. 8544; 1:1,000; CST),
p38 (cat. no. 14451; 1:1,000; CST), p-p38 (cat. no. 4511; 1:1,000;
CST), mTOR (cat. no. ab2732; 1:1,000; Abcam), TSC (cat. no.
ab200728; 1:1,000; Abcam), and p62 (cat. no. ab109012; 1:500;
Abcam) primary antibodies as well as mouse anti-human GAPDH (cat.
no. ab8245; 1:500; Abcam) and β-actin (cat. no. ab5694; 1:1,000;
Abcam) primary antibodies. The membrane was then thoroughly washed
three times with PBST (including 0.1% v/v Tween-20) for 5 min each
time. Next, PVDF membranes were incubated with HRP-conjugated
secondary antibodies (anti-rabbit ab205718 and anti-mouse ab205719;
1:4,000; Abcam) at room temperature for 60 min. Subsequently, the
membranes were washed multiple times with PBST and then developed
using an enhanced chemiluminescence detection kit (Sigma-Aldrich;
Merck KGaA) for imaging. Image Lab V3.0 software (Bio-Rad
Laboratories, Inc.) was used to obtain and analyze imaging data.
The relative expression of the target protein was expressed as the
ratio of GAPDH or β-actin.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from NSCLC cells using
TRIzol® (Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions (26). Collected NSCLC cells were lysed by
1 ml of TRIzol (Thermo Fisher Scientific, Inc.). Following lysis,
total RNA was extracted using the phenol-chloroform method
(27). The purity of RNA was
determined by UV A260/A280 spectrophotometry (Nanodrop ND2000;
Thermo Fisher Scientific, Inc.). cDNA was then obtained by reverse
transcription from 1 µg RNA using miScript II RT kit (Qiagen GmbH)
and stored at −20°C. RT-qPCR was performed using SYBR Green PCR
kit. The reaction system consisted of 10 µl RT-qPCR-mix, 0.5 µl
forward primer (human PEDF forward, 5′-ATTCCCGATGAGATCAGCA-3′; and
human GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′), 0.5 µl reverse
primer (human PEDF reverse, 5′-CTTAGGGTCCGACATCATGG-3′; and human
GAPDH reverse, 5′GCCCAATACGACCAAATCC-3′), 2 µl cDNA and 7 µl double
distilled water (ddH2O). The reaction protocol was as
follows: Initial denaturation at 95°C for 10 min, and 40 cycles at
95°C for 1 min and 60°C for 30 sec. Analysis of relative gene
expression data by RT-qPCR was 2−ΔΔCq method (28).
PEDF lentiviral vector construction
and cell transfection assays
Recombinant lentivirus was prepared as previously
described (29). Briefly,
lentiviral plasmids were transfected into 293SF cells (cat. no.
CRL3249; ATCC) with PEI as the transfection reagent. Plasmids were
constructed and purified by chromatography using maxiprep plasmid
purification kit (Qiagen GmbH). Lentivirus (LVs) was purified by
PEG6000. The assay was performed following the manufacturer's
instructions (30). Various
concentration steps eventually resulted in a titer of 1,010 IU/ml.
Cells and LVs were co-cultured to construct stable cell lines with
a multiplicity of infection of 10:1.
Autophagy monitoring assay
A tandem GFP-red fluorescent protein (RFP)-LC3
adenovirus construct obtained from Hanbio Biotechnology Co., Ltd.
was used in this study. This tandem GFP-RFP-LC3 construct utilizes
the pH difference between acidic and neutral autophagosomes, and
the difference in pH sensitivity exhibited by GFP and RFP to
monitor the progression from autophagosomes to autolysosomes. In
brief, for image-based autophagy analysis, NSCLC cells were
infected with tandem GFP-RFP-LC3 adenovirus for 2 h and then were
cultured with normal medium for 24 h, and then the cells were
