Introduction
Comprehensive genomic profiling (CGP) using
next-generation sequencing (NGS) has become a clinical practice for
the effective selection of treatments based on druggable genomic
variants. Identifying somatic variants of tumors using tissue is
widely implemented, whereas circulating tumor DNA (ctDNA) detection
is a noninvasive method for assessing genomic profiles using
peripheral blood (1,2). ctDNA constitutes only a minor fraction
of the cell-free DNA (cfDNA) circulating in cancer patients,
complicating ctDNA detection (3,4).
Genomic profiling using ctDNA has the potential to predict
recurrence, survival and response to therapy (5,6).
However, advanced tools are required to accurately personalize
treatment decisions following the detection of somatic variants and
monitoring of tumor clone and subclone evolution (7).
Tumor heterogeneity is associated with the treatment
outcomes of metastatic cancers (8);
however, only a small number of studies have examined whether tumor
heterogeneity in ctDNA can be used to predict treatment outcomes
(9). Recent studies have
demonstrated that the number of somatic variants in ctDNA can be
used to noninvasively assess tumor heterogeneity (10–13).
Therefore, multiple ctDNA analyses following tumor resection may
capture molecular and genetic heterogeneity by identifying somatic
variants for targeted therapy and monitoring tumor evolution and
resistance in real time (14).
Ion Torrent™ Genexus™ Integrated Sequencer (Genexus,
Thermo Fisher Scientific, Inc.) was developed to automate all
targeted NGS workflows from library construction using either
nucleic acids of formalin-fixed paraffin-embedded (FFPE) tissues or
cell-free total nucleic acids (cfTNA) of plasma to data analyses
and delivers results within 24 h. Therefore, Genexus enables
clinicians to provide patients with somatic variant information
leading to more efficient cancer treatment (15,16).
This study, thus, aimed to evaluate the concordance
rate of somatic variants between resected tumor tissue and ctDNAs
at post-resection in patients with various types of solid tumor by
using Genexus. Furthermore, the presence of recurrence was
evaluated at one year post-resection.
Patients and methods
Patients and samples
The study was approved by the Institutional Review
Board of Kurume University Hospital (approval no. 2022008) and was
registered in the Japan Registry of Clinical Trials (April 10,
2023; no. 072230003). All methods were performed in accordance with
relevant guidelines and regulations. Among the patients scheduled
for curative surgery at Kurume University Hospital (Kurume, Japan)
in April and May 2023, patients with breast, lung, pancreas, and
head and neck cancer were selected in the order of the earliest
scheduled surgery, aiming to enroll 2 patients per cancer type. All
of the candidates were asked to participate in this study and the
patients who agreed to this study were enrolled. A total of eight
patients with resectable solid tumors who agreed to participate in
this study were enrolled: Two patients with breast cancer, two with
lung cancer, two with pancreatic cancer and two with head and neck
cancer. Patients were excluded if they had synchronous and
metachronous advanced cancers or severe complications. Participants
were fully informed of the purpose and procedures of the study and
had adequate time to ask questions and contemplate their voluntary
participation. Written informed consent was obtained from all
patients prior to enrollment. Detailed patient information is
presented in Table I and Fig. 1A. Blood samples were obtained from
the enrolled patients before surgery. Radical resection was
performed and the resected tumor tissues were diagnosed by
pathologists at Kurume University Hospital (Kurume, Japan).
Following pathological diagnosis, DNA and RNA were extracted from
FFPE tumor blocks. Blood samples were also collected in the first
(between fourth to fifth week) and second (between eighth to nineth
week) postoperative months, and ctDNA was promptly extracted.
Tissue samples were analyzed using the Genexus Oncomine
Comprehensive Assay v3 (OCAv3), while blood samples were analyzed
using the Genexus Oncomine Precision Assay (OPA). The gene list for
each test is provided in Table
SI.