treated and imaged for GFP and RFP using fluorescence microscopy.
The cells were then observed using a fluorescence microscope
(Olympus Corporation) or confocal laser scanning microscope
(Olympus Corporation). Image-Pro Plus (Media Cybernetics, Inc.)
analyzed the co-localization rates and intensity of
LC3/Mito-tracker Red (cat. no. M7512; Invitrogen™; Thermo Fisher
Scientific, Inc.).
Statistical analysis
Data are presented as the mean ± SEM. Data were
measured using a two-tailed unpaired Student's t-test for
comparison between two groups and one-way ANOVA for multiple
comparisons, followed by a Student-Newman-Keuls test. Statistical
analysis was performed using PASW Statistics 21 (IBM Corp.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
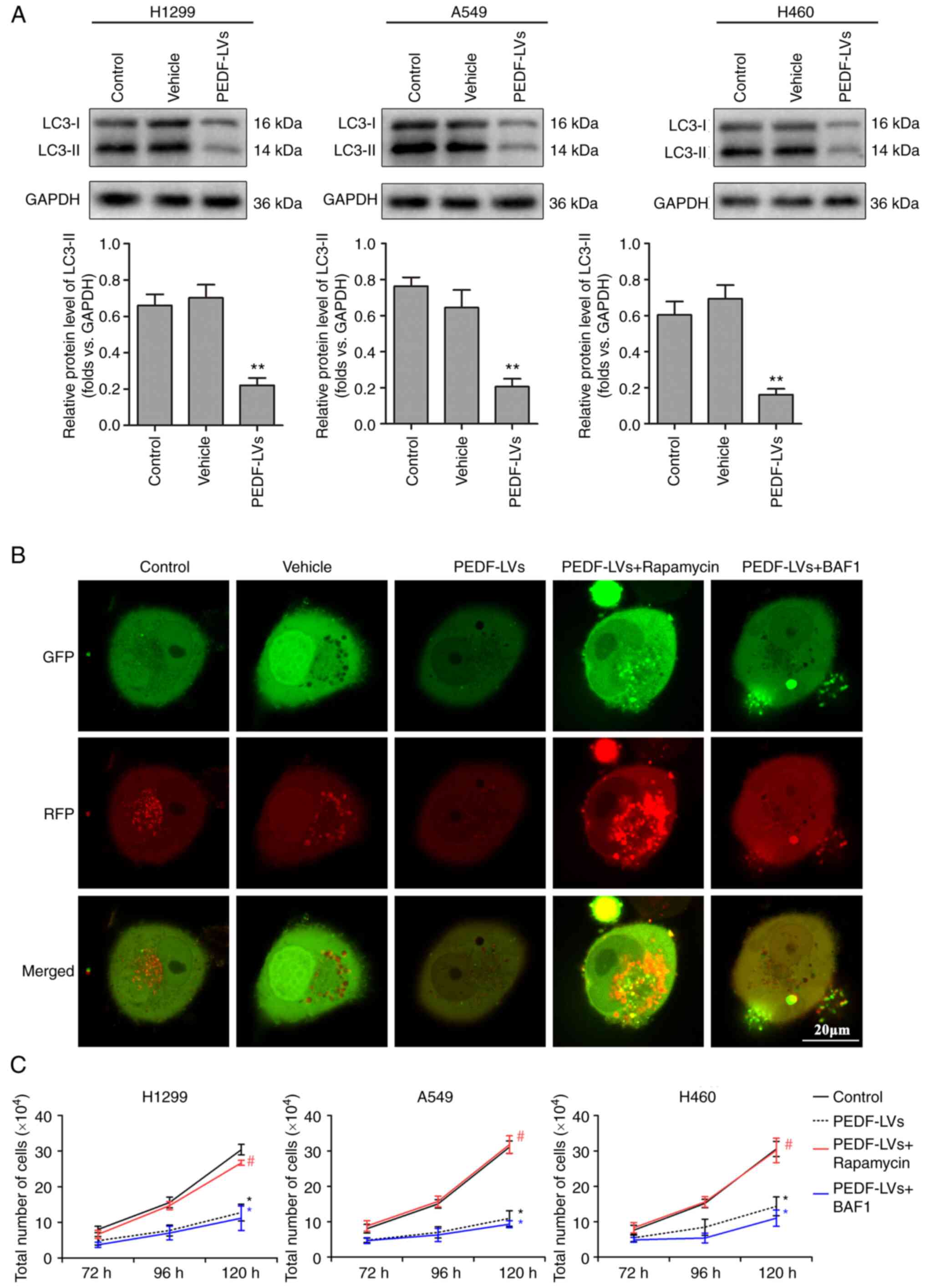
PEDF inhibits NSCLC proliferation
To detect the biological function of PEDF on the
cellular level in lung cancer, three NSCLC cells (H1229, A549 and
H460) and two normal tissue cells (HBE and fibroblasts) were
cultured under the same conditions. The expression of PEDF was then
detected by WB, and it was found that, compared with normal cells,
the expression and mRNA levels of PEDF were significantly decreased
in all three NSCLC cell lines (Fig. 1A
and B). To confirm this, PEDF overexpression was induced in
H1299, A549 and H460 cells and a comparative analysis of the
control and vehicle groups was performed. The results revealed
increased expression of PEDF protein (Fig. 1C) and mRNA (Fig. 1D) via WB and quantitative PCR in
H1229, A549 and H460 cell lines transfected with
PEDF-overexpressing lentiviral vectors. In addition, it was
determined that PEDF overexpression could significantly reduce the
proliferation of NSCLC cells compared with normal cells (Fig. 1E).
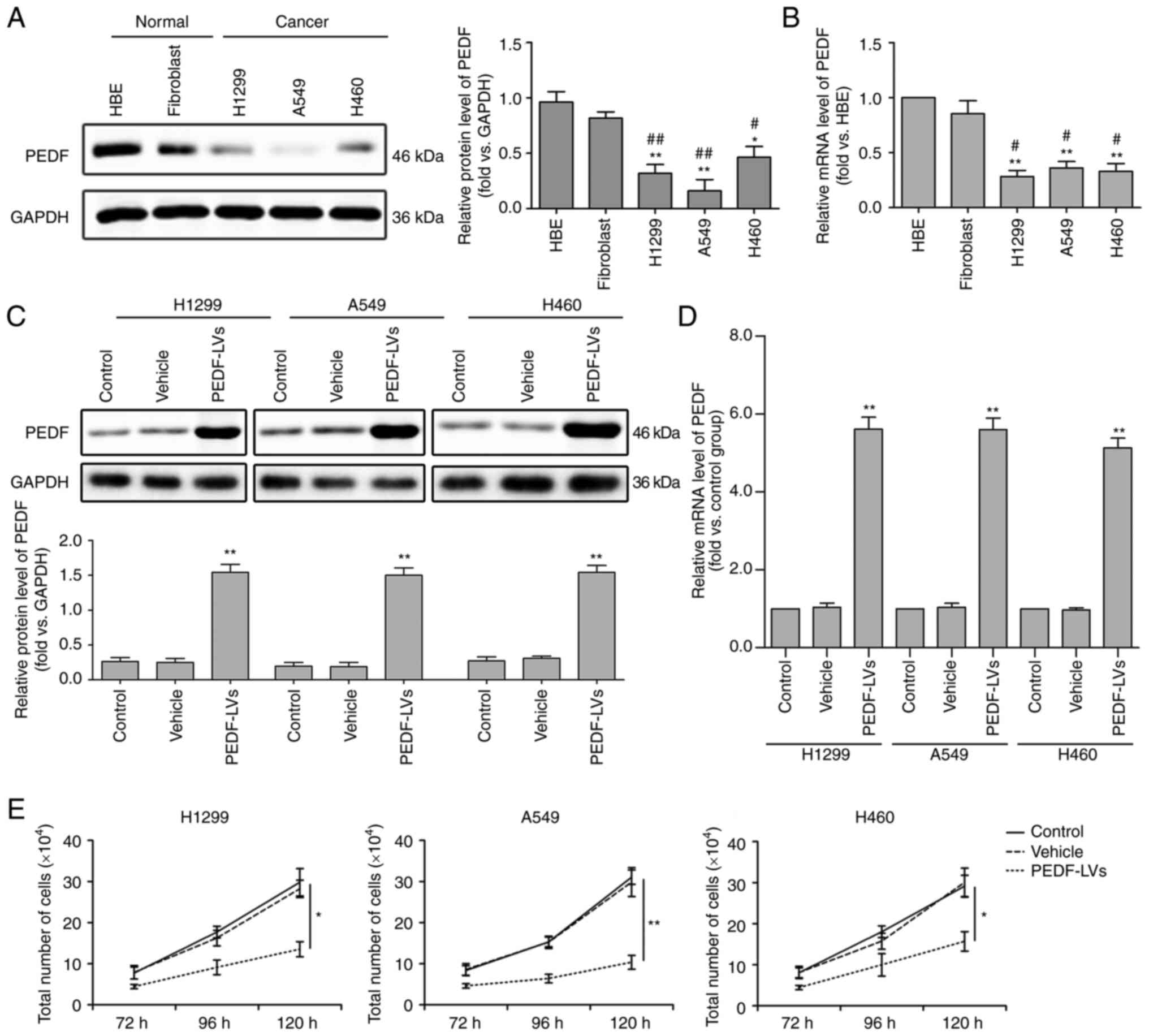 | Figure 1.Expression of PEDF in normal and
cancer tissues and the effect of PEDF expression on cell
proliferation. Three types of NSCLC cell lines H460, A549 and H1299
were assessed, and HBE and fibroblast cell lines were used as
normal controls. (A and B) WB was used to determine the expression
of PEDF in NSCLC cells. Detection of the mRNA expression of PEDF
using RT-qPCR. *P<0.05 and **P<0.01 vs. the HBE group; and
#P<0.05 and ##P<0.01 vs. the fibroblast
group. (C and D) Decreased expression of the protein and mRNA of
PEDF was identified by WB and RT-qPCR in H460, A549 and H1299 cell
lines transfected with PEDF-overexpressing lentiviral vectors. (E)
Following overexpression of PEDF, the number of NSCLC cells was
detected at various time-points. *P<0.05 and **P<0.01 vs. the
control group. Data are presented as the mean ± standard error of
the mean. PEDF, pigment epithelium-derived factor; NSCLC, non-small
cell lung cancer; HBE, human bronchial epithelial; WB, western
blotting; RT-qPCR, reverse transcription-quantitative PCR; LVs,
lentivirus. |
In addition, the expression of ERK, p-ERK, p38 and
p-p38 was examined. The results revealed that the expression of
p-ERK and p-p38 was decreased, which indicated that the
proliferation of NSCLC cells was inhibited (Fig. 2A). Similarly, the results of the
CCK-8 and LDH assays showed that PEDF overexpression resulted in a
distinct decrease in cell viability and a notable increase in
cytotoxicity (Fig. 2B and C). The
confocal microscopy results showed that PEDF overexpression clearly
increased the aggregation of cancer cells (Fig. 2D). Collectively, it was
demonstrated that PEDF expression inhibited the proliferation of
NSCLC cells.
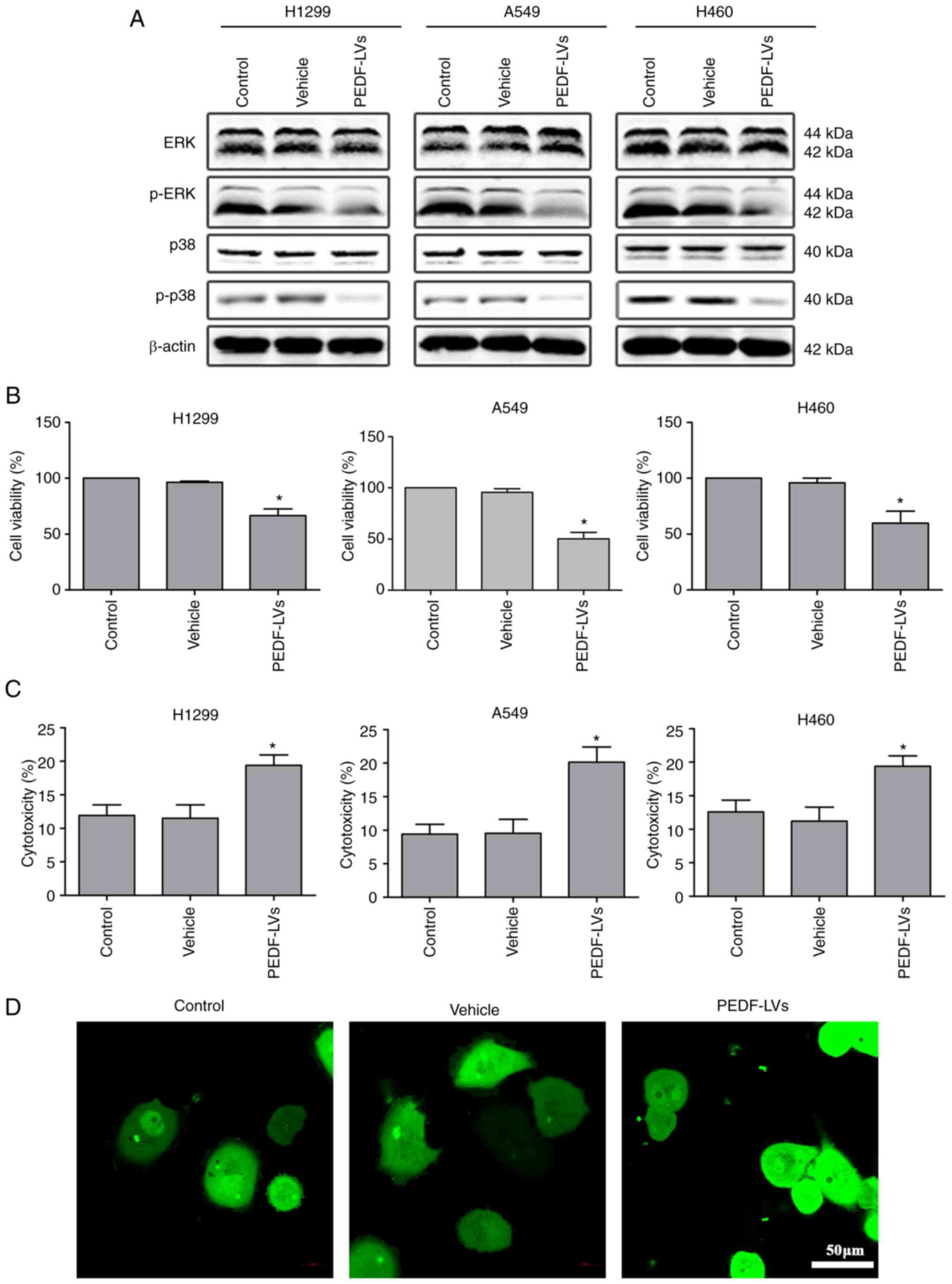 | Figure 2.Effects of PEDF overexpression on
viability and cytotoxicity of NSCLC cells. (A) Western blotting was
used to detect the expression of cyclins (ERK, p-ERK, p38, p-p38)
in NSCLC cells, assessing cell proliferation compared with the
control and vehicle group. (B) A CCK-8 assay was used to assess the
inhibitory effects of PEDF-LVs on the viability of NSCLC cells. (C)
An LDH assay was used to assess the cytotoxicity of PEDF in NSCLC
cells. (D) A549 cells were analyzed for cell-cell adhesion by
confocal microscopy (green, cytoplasm); scale bar, 50 µm.
*P<0.05 vs. the control group. Data are presented as the mean ±
standard error of the mean. PEDF, pigment epithelium-derived
factor; NSCLC, non-small cell lung cancer; ERK, extracellular
signal-regulated kinase; p-, phosphorylated; CCK-8, Cell Counting
Kit-8; LDH, lactate dehydrogenase; LVs, lentivirus. |
PEDF reduces NSCLC proliferative
activity by negatively regulating autophagy
Abnormal autophagy is closely associated with the
presence of tumors, neurodegenerative diseases and metabolic and
immune diseases (31). LC3-I is
activated by APG7L/ATG7, translocates with ATG3 and is coupled with
fatty acyl ethanolamine (PE) to form the membrane-bound form
LC3-II, which can attach to the membrane of autophagosomes and is
the structural protein of autophagosomes (32). LC3-II is often considered a marker
of autophagosomes (33). In the
previous experiment, it was demonstrated that PEDF overexpression
markedly inhibited the proliferation of NSCLC cells, which was
hypothesized may be influenced by autophagy. The expression of LC3
was therefore examined. The results revealed that PEDF
overexpression downregulated the autophagy marker LC3 compared with
the vehicle group, indicating that PEDF negatively regulates
autophagy in NSCLC cells (Fig.