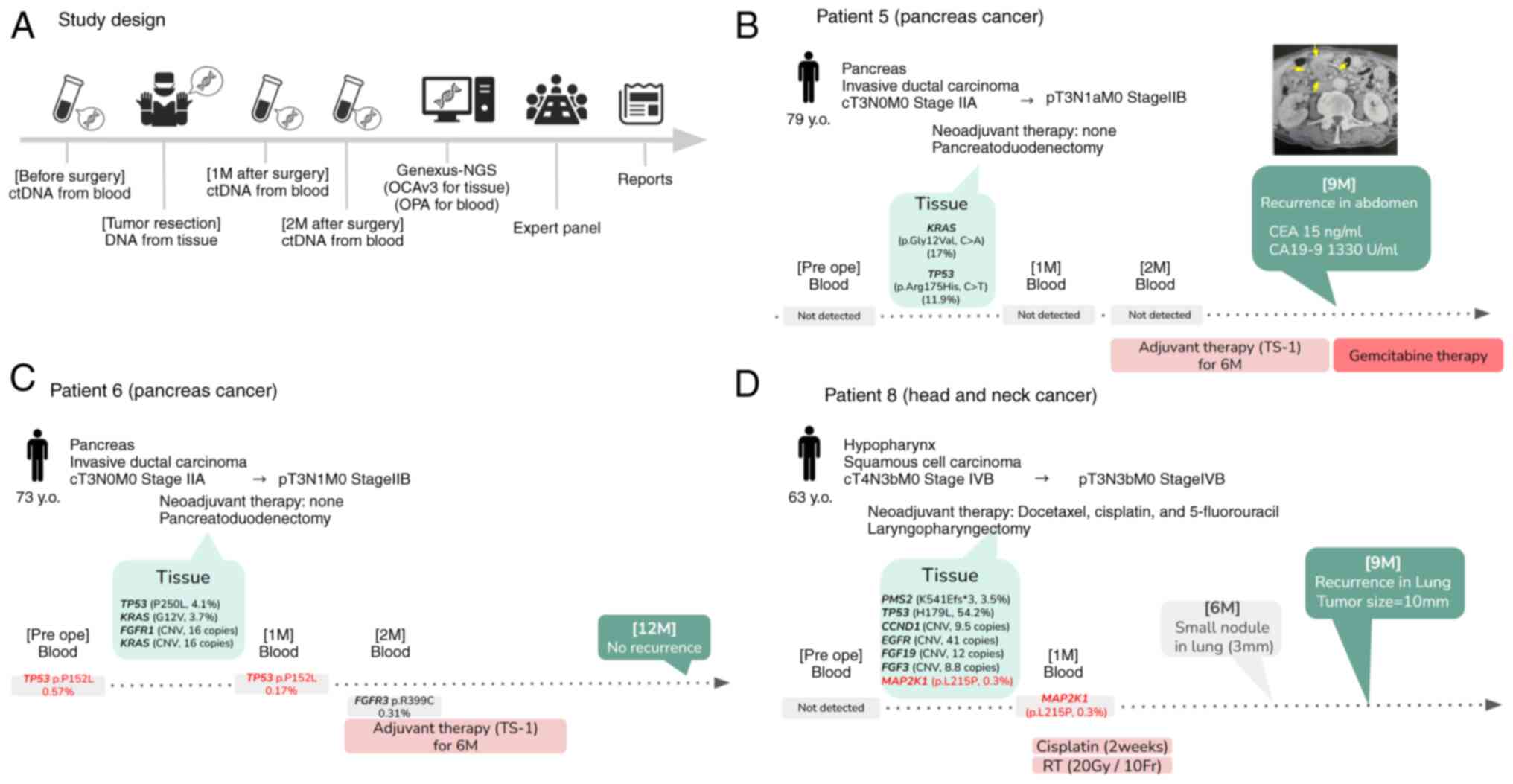 | Figure 1.Study design and clinical course of
the cases. (A) Blood samples were obtained before surgery and 1 and
2 months after surgery. A tissue sample was obtained during
surgery. All samples were analyzed using Genexus-NGS (OPA for blood
and OCAv3 for tissue samples). (B-D) Clinical information and
somatic variants of (B) patient 5, (C) patient 6 and (D) patient 8.
The CT image on the upper right in B shows the recurrence (yellow
arrows) in the abdomen at nine months postoperatively. M, month(s);
y.o., years old; NGS, next-generation sequencing; ctDNA,
circulating tumor DNA; RT, radiation therapy; Pre ope,
preoperative; OCAv3, Genexus Oncomine Comprehensive Assay v3; OPA,
Genexus Oncomine Precision Assay; TS-1,
tegafur/gimeracil/oteracil. |
 | Table I.Patients' characteristics. |
Table I.
Patients' characteristics.
| Patient no. | Age, years | Sex | Primary tumor | Histological
findings | Clinical
staging | Preoperative
therapy | Surgical
procedure | Disease status at
testing | Postoperative
therapy |
|---|
| 1 | 57 | Female | Breast | Invasive ductal
carcinoma | cT1cN0M0 Stage
I | Anastrozole
(2M) | Skin-sparing
mastectomy | pTisN0M0 Stage
0 | None |
| 2 | 72 | Female | Breast | Invasive ductal
carcinoma | cT2N1M0 Stage
IIB | Dose-dense PTX (4
cycles) | Partial
mastectomy | pT1N0M0 Stage
0 | None |
| 3 | 71 | Female | Lung | Papillary
adenocarcinoma | cT2aN0M0 Stage
IB | None | Lower
lobectomy | pT1cN0M0 Stage
IA3 | None |
| 4 | 76 | Male | Lung | Squamous cell
carcinoma, | cT2N0M0 Stage
IB | None | Upper
lobectomy | pT1bN0M0 Stage
IA2 | None |
| 5 | 79 | Male | Pancreas | Invasive ductal
carcinoma | cT3N0M0 Stage
IIA | None |
Pancreatoduodenectomy | pT3N1aM0
StageIIB | TS-1 (6
cycles) |
| 6 | 73 | Male | Pancreas | Invasive ductal
carcinoma | cT3N0M0 Stage
IIA | None |
Pancreatoduodenectomy | pT3N1M0 Stage
IIB | TS-1 (6
cycles) |
| 7 | 29 | Female | Parotid gland | Secretory
carcinoma | cT2aN0M0 Stage
I | None | Partial
parotidectomy | pT3N0M0 Stage
III | None |
| 8 | 63 | Male | Hypopharynx | Squamous cell
carcinoma | cT4N3bM0 Stage
IVB | TPF (2 cycles) |
Laryngopharyngectomy | pT3N3bM0 Stage
IVB | Cisplatin (2 weeks)
plus radiation (20 Gy/10 Fr) |
Samples, DNA extraction and
quantification
DNA and RNA from FFPE tissue specimens were
extracted using a Maxwell RSC Instrument (cat. no. AS4500; Promega,
Corp.), Maxwell RSC FFPE Plus DNA kit (cat. no. AS1720; Promega,
Corp.) and Maxwell RSC RNA FFPE kit (cat. no. AS1440; Promega,
Corp.). Whole blood for plasma analysis was processed within 15 min
of collection. To obtain blood plasma, 14 ml of whole blood with
EDTA-2Na was cooled and centrifuged (4°C, 2,000 × g, 10 min) twice.