3A). Mitochondrial autophagy was detected using a tandem
GFP-RFP-LC3 adenovirus plasmid, which represents autophagosome
formation. When autophagosomes fuse with lysosomes to form
autolysosomes, GFP molecules are degraded from the tandem proteins,
but RFP-LC3 remains punctate (34). As shown in Fig. 3B, cells with PEDF overexpression
transfected with the GFP-RFP-LC3 plasmid exhibited low LC3
expression. However, autophagy activator rapamycin reversed this
outcome, whereas autophagy inhibitor Myb-like DNA-binding protein
BAS1 had no impact on the inhibition of autophagy caused by PEDF
overexpression (Fig. 3B). These
results indicated that PEDF overexpression affected the expression
of autophagic proteins and decreased the formation of phagosomes.
Furthermore, it was explored whether the inhibition of NSCLC
proliferation by PEDF is related to its negative regulation of
autophagy. Following PEDF overexpression, NSCLC cells were treated
with rapamycin or BAF1 in culture for 120 h, respectively. The
autophagy activator rapamycin reversed the inhibitory effect of
PEDF on the proliferative activity of NSCLC cells, whereas the
autophagy inhibitor BAF1 had no effect (Fig. 3C). The aforementioned results
indicated that PEDF inhibits the proliferative activity of NSCLC
cells by reducing autophagy.
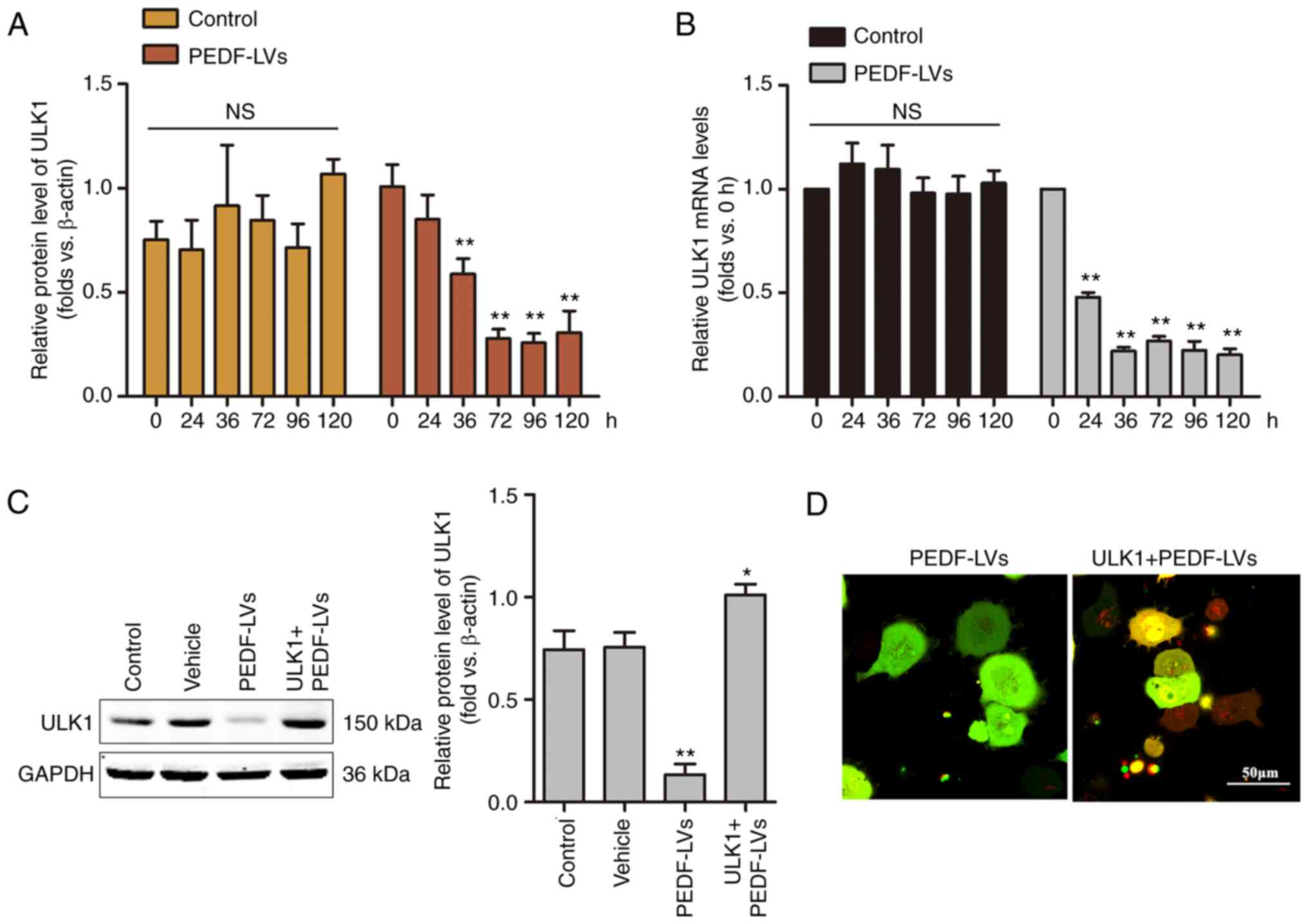
ULK1 is critical for PEDF-induced
autophagy in NSCLC
Among the molecular mechanisms of macroautophagy,
ULK1, which is homologous to the yeast autophagy gene Atg1, is a
key autophagy-initiating kinase that integrates cellular nutrients
and energy and regulates the induction of autophagy (35). To investigate the role of ULK1 on
PEDF-induced autophagy, a time-course analysis of the mRNA and
protein expression of ULK1 was performed using RT-qPCR and WB,
respectively. As demonstrated in Fig.
4A and B, the mRNA and protein levels of ULK1 were decreased
within 36 h and remained low to 120 h. The results demonstrated
that the overexpression of PEDF induced a significant decrease in
the protein expression of ULK1 in NSCLC cells, suggesting that PEDF
plays a key role in regulating autophagy through the involvement
ULK1 signaling. To verify the hypothesis that ULK1 is involved in
mediating PEDF-induced autophagy in NSCLC cells, a
ULK1-overexpressing lentivirus was constructed, and the effect was
confirmed by WB (Fig. 4C). In the
present study, following the overexpression of ULK1 and
co-expression of ULK1 with PEDF in NSCLC cells, confocal microscopy
results revealed a punctiform distribution of autophagosomes in
cells overexpressing PEDF, while autophagosomes in cells
co-expressing PEDF + ULK1 showed a diffuse distribution with a
noticeable increase in red spots (Fig.
4D). The results demonstrated that PEDF overexpression
suppressed ULK1 expression in NSCLC cells, indicating that PEDF
plays a key role in regulating autophagy through the involvement of
ULK1 signaling.
Effects of PEDF on the expression of
genes downstream of the AMPK pathway in NSCLC cells
The ULK1-mediated upstream regulation of autophagy
consists of three main signaling pathways, namely the AMPK,
PI3K/Akt and MAPK/ERK1/2 signaling pathways (36–38).