cfTNA, which included both DNA and RNA, was extracted using a
Maxwell RSC Instrument with a Maxwell RSC miRNA Plasma and Serum
Kit (cat. no. AS1680; Promega, Corp.). DNA and RNA concentrations
were measured using the QuantiFluor ONE dsDNA System (cat. no.
E4871; Promega, Corp.) and the QuantiFluor RNA System (cat. no.
E3310; Promega, Corp.), respectively. Nucleic acid >1.1 ng/µl
for DNA and >0.95 ng/µl for RNA extracted from tissues, and
>1.33 ng/µl from blood were recommended as the cutoffs for
further processes. The DNA and RNA fragment lengths were evaluated
using an Agilent 4200 TapeStation system (Agilent Technologies,
Inc.).
Genexus sequencing
The Ion Torrent Genexus Integrated Sequencer (Thermo
Fisher Scientific, Inc.) is a fully automated NGS system that
integrates library preparation, including cDNA synthesis, template
preparation, sequencing and data analysis of purified and
quantified nucleic acids (DNA, RNA and cfTNA). In this study, DNA
and RNA derived from FFPE tissues were used as samples for OCAv3,
and cfTNA for OPA. A total of 25 µl of DNA (>1.1 ng/µl) and RNA
(>0.95 ng/µl) for OCAv3 and 20 µl of cfTNA (>1.33 ng/µl) for
OPA were recommended for library preparation. After assigning the
assay to the Genexus Integrative System (https://assets.thermofisher.cn/TFS-Assets/LSG/manuals/MAN0017910_GenexusIntegratedSequencer_UG.pdf),
the sample type and sequence run settings were specified and they
were loaded onto sample plates. The Multiplex I cfDNA Reference
Standard Set (HD780; Horizon Diagnostics) was used to evaluate the
performance of OCAv3 and OPA (Table
SI) (17).
Genexus variant analyses
Ion Torrent™ Genexus™ Software ver. 6.6.2.1 (Thermo
Fisher Scientific, Inc.) supports the Ion Torrent™ Genexus™
Integrated Sequencer (Thermo Fisher Scientific, Inc.) workflow for
research use purposes from sample preparation through library
preparation, template preparation, and sequencing. During and after
sequencing, the software generates base calls, trims reads and
determines quality values (primary analysis), then aligns reads,
calls variants, and generates reports (secondary analysis)
(https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0024953_IonTorrentGenexusSoftware6.6RUO_UG.pdf).
The sequencing data were mapped to the standard reference genome,
the human genome assembly 19, using the software and aligned using
the torrent mapping alignment program. After the initial mapping,
somatic variants were identified using the Torrent Variant Caller.
The variants were selected using a built-in variant filter. The
four major classes of variants evaluated were single nucleotide
variants, insertions and deletions, copy number amplifications and
gene fusions. Variants with a low frequency of <0.3%, an allele
count of ≤5, and a C-T or G-A change were excluded from the results
as potential deaminations for the OPA of the liquid samples
(17).
Statistical analysis
The concordance rate was calculated for the 45 genes
in common between the genes in OCA-V3 and OPA (Table SI). The concordance rate for each
case was calculated as the number of concordant genes regardless of
the presence of somatic variants/45 genes. In addition, the number
of genes with somatic variants/45 genes was also calculated using
JMP 16 software (SAS Institute).
Results
Patients' treatment course
A total of eight patients were enrolled in this
study: Two patients with breast cancer, two with lung cancer, two
with pancreatic cancer and two with head and neck cancer (Table I). Patient 1 was a 57-year-old
female with cT1cN0M0 stage I breast cancer who received anastrozole
as preoperative therapy (daily oral anastrozole 1mg for 2 months).
Following mastectomy, the final stage was pTisN0M0, stage 0. No
postoperative therapy was administered. Patient 2 was a 72-year-old
female with cT2N1M0, stage IIB breast cancer who received
paclitaxel as preoperative therapy (dose-dense paclitaxel,
paclitaxel 175 mg/m2 every 2 weeks for 4 cycles). A
mastectomy was performed and the final stage was pT1N0M0, stage 0.
No postoperative therapy was administered. Patient 3 was a
71-year-old female with cT2N1M0, stage IIB lung adenocarcinoma.
Following pulmonary lobectomy, the final stage was pT1cN0M0, stage
IA3. No pre- and postoperative therapy was administered. Patient 4,
a 76-year-old male, had cT2N1M0, stage IIB lung squamous cell
carcinoma. Pulmonary lobectomy was performed and the final stage
was pT1bN0M0, stage IA2. No pre- and postoperative therapy was
administered. Patient 5 (Fig. 1B)
was a 79-year-old male with cT3N0M0, stage IIA pancreatic cancer.
Following pancreatoduodenectomy, the final stage was pT3N1aM0 stage
IIB. TS-1 (tegafur/gimeracil/oteracil) was administered as adjuvant
chemotherapy (6 cycles of daily oral TS-1 for 14 days followed by a
14-day break). Patient 6 (Fig. 1C),
a 73-year-old male with cT2N1M0 stage IIB lung squamous cell
carcinoma underwent pulmonary lobectomy, and the final stage was
pT3N1M0, stage IIB. TS-1 was administered as adjuvant chemotherapy
(6 cycles of daily oral TS-1 for 14 days followed by a 14-day
break). Patient 7 was a 29-year-old female with cT2aN1M0 stage I
secretory carcinoma. Partial parotidectomy was performed and the
final stage was pT3N0M0, stage III. No pre- and postoperative
therapy was administered. Patient 8 (Fig. 1D), a 63-year-old female, had
cT2aN1M0 stage I squamous cell hypopharyngeal cancer and received
preoperative therapy with docetaxel, cisplatin and 5-fluorouracil
(2 cycles of TPF; cisplatin, fluorouracil, and docetaxel).
Laryngopharyngectomy was performed and the final stage was
pT3N3bM0, stage IVB. Cisplatin plus radiation (20 Gy/10Fr) was
administered postoperatively [weekly cisplatin with daily radiation
(66 Gy/33Fr) was planned, but only 2 weeks had implemented due to
patient refusal of treatment].
Somatic variants in tissue and
blood
All eight paired tissue samples and three blood
samples (prior to resection and 1 and 2 months after resection)
were analyzed using Genexus. A total of 26 somatic variants were
detected in the resected tumor tissues (Table II). The overall concordance rate in
all genes between tissue and postoperative blood was 94.2%
(91.1–97.8%), whereas the overall concordance rate in genes with
somatic variants was 4.76% (0.0–33.3%), but the concordance rate in
genes with somatic variants was low (4.76%). Actionable mutations,
ETV6-NTRK3 fusion and EML4-ALK fusion, were found in two patients
(Patients #3 and #7). The two somatic variants were concordant
between the two patients (TP53 variant in patient 6 and
MAP2K1 variant in patient 8). In patient 6 (Fig. 1C), the TP53 p.P152L variant
was concordant in the blood samples obtained before surgery and
that obtained 1 month after surgery. The TP53 p.P152L
variant was detected in ctDNA before surgery. The same variant
(TP53 p.P152L) was found 1 month postresection but was not
detected 2 months after resection and in the resected tissue
samples, and the patient did not experience recurrence at 1 year
after surgery. In patient 8 (Fig.
1D), the MAP2K1 p.L215P variant was concordant in
tissues obtained during surgery and in the blood sample obtained 1
month after surgery. The MAP2K1 p.L215P variant was
identified in a tissue sample. The same variant (MAP2K1
p.L215P) was identified 1 month after resection but was not
detected in ctDNA prior to surgery and at two months
post-resection. The patient was diagnosed with lung recurrence [a
10-mm tumor identified via chest computed tomography (CT)] 9 months
post-operatively. A retrospective review of the chest CT image
obtained 6 months after surgery revealed the presence of a small
pulmonary nodule (measuring 3 mm on the chest CT scan) in the same
region. In patient 5 (Fig. 1B), the
somatic variants KRAS G12V and TP53 R175H were
identified in the tissue samples. Somatic variants were not
identified in the ctDNA at 1 and 2 months postresection. However,
CA19-9 levels were elevated 9 months post-surgery and recurrence in
the abdomen was observed on the CT scan.
 | Table II.Somatic variants from surgical tissue
and pre- and post-operative blood. |
Table II.