AMPK is a known upstream regulator of ULK1 that phosphorylates and
activates ULK1 at multiple sites in a cross-talk manner (37). The results of WB revealed that the
overexpression of PEDF decreased the protein expression of AMPK
compared with the empty vector control or DMSO control group
(Fig. 5A and B). However, it had
no effect on the protein expression of PI3K and MAPK. The effect of
PEDF on PI3K and MAPK is dual, where PEDF inhibits them under
hypoxia but has no effect under normoxia (39–42).
Therefore, it was hypothesized that PEDF inhibits autophagy by
downregulating the AMPK signaling pathway. Subsequently, the
association between PEDF and the autophagy-associated AMPK/ULK1
signaling pathway was investigated. The data obtained using WB
revealed that PEDF reduced ULK1 expression through inhibiting AMPK,
and AMPK overexpression reversed the effects of PEDF (Fig. 5C and D). The expression of mTOR,
which is a negative regulator of ULK1, was also assessed. AMPK was
able to inhibit mTOR directly and indirectly by activating TSC
(43). As shown in Fig. 5E, PEDF markedly supressed TSC
expression and increased mTOR expression, while overexpression of
AMPK reversed these effects. In addition, PEDF overexpression
significantly decreased p62 and LC3-II expression, but AMPK
overexpression markedly increased their expression (Fig. 5C and D). Furthermore, PEDF
significantly inhibited NSCLC cell proliferation and viability,
while AMPK overexpression reversed this effect (Fig. 5F and G). These results indicated
that PEDF regulates the proliferative activity and autophagy of
NSCLC cells through the AMPK/ULK1 pathway.
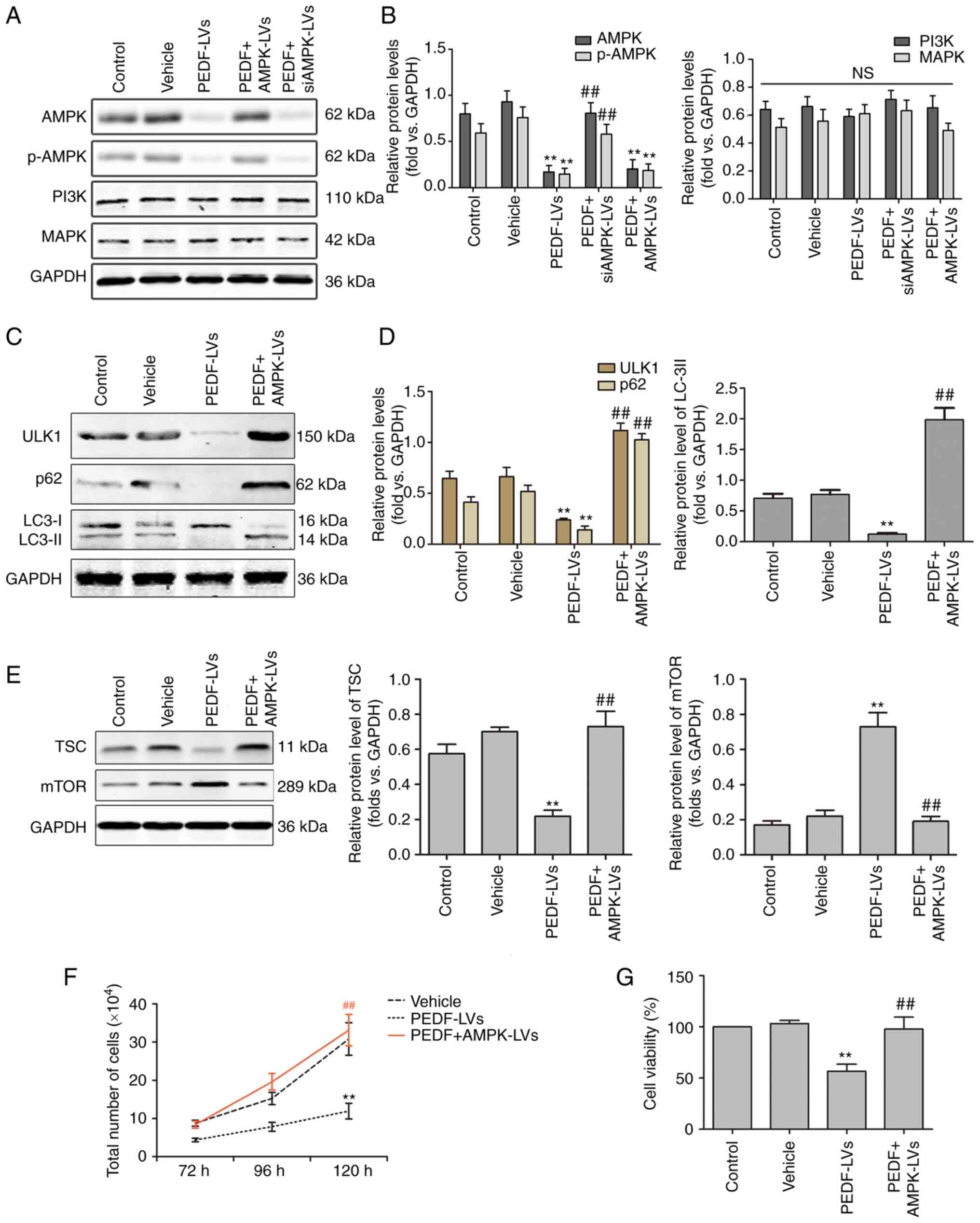 | Figure 5.PEDF regulates autophagy via the
AMPK/ULK1 signaling pathway in NSCLC cells. (A and B) Western
blotting was used to determine the expression of AMPK, p-AMPK,
PI3K, and MAPK. PEDF downregulates total and p-AMPK, which could be
reversed by AMPK-overexpressing lentiviral delivery to cells; n=4.
(C and D) Protein levels of ULK1, p62, LC3-I, and LC3-II in NSCLC
cells treated with PEDF-LVs with or without AMPK overexpression
lentivirus; n=4. (E) Protein levels of TSC and mTOR treated with
PEDF-LVs with or without AMPK overexpression lentivirus; n=4. (F)
The number of cells cultured for 72–120 h with PEDF-LVs with or
without AMPK-LVs intervention was assessed; n=4. (G) Cell viability
assay; n=3. **P<0.01 vs. the control group; and
##P<0.01 vs. the PEDF-LVs group. PEDF, pigment
epithelium-derived factor; AMPK, adenosine
5′-monophosphate-activated protein kinase; ULK1, Unc-51 like
autophagy-activated kinase 1; NSCLC, non-small cell lung cancer;
p-, phosphorylated; PKCα, anti-protein kinase C α; LC3,
microtubule-associated protein light chain 3; LV, lentivirus. |
Discussion
In the present study, it was found that PEDF
inhibited autophagy in lung cancer cell lines by reducing the
expression and activation of AMPK. Of note, in a previous study
(23), it was found that PEDF also
inhibited AMPK levels in hypoxic cardiomyocytes, whereas autophagy
was markedly increased, which was inconsistent with the findings of
the present study obtained from lung cancer cell lines. In another
study, the mechanism through which PEDF can induce a significant
increase and activation of PKCα in hypoxic cardiomyocytes, thereby
displacing AMPK to activate the ULK1 signaling pathway and inducing
higher levels of autophagy, was elucidated (23). By contrast, PEDF had no effect on
the expression of PKCα in lung cancer cell lines (data not shown).