Somatic variants from surgical tissue
and pre- and post-operative blood.
|
|
|
|
|
| Ope (tissue) |
|
|
|
|
|
|
|
|---|
|
|
| Pre-ope
(blood) |
| Post-ope 1M
(blood) | Post-ope 2M
(blood) | Recurrence at one
year after resection |
|---|
|
|
|
| Gene | Variant | Allele freq, # of
leads, # of copies |
|
|
|---|
| Patient | Tumor | Gene | Variant | Allele freq, % | Gene | Variant | Allele freq | Gene | Variant | Allele freq, % |
|---|
| 1 | Breast |
| ND |
| PIK3CA | H1047R | 5.8% |
| ND |
|
| ND |
| ND |
|
|
|
|
|
|
ESR1-CCDC170 | Fusion | 88 reads |
|
|
|
|
|
|
|
|
|
|
|
|
| KRAS | CNV | 64 copies |
|
|
|
|
|
|
|
| 2 | Breast |
| ND |
|
| ND |
|
| ND |
|
| ND |
| ND |
| 3 | Lung |
| ND |
|
EML4-ALKa | Fusion | 189058 reads |
| ND |
|
| ND |
| ND |
|
|
|
|
|
|
EML4-ALKa | Fusion | 39 reads |
|
|
|
|
|
|
|
|
|
|
|
|
| MDM2 | CNV | 23 copies |
|
|
|
|
|
|
|
|
|
|
|
|
|
| ND |
|
TRIM24-BRAF | Fusion | 243 reads |
|
|
|
|
| 4 | Lung |
| ND |
| PMS2 | K541EQfs*3 | 16.7% |
| ND |
|
| ND |
| ND |
|
|
|
|
|
| EGFR | CNV | 16 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| FGF19 | CNV | 11 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| KRAS | CNV | 17 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| PIK3CA | CNV | 10 copies |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ND |
| PIK3CA | R108C | 0.39 |
|
|
|
|
|
|
|
|
|
|
|
|
| PIK3CA | G106V | 0.30 |
|
| 5 | Pancreas |
| ND |
| KRAS | G12V | 17.0% |
| ND |
|
| ND |
| Recurrence |
|
|
|
|
|
| TP53 | R175H | 11.9% |
|
|
|
|
|
| (intra- |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| abdomen) |
| 6 | Pancreas |
|
|
| TP53 | P250L | 4.1% |
|
|
|
|
|
| ND |
|
|
|
|
|
| KRAS | G12V | 3.7% |
|
|
|
|
|
|
|
|
|
|
|
|
| FGFR1 | CNV | 16 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| KRAS | CNV | 16 copies |
|
|
|
|
|
|
|
|
|
| TP53 | P152L | 0.57 |
| ND |
| TP53 | P152L | 0.17% |
|
|
|
|
|
|
|
| ND |
|
| ND |
|
| ND |
| FGFR3 | R399C | 0.31 |
|
| 7 | Parotid |
| ND |
|
ETV6-NTRK3a | Fusion | 347801 copies |
| ND |
|
| ND |
| ND |
|
| gland |
|
|
|
EML4-ALKa | Fusion | 5403 copies |
|
|
|
|
|
|
|
|
|
|
|
|
|
| ND |
|
UBN2-BRAF | Fusion | 135 copies |
|
|
|
|
| 8 | Hypopharynx |
| ND |
| PMS2 | K541Efs*3 | 3.5% |
| ND |
|
| ND |
| Recurrence |
|
|
|
|
|
| TP53 | H179L | 54.2% |
|
|
|
|
|
| (lung) |
|
|
|
|
|
| CCND1 | CNV | 9.5 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| EGFR | CNV | 41 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| FGF19 | CNV | 12 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| FGF3 | CNV | 8.8 copies |
|
|
|
|
|
|
|
|
|
|
|
|
| MAP2K1 | L215P | 0.30% | MAP2K1 | L215P | 0.08% |
|
|
|
|
Discussion
The present findings demonstrate that the overall
concordance rate in all genes between tissue and postoperative
blood was high (94.2%), but the concordance rate in genes with
somatic variants was low (4.76%). However, in one patient (Patient
#8) with somatic variants, which corresponded to the tissue and
postoperative blood samples, distant recurrence was identified
during subsequent surveillance. This finding suggests that
detecting a somatic variant in postoperative ctDNA matching the
same variant in the tissue may predict the occurrence of metastatic
relapse in localized cancer. However, sensitivity must be
validated, as recurrences were also noted in cases in which no
somatic variants were identified in the postoperative blood. This
pilot study, with its limited sample size, serves as a preliminary
investigation into the potential benefits of multiple CGP testing.
Subsequent studies with larger sample sizes and more extensive
research designs are necessary.
Regarding the concordance of somatic variants
between tissue and blood, high concordance has been reported
between tumor tissue NGS and cfDNA in various studies: EGFR
alterations in non-small cell lung cancer; multiple genes (KRAS,
TP53, APC, FBXW7, SMAD4) in pancreaticobiliary cancers;
KRAS variants in exons 12–13 in colorectal cancer;
BRAF V600E and KIT variants in melanoma; and BRAF,
EGFR, KRAS and PIK3CA across a variety of advanced
cancers (7,18–21).
In a study investigating the clinical utility of
ctDNA in plasma in patients with locally advanced esophageal cancer
(n=11) who received neoadjuvant chemotherapy followed by
esophagectomy, somatic variants from the primary tumor and ctDNA
after resection were analyzed. Patients with the TP53
somatic variant in both ctDNA and resected tissues had a 1-year
recurrence-free survival rate of 90% compared to 0% in patients
with ctDNA negativity (22,23). This suggests that identification of
the same variant in the resected tissue and ctDNA following
resection is a valuable indicator for predicting recurrence. In the
present study, a case of recurrence showing somatic variant
matching in the tissue with the postoperative blood was
encountered, indicating that detecting a somatic variant in
postoperative ctDNA and identifying the same variant in the tissue
may predict recurrence. However, the present study also included a
case of recurrence with no somatic variants in the postoperative
blood, suggesting that the specificity of predicting recurrence
based on variants in postoperative blood may be high, but without
high sensitivity. However, the present study showed that the
concordance rate in the genes with somatic variants was low, but
the overall concordance rate in all genes was high. Potential
explanations for the lack of concordance include spatial and
temporal tumor heterogeneity, differences in sampling intervals and
potential germline DNA contamination.
In the present study, a major cause of discordance
could be heterogeneity. A study comparing tumor tissue NGS
(FoundationOne) and ctDNA (Guardant360) showed significant
discordance between the same NGS panels in a limited number of
patients with diverse solid tumors, including those of the breast,
lung, pancreas and salivary glands (24). Additionally, advanced-stage
urothelial cancer studies have shown significant discordance
between clinically available NGS panels, even when collected at
approximately the same time (25).
A major cause of discordance could, in part, be differences in
tumor type, timing of specimen collection, intratumoral
heterogeneity, clonal evolution, discrete gene alteration types and
assays performed (24–26). Furthermore, in a study with multiple
CGP, the concordance rate was lower in cases with longer intervals
between CGPs. EGFR mutation was analyzed in patients with lung
cancer and it was found that increasing timing intervals between
tumor and cfDNA sampling from <2 weeks to >6 months led to
significantly lower concordance (P=0.038) (27). In a similar study on lung cancer,
the overall concordance of EGFR mutations between tissue and blood
varied depending on sampling time; concordance for time intervals
of 0.8 and >0.8 months were 88.2 and 64.7%, respectively
(28). The tissue and blood
intervals in this study were 1 and 2 months. Considering the
previous studies, the relatively long intervals in the present
study may be associated with the low concordance rate.
Additionally, the use of fresh frozen samples for tumor tissue
instead of FFPE marginally increased the concordance rate from 57.1
to 66.7%, suggesting fragmentation of DNA in FFPE processing may be
significant, particularly when the detection assay relies on
amplicon-based amplification (29).
The use of FFPE for the analysis of tissues in this study may also
be associated with the low concordance rate. To conclude, the
reasons for the low concordance rate between tissue and blood in
this study were considered to be the presence of heterogeneity, the
different collection time between tissue and blood, and nucleic
acid extraction using FFPE.
The major limitation of the present study is that
the number of samples and types of tumors were limited. The type of
cancer and biopsy site can influence the results of the tests to
identify variants. Pancreatic, ovarian, colorectal, breast,
bladder, gastroesophageal, melanoma and hepatocellular carcinomas
are more likely to possess detectable cfDNA than are primary brain,
renal, prostate and thyroid cancers (30). Additionally, the potential
limitations of ctDNA may be related to either quantity, such as the
amount of ctDNA accessible in the peripheral blood, or quality,
such as the purity of noncancerous cells in the tumor
microenvironment, which may complicate cfDNA assays (31). Moreover, selection bias might be
included in this study, because we have selected the enrolled
patients. As this is a preliminary study with a limited sample
size, subsequent studies with larger sample sizes and more
extensive research designs are warranted.
In conclusion, the present study demonstrated that
the overall concordance rate in all genes between tissue and
postoperative blood was high, but the concordance rate in genes
with somatic variants was low. Subsequent surveillance revealed
that a single case in which matched somatic variants identified in
tissue samples and postoperative blood samples exhibited distant
recurrence. These findings indicate the potential of using
postoperative ctDNA for recurrence detection. However, further
studies on sensitivity assessment are warranted with a large sample
size because the present study also included a case of recurrence
despite the absence of somatic variants in the postoperative blood.
The early detection of recurrence and the initiation of treatment
can be facilitated by the detection of somatic variants that
correspond to resected tissue.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data derived from the Ion Torrent™ Genexus™
Sequencer are available in the DNA Data Bank of Japan (DDBJ;
http://www.ddbj.nig.ac.jp) as Bioproject
(no. PRJDB20670; http://ddbj.nig.ac.jp/search/entry/bioproject/PRJDB20670)
and DRA (no. DRA021249; http://ddbj.nig.ac.jp/public/ddbj_database/dra/fastq/DRA021/DRA021249/).