This may be the reason why PEDF inhibits autophagy rather than
activates it in lung cancer cell lines. It was then demonstrated
that PEDF exerts an inhibitory function by suppressing
intracellular autophagy in NSCLC cells. Finally, the signaling
pathways involved in autophagy were explored, and it was found that
PEDF reduces the occurrence of autophagy by blocking the AMPK/ULK1
signaling pathway.
According to lung cancer statistics, NSCLC accounts
for ~85% of reported lung cancer cases, and nearly 80% of patients
with NSCLC are diagnosed at an advanced stage (2). Patients with advanced NSCLC often
lose the opportunity of surgery due to extensive metastasis.
Radiotherapy and chemotherapy can easily induce drug resistance in
tumor cells, resulting in a very poor prognosis for patients. In
recent years, the role of angiogenesis in tumor development, growth
and metastasis has been a research hotspot. Of note, PEDF molecules
have been revealed to inhibit various malignant phenotypes of NSCLC
and have emerged as potential tumor therapeutic targets. In a study
by Zhang et al, PEDF was reduced at both the protein and
mRNA levels in NSCLC tumors compared with normal lung tissue. This
decrease was associated with an increase in microvessel density in
tumors. The increased microvessel density in tumors was associated
with a significant correlation between TNM stage, tumor size and
overall survival. This suggests that PEDF is an important factor in
the development of NSCLC and may have a prognostic value for
patients with NSCLC (19). Chen
et al (20) also
demonstrated that reduced levels of PEDF in lung cancer tissue was
significantly correlated with lymph node metastasis and poor
overall prognosis in patients with lung cancer. PEDF inhibited the
growth and motility of lung cancer cells and was significantly
correlated with the clinical outcome of patients. In the present
study, it was found that PEDF overexpression in NSCLC significantly
decreased autophagy marker proteins p62 and ULK1, and inhibited the
proliferative capacity and viability of NSCLC cells. AMPK is a key
molecule that regulates biological energy metabolism and autophagy.
Current studies have suggested that AMPK plays an essential role in
regulating cellular energy homeostasis in eukaryotic cells,
including tumor cells, and restoring the normal function of the
liver and other tissues in diabetic patients (44). Once activated, AMPK is involved in
the regulation of four major types of metabolism in mammals:
Protein metabolism, lipid metabolism, carbohydrate metabolism, as
well as autophagy and mitochondrial homeostasis (45). Activated AMPK induces the
phosphorylation of ULK1, leading to the activation of autophagy.
Thus, the question is raised of whether there is an association
between PEDF and AMPK. A study by Qiu et al (25) found that PEDF promoted proteasomal
degradation of AMPK and subsequently reduced ATP production. Yang
et al (46) reported that
metformin inhibited PEDF expression and secretion in adipocytes and
hepatocytes by promoting AMPK phosphorylation. However, as no
correlation between lung cancer-derived AMPK functional activity
and PEDF expression has been reported, further studies are required
to fully elucidate this mechanism. The results of the present study
demonstrated that PEDF clearly downregulated AMPK expression,
indicating that PEDF suppressed the AMPK-related signaling
pathways. The present study also explored the role of the
downstream factor AMPK in autophagy and the results revealed that
PEDF reduced ULK1 expression through AMPK, thus suggesting that
PEDF blocked ULK1-induced autophagy. Of note, in the present study
it was found that rapamycin promoted the growth of NSCLC cells, but
some previous studies have revealed the opposite results. Chen
et al (47) showed that
rapamycin greatly enhanced dasatinib-induced cell growth inhibition
and cell cycle G1 arrest in human lung adenocarcinoma A549 cells,
without affecting apoptosis. Sun et al (48) demonstrated that a combination of
rapamycin and trametinib could more effectively inhibit NSCLC cell
viability and proliferation. The reason for this discrepancy
between the results of the present study and those of the
aforementioned two studies may be that rapamycin used alone to
treat cells mainly activates the autophagy pathway, while
synergistic action with other anticancer drugs affects other
pathways, thus leading to different results.
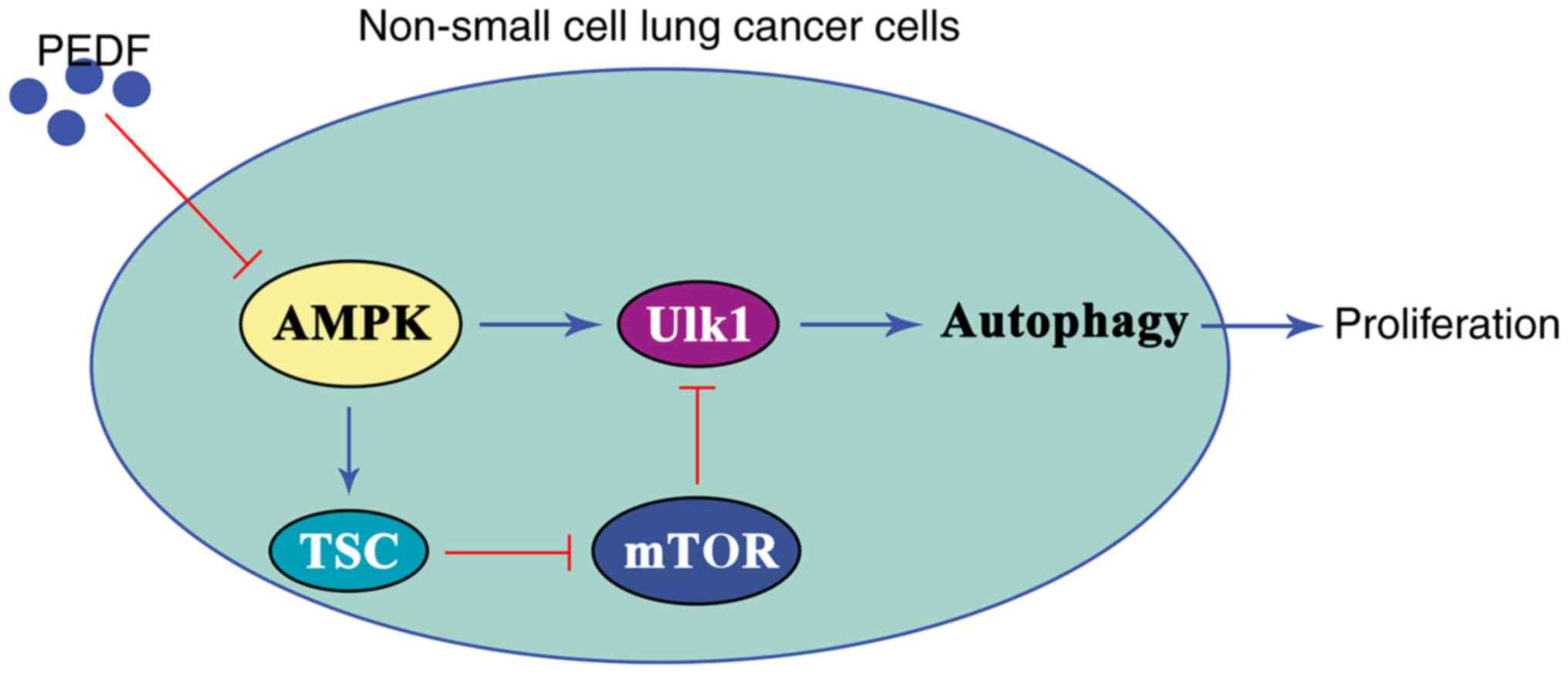
In conclusion, the results of the present study
suggested that PEDF exerts an anticancer effect by inhibiting
autophagy induced by the AMPK/ULK1 signaling pathway in lung cancer
cells. Specifically, PEDF inhibited the expression and activation
of AMPK, leading to inactivation of ULK1 and ultimately inducing
autophagy inhibition and suppressed cell proliferation in NSCLC
cells (Fig. 6). The overexpression
of PEDF inhibited the proliferation and viability of NSCLC cells
and significantly reduced the metastatic potential of NSCLC cells.