Authors' contributions
YN and KF designed the study and wrote the protocol.
KF, YW, TS, SN, KY, HU, MN and YN contributed to the design. NM,
JA, HA, YI, KI and FY extracted DNA and implemented NGS. KF and YN
managed the literature search and analyses. RS, SO, YO, TO, KO, MK,
RK, HK, AK, RT, RU, UT, KH, TH, SH, MM and DM included the patients
and performed the clinical assessments. KF, NM and YN checked and
confirmed the authenticity of the raw data. KF and NM wrote the
first draft of the manuscript. YN and TS supervised the whole
process and critically reviewed the article. All authors read,
reviewed and approved the final version of the manuscript.
Ethics approval and consent to
participate
The study was approved by Kurume University
Hospital's Institutional Review Board (Kurume, Japan; approval no.
2022008) and was registered in the Japan Registry of Clinical
Trials (April 10, 2023; no. 072230003). All methods were carried
out in accordance with relevant guidelines and regulations.
Participants of this study were fully informed of the purpose and
procedures of the study and had adequate time to ask questions and
contemplate their voluntary participation. Written informed consent
was obtained from all patients before enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
cfTNA
|
cell-free total nucleic acid
|
|
CGP
|
comprehensive genomic profiling
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
Genexus
|
Ion Torrent™ Genexus™ Sequencer
|
|
NGS
|
next-generation sequencing
|
|
OCAv3
|
Genexus Oncomine Comprehensive Assay
v3
|
|
OPA
|
Genexus Oncomine Precision Assay
|
References
|
1
|
Heitzer E, Haque IS, Roberts CES and
Speicher MR: Current and future perspectives of liquid biopsies in
Genomics-driven oncology. Nat Rev Genet. 20:71–88. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tivey A, Church M, Rothwell D, Dive C and
Cook N: Circulating tumour DNA-looking beyond the blood. Nat Rev
Clin Oncol. 19:600–612. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kotani D, Oki E, Nakamura Y, Yukami H,
Mishima S, Bando H, Shirasu H, Yamazaki K, Watanabe J, Kotaka M, et
al: Molecular residual disease and efficacy of adjuvant
chemotherapy in patients with colorectal cancer. Nat Med.
29:127–134. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ignatiadis M, Sledge GW and Jeffrey SS:
Liquid biopsy enters the clinic-implementation issues and future
challenges. Nat Rev Clin Oncol. 18:297–312. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cristofanilli M, Hayes DF, Budd GT, Ellis
MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC,
et al: Circulating tumor cells: A novel prognostic factor for newly
diagnosed metastatic breast cancer. J Clin Oncol. 23:1420–1430.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cristofanilli M: Circulating tumor cells,
disease progression, and survival in metastatic breast cancer.
Semin Oncol. 33 (Suppl 9):S9–S14. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chae YK, Davis AA, Jain S, Santa-Maria C,
Flaum L, Beaubier N, Platanias LC, Gradishar W, Giles FJ and
Cristofanilli M: Concordance of genomic alterations by
Next-generation sequencing in tumor tissue versus circulating tumor
DNA in breast cancer. Mol Cancer Ther. 16:1412–1420. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dagogo-Jack I and Shaw AT: Tumour
heterogeneity and resistance to cancer therapies. Nat Rev Clin
Oncol. 15:81–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma F, Guan Y, Yi Z, Chang L, Li Q, Chen S,
Zhu W, Guan X, Li C, Qian H, et al: Assessing tumor heterogeneity
using ctDNA to predict and monitor therapeutic response in
metastatic breast cancer. Int J Cancer. 146:1359–1368. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Siravegna G, Mussolin B, Buscarino M,
Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu
A, Lauricella C, et al: Clonal evolution and resistance to EGFR
blockade in the blood of colorectal cancer patients. Nat Med.
21:8272015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murtaza M, Dawson SJ, Pogrebniak K, Rueda
OM, Provenzano E, Grant J, Chin SF, Tsui DWY, Marass F, Gale D, et
al: Multifocal clonal evolution characterized using circulating
tumour DNA in a case of metastatic breast cancer. Nat Commun.
6:87602015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rothé F, Laes JF, Lambrechts D, Smeets D,
Vincent D, Maetens M, Fumagalli D, Michiels S, Drisis S, Moerman C,
et al: Plasma circulating tumor DNA as an alternative to metastatic
biopsies for mutational analysis in breast cancer. Ann Oncol.
25:1959–1965. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diaz LA Jr and Bardelli A: Liquid
biopsies: Genotyping circulating tumor DNA. J Clin Oncol.