However, the results of the present study only provide a reference
point rather than the final conclusion. The reason for this is that
the regulation of autophagy relies on a set of complex and complete
signaling networks, and its mechanism still needs to be verified by
a large number of experiments. In future studies, animal
experiments should be performed to further validate the results of
the present study at the in vivo level, and bioinformatics
analysis is also required to elucidate the relevant mechanisms.
Acknowledgements
We would like to thank Dr Jingjun Han from the
Eighth Affiliated Hospital of Sun Yat-sen University (Shenzhen,
China) for the donation of A549 and H460 non-small cell lung cancer
cell lines.
Funding
The present study was supported by Futian Healthcare Research
Project (No. FTWS2020054) and the Research Fund of the Eighth
Hospital of Sun Yat-sen University (No. GCCRCYJ026).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HM, HH, HL, YZ and BJ conceived and designed the
study. HM performed the confocal microscopy fluorescence imaging,
and was a major contributor in writing the manuscript. HH performed
the western blotting and the culture of the cells, and was a minor
contributor in writing the manuscript. HL was a major contributor
in the statistical analysis of data. YL performed the western
blotting, assessed the activity of cells, and was a minor
contributor in writing the manuscript. DL performed the cell
culture, and was a minor contributor in the statistical analysis of
data. ML performed the western blotting and the culture of the
cells, and was a minor contributor in writing the manuscript. BJ
performed some of the experiments in this study, such as the PCR,
and drafted the work and revised it critically for important
intellectual content, as well as provided experimental technical
support. YZ drafted the work and revised it critically for
important intellectual content, and provided final approval of the
version to be published. YZ and BJ confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chung SJ, Nagaraju GP, Nagalingam A,
Muniraj N, Kuppusamy P, Walker A, Woo J, Győrffy B, Gabrielson E,
Saxena NK and Sharma D: ADIPOQ/adiponectin induces cytotoxic
autophagy in breast cancer cells through STK11/LKB1-mediated
activation of the AMPK-ULK1 axis. Autophagy. 13:1386–1403. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thakur SK, Singh DP and Choudhary J: Lung
cancer identification: A review on detection and classification.
Cancer Metastasis Rev. 39:989–998. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng M: Classification and pathology of
lung cancer. Surg Oncol Clin N Am. 25:447–468. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jonna S and Subramaniam DS: Molecular
diagnostics and targeted therapies in non-small cell lung cancer
(NSCLC): An update. Discov Med. 27:167–170. 2019.PubMed/NCBI
|
|
5
|
Ferro F, Servais S, Besson P, Roger S,
Dumas JF and Brisson L: Autophagy and mitophagy in cancer metabolic
remodelling. Semin Cell Dev Biol. 98:129–138. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kocaturk NM, Akkoc Y, Kig C, Bayraktar O,
Gozuacik D and Kutlu O: Autophagy as a molecular target for cancer
treatment. Eur J Pharm Sci. 134:116–137. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Poillet-Perez L, Xie X, Zhan L, Yang Y,
Sharp DW, Hu ZS, Su X, Maganti A, Jiang C, Lu W, et al: Autophagy
maintains tumour growth through circulating arginine. Nature.
563:569–573. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Katheder NS, Khezri R, O'Farrell F,
Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen
T, Juhász G, et al: Microenvironmental autophagy promotes tumour
growth. Nature. 541:417–420. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhardwaj M, Leli NM, Koumenis C and
Amaravadi RK: Regulation of autophagy by canonical and
non-canonical ER stress responses. Semin Cancer Biol. 66:116–128.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou XJ, Klionsky DJ and Zhang H:
Podocytes and autophagy: A potential therapeutic target in lupus
nephritis. Autophagy. 15:908–912. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang C, Hu Q and Shen HM: Pharmacological
inhibitors of autophagy as novel cancer therapeutic agents.
Pharmacol Res. 105:164–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang L, Qiang P, Yu J, Miao Y, Chen Z, Qu
J, Zhao Q, Chen Z, Liu Y, Yao X, et al: Identification of compound
CA-5f as a novel late-stage autophagy inhibitor with potent
anti-tumor effect against non-small cell lung cancer. Autophagy.
15:391–406. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi TT, Yu XX, Yan LJ and Xiao HT:
Research progress of hydroxychloroquine and autophagy inhibitors on
cancer. Cancer Chemother Pharmacol. 79:287–294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He X, Cheng R, Benyajati S and Ma JX: PEDF
and its roles in physiological and pathological conditions:
Implication in diabetic and hypoxia-induced angiogenic diseases.
Clin Sci (Lond). 128:805–823. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Gao H, Dong H, Wang W, Xu Z, Wang G,
Liu Y, Wang H, Ju W, Qiao J, et al: PEDF reduces malignant cells
proliferation and inhibits the progression of myelofibrosis in
myeloproliferative neoplasms. Biochem Pharmacol. 199:1150132022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yin J, Park G, Kim TH, Hong JH, Kim YJ,
Jin X, Kang S, Jung JE, Kim JY, Yun H, et al: Pigment
epithelium-derived factor (PEDF) expression induced by EGFRvIII
promotes self-renewal and tumor progression of glioma stem cells.
PLoS Biol. 13:e10021522015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma R, Chu X, Jiang Y and Xu Q: Pigment
epithelium-derived factor, an anti-VEGF factor, delays ovarian
cancer progression by alleviating polarization of tumor-associated
macrophages. Cancer Gene Ther. 4:10.1038/s41417–022-00447-4.
2022.
|
|
18
|
Abooshahab R, Al-Salami H and Dass CR: The
increasing role of pigment epithelium-derived factor in metastasis:
From biological importance to a promising target. Biochem
Pharmacol. 193:1147872021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Chen J, Ke Y, Mansel RE and Jiang
WG: Expression of pigment epithelial derived factor is reduced in
non-small cell lung cancer and is linked to clinical outcome. Int J
Mol Med. 17:937–944. 2006.PubMed/NCBI
|
|
20
|
Chen J, Ye L, Zhang L and Jiang WG: The
molecular impact of pigment epithelium-derived factor, PEDF, on
lung cancer cells and the clinical significance. Int J Oncol.
35:159–166. 2009.PubMed/NCBI
|
|
21
|
Yamagishi SI, Koga Y, Sotokawauchi A,
Hashizume N, Fukahori S, Matsui T and Yagi M: Therapeutic potential
of pigment epithelium-derived factor in cancer. Curr Pharm Des.
25:313–324. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He J, Liu J, Huang Y, Tang X, Xiao H, Liu
Z, Jiang Z, Zeng L, Hu Z and Lu M: OM-MSCs alleviate the golgi
apparatus stress response following cerebral ischemia/reperfusion
injury via the PEDF-PI3K/Akt/mTOR signaling pathway. Oxid Med Cell
Longev. 2021:48050402021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miao H, Qiu F, Huang B, Liu X, Zhang H,
Liu Z, Yuan Y, Zhao Q, Zhang H, Dong H and Zhang Z: PKCα replaces
AMPK to regulate mitophagy: Another PEDF role on ischaemic
cardioprotection. J Cell Mol Med. 22:5732–5742. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan Y, Liu X, Miao H, Huang B, Liu Z,
Chen J, Quan X, Zhu L, Dong H and Zhang Z: PEDF increases
GLUT4-mediated glucose uptake in rat ischemic myocardium via
PI3K/AKT pathway in a PEDFR-dependent manner. Int J Cardiol.
283:136–143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qiu F, Zhang H, Yuan Y, Liu Z, Huang B,
Miao H, Liu X, Zhao Q, Zhang H, Dong H and Zhang Z: A decrease of
ATP production steered by PEDF in cardiomyocytes with
oxygen-glucose deprivation is associated with an AMPK-dependent
degradation pathway. Int J Cardiol. 257:262–271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Valadares AC, Gorki H, Liebold A and
Hoenicka M: Extraction of total RNA from calcified human heart
valves for gene expression analysis. J Heart Valve Dis. 26:185–192.
2017.PubMed/NCBI
|
|
27
|
Brena RM, Auer H, Kornacker K and Plass C:
Quantification of DNA methylation in electrofluidics chips
(Bio-COBRA). Nat Protoc. 1:52–58. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang H, Hui H, Li Z, Pan J, Jiang X, Wei
T, Cui H, Li L, Yuan X, Sun T, et al: Pigment epithelium-derived
factor attenuates myocardial fibrosis via inhibiting
endothelial-to-mesenchymal transition in rats with acute myocardial
infarction. Sci Rep. 7:419322017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kutner RH, Zhang XY and Reiser J:
Production, concentration and titration of pseudotyped HIV-1-based
lentiviral vectors. Nat Protoc. 4:495–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo F, Sandhu AF, Rungratanawanich W,
Williams GE, Akbar M, Zhou S, Song BJ and Wang X: Melatonin and
autophagy in aging-related neurodegenerative diseases. Int J Mol
Sci. 21:71742020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Runwal G, Stamatakou E, Siddiqi FH, Puri
C, Zhu Y and Rubinsztein DC: LC3-positive structures are prominent
in autophagy-deficient cells. Sci Rep. 9:101472019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Streeter A, Menzies FM and Rubinsztein DC:
LC3-II tagging and western blotting for monitoring autophagic
activity in mammalian cells. Methods Mol Biol. 1303:161–170. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Negoita F, Blomdahl J, Wasserstrom S,
Winberg ME, Osmark P, Larsson S, Stenkula KG, Ekstedt M, Kechagias
S, Holm C and Jones HA: PNPLA3 variant M148 causes resistance to
starvation-mediated lipid droplet autophagy in human hepatocytes. J
Cell Biochem. 120:343–356. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zachari M and Ganley IG: The mammalian
ULK1 complex and autophagy initiation. Essays Biochem. 61:585–596.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deng R, Zhang HL, Huang JH, Cai RZ, Wang
Y, Chen YH, Hu BX, Ye ZP, Li ZL, Mai J, et al: MAPK1/3
kinase-dependent ULK1 degradation attenuates mitophagy and promotes
breast cancer bone metastasis. Autophagy. 17:3011–3029. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hung CM, Lombardo PS, Malik N, Brun SN,
Hellberg K, Van Nostrand JL, Garcia D, Baumgart J, Diffenderfer K,
Asara JM and Shaw RJ: AMPK/ULK1-mediated phosphorylation of Parkin
ACT domain mediates an early step in mitophagy. Sci Adv.
7:eabg45442021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang M, Tombran-Tink J, Yang S, Zhang X,
Li X and Barnstable CJ: PEDF is an endogenous inhibitor of VEGF-R2
angiogenesis signaling in endothelial cells. Exp Eye Res.
213:1088282021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim JE, Park H, Jeong MJ and Kang TC:
Epigallocatechin-3-Gallate and PEDF 335 peptide, 67LR activators,
attenuate vasogenic edema, and astroglial degeneration following
status epilepticus. Antioxidants (Basel). 9:8542020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li F, Song N, Tombran-Tink J and Niyibizi
C: Pigment epithelium-derived factor enhances differentiation and
mineral deposition of human mesenchymal stem cells. Stem Cells.
31:2714–2723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ma B, Zhou Y, Liu R, Zhang K, Yang T, Hu
C, Gao Y, Lan Q, Liu Y, Yang X and Qi H: Pigment epithelium-derived
factor (PEDF) plays anti-inflammatory roles in the pathogenesis of
dry eye disease. Ocul Surf. 20:70–85. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Vessey KA, Jobling AI, Tran MX, Wang AY,
Greferath U and Fletcher EL: Treatments targeting autophagy
ameliorate the age-related macular degeneration phenotype in mice
lacking APOE (apolipoprotein E). Autophagy. 18:2368–2384. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Carling D: AMPK signalling in health and
disease. Curr Opin Cell Biol. 45:31–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Garcia D and Shaw RJ: AMPK: Mechanisms of
cellular energy sensing and restoration of metabolic balance. Mol
Cell. 66:789–800. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang S, Lv Q, Luo T, Liu L, Gao R, Chen S,
Ye P, Cheng Q and Li Q: Metformin inhibits expression and secretion
of PEDF in adipocyte and hepatocyte via promoting AMPK
phosphorylation. Mediators Inflamm. 2013:4292072013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen B, Xu X, Luo J, Wang H and Zhou S:
Rapamycin enhances the anti-cancer effect of dasatinib by
suppressing Src/PI3K/mTOR pathway in NSCLC cells. PLoS One.
10:e01296632015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun CY, Li YZ, Cao D, Zhou YF, Zhang MY
and Wang HY: Rapamycin and trametinib: A rational combination for
treatment of NSCLC. Int J Biol Sci. 17:3211–3223. 2021. View Article : Google Scholar : PubMed/NCBI
|