32:579–586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murtaza M, Dawson SJ, Tsui DWY, Gale D,
Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong AS,
et al: Non-invasive analysis of acquired resistance to cancer
therapy by sequencing of plasma DNA. Nature. 497:108–112. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Low SKK, Uchibori K, Hayashi R, Chan T,
Ariyasu R, Kitazono S, Yanagitani N, Nishio M and Nishio Y:
Evaluation of Genexus system that automates specimen-to-report for
cancer genomic profiling within a day using liquid biopsy. J Clin
Orthod. 38 (Suppl 15):S3538. 2020.
|
|
16
|
Casuga I, Chan F, Huynh M, Govoni GR,
Zochowski K, Jayaweera J and Au-Young J: Abstract 2944: Rapid and
accurate variant calling of FFPE samples with the Genexus System.
Cancer Res. 82:2944. 2022. View Article : Google Scholar
|
|
17
|
Guo F, Lang Y, Long G, Liu Z, Jing G, Zhou
Y, Zhang B and Yu S: Ion Torrent TM Genexus
TM Integrated Sequencer and ForeNGS Analysis Software-An
automatic NGS-STR workflow from DNA to profile for forensic
science. Forensic Sci Int Genet. 61:1027532022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong
V, Sandoval-Tan J, Ladrera G, Thongprasert S, Srimuninnimit V, Liao
M, et al: Detection and dynamic changes of EGFR mutations from
circulating tumor DNA as a predictor of survival outcomes in NSCLC
patients treated with First-line intercalated erlotinib and
chemotherapy. Clin Cancer Res. 21:3196–3203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zill OA, Greene C, Sebisanovic D, Siew LM,
Leng J, Vu M, Hendifar AE, Wang Z, Atreya CE, Kelley RK, et al:
Cell-Free DNA Next-generation sequencing in pancreatobiliary
carcinomas. Cancer Discov. 5:1040–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim ST, Lee WS, Lanman RB, Mortimer S,
Zill OA, Kim KM, Jang KT, Kim SH, Park SH, Park JO, et al:
Prospective blinded study of somatic mutation detection in
cell-free DNA utilizing a targeted 54-gene next generation
sequencing panel in metastatic solid tumor patients. Oncotarget.
6:40360–40369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Janku F, Angenendt P, Tsimberidou AM, Fu
S, Naing A, Falchook GS, Hong DS, Holley VR, Cabrilo G, Wheler JJ,
et al: Actionable mutations in plasma cell-free DNA in patients
with advanced cancers referred for experimental targeted therapies.
Oncotarget. 6:12809–12821. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morimoto Y, Matsuda S, Kawakubo H,
Nakamura K, Kobayashi R, Hisaoka K, Okui J, Takeuchi M, Aimono E,
Fukuda K, et al: ASO visual abstract: Tumor burden monitoring with
circulating tumor DNA during treatment in patients with esophageal
squamous cell carcinoma. Ann Surg Oncol. 30:37592023. View Article : Google Scholar
|
|
23
|
Morimoto Y, Matsuda S, Kawakubo H,
Nakamura K, Kobayashi R, Hisaoka K, Okui J, Takeuchi M, Aimono E,
Fukuda K, et al: Tumor burden monitoring with circulating tumor DNA
during treatment in patients with esophageal squamous cell
carcinoma. Ann Surg Oncol. 30:3747–3756. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kuderer NM, Burton KA, Blau S, Rose AL,
Parker S, Lyman GH and Blau CA: Comparison of 2 commercially
available Next-generation sequencing platforms in oncology. JAMA
Oncol. 3:996–998. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barata PC, Koshkin VS, Funchain P, Sohal
D, Pritchard A, Klek S, Adamowicz T, Gopalakrishnan D, Garcia J,
Rini B and Grivas P: Next-generation sequencing (NGS) of cell-free
circulating tumor DNA and tumor tissue in patients with advanced
urothelial cancer: A pilot assessment of concordance. Ann Oncol.
28:2458–2463. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jahangiri L and Hurst T: Assessing the
concordance of genomic alterations between circulating-free DNA and
tumour tissue in cancer patients. Cancers (Basel). 11:19382019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thompson JC, Yee SS, Troxel AB, Savitch
SL, Fan R, Balli D, Lieberman DB, Morrissette JD, Evans TL, Bauml
J, et al: Detection of therapeutically targetable driver and
resistance mutations in lung cancer patients by next-generation
sequencing of Cell-free circulating tumor DNA. Clin Cancer Res.
22:5772–5782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schwaederlé MC, Patel SP, Husain H, Ikeda
M, Lanman RB, Banks KC, Talasaz A, Bazhenova L and Kurzrock R:
Utility of genomic assessment of blood-derived circulating tumor
DNA (ctDNA) in patients with advanced lung adenocarcinoma. Clin
Cancer Res. 23:5101–5111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo Q, Wang J, Xiao J, Wang L, Hu X, Yu W,
Song G, Lou J and Chen J: Heterogeneous mutation pattern in tumor
tissue and circulating tumor DNA warrants parallel NGS panel
testing. Mol Cancer. 17:1312018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and Late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aran D, Sirota M and Butte AJ: Systematic
pan-cancer analysis of tumour purity. Nat Commun. 6:89712015.
View Article : Google Scholar : PubMed/NCBI
|















