Introduction
Breast cancer exhibits geographical variation as
regards incidence. In the US alone, in 2018, an estimated 266,120
women were diagnosed with breast cancer and an additional 40,920
women succumbed to the disease (1). By the year 2030, it has been
projected that there will be 294,000 new cases, thus making breast
cancer a growing public health concern (1). In Kashmir (North India), as per
population-based cancer registries (HCR) under the National Cancer
Registry (NCR) programme maintained in SMHS and in the Regional
Cancer Centre (RCC) SKIMS, cancer of the breast is the 2nd leading
type of cancer among females with an incidence of 16.3/100,000
population/year, which is less than what is reported for other
states in India, such as Mumbai, Delhi, Bangalore, Bhopal,
Ahmadabad, Kolkata and Chennai, where breast cancer is leading type
of cancer among women with an incidence of 25.8 per 100,000
individuals (2-4).
Breast cancer, although a heterogeneous disease, exhibits a high
cure rate, possibly due to increased screening, early detection and
the use of targeted anticancer therapies. Although a number of risk
factors are associated with the disease, such as early menarche,
late menopause, late-age marriages and nulliparity, the genesis of
the disease involves a number of genetically predisposed genes
(BRCA genes), tumour suppressors (TP53, Pten, p16) and oncogenes of
the mitogen-activated protein kinase (MAPK), phosphoinositide
3-kinase (PI3K) and insulin-like growth factor 1 (IGF-1) signalling
pathways (5).
BRAF is a member of the Raf-Ras-MAP kinase family
proteins, weighing 75-100 kDa. It is the most important activator
of MEK kinase in the Ras/Raf/MEK/ERK pathway (6). BRAF mutation is a typical cause of
aberrant extracellular-signal-regulated kinase (ERK) signalling
(7). BRAF mutation was first
reported in 2002 and 90% of the mutations found are missense
mutations at nucleotide 1796, which result in a valine to glutamic
acid substitution at codon 600(7). BRAFV600E mutation has been reported
at different frequencies in various neoplasms, such as malignant
melanoma (40-70%), colorectal carcinoma (5-22%), thyroid papillary
carcinoma (36-53%), glioma (11%), ovary serous carcinoma (30%),
lung adenocarcinoma (4%) and hairy cell leukaemia (100%) (8). It has been previously demonstrated
that 10% of breast cancer cell lines harbour BRAF mutations
(9), suggesting the possible
presence of BRAF mutations in breast cancer tissues as well. At
least to the best of our knowledge, only a few studies on BRAF
mutation detection in breast cancer tissues have been performed to
date using sequencing methods (10,11). The Sanger sequencing method is the
gold standard method for the detection of mutations; however, this
method not highly sensitive, costly and requires expensive
equipment and expertise (12). To
overcome these drawbacks, in this study, we utilized previously
developed highly sensitive mutation allele-specific multiplex PCR
(MASMP) as an effective surrogate method for evaluating BRAFV600E
mutations in breast cancer patients (13).
The aim of this study was to use MASMP as a simple
and sensitive screening tool for determining the frequency of
BRAFV600E mutation, its association with immune markers [oestrogen
receptor (ER), progesterone receptor (PR) and human epidermal
growth factor receptor (HER)] and indicators of prognosis and
outcome in patients with breast cancer from the Kashmir valley in
India, since such data from this region are not available in the
literature; we also discuss its clinical implications.
Materials and methods
Patient selection, histological and
immune marker evaluation
In this study, patients attending the Medical
Oncology Department at the SMHS Hospital of Government Medical
College (GMC), Srinagar, Jammu and Kashmir, India, between
February, 2015 and December, 2018 for breast cancer management,
were recruited in this study, with prior informed consent. A total
of 50 surgical tumour specimens and an equal number of adjacent
normal tissues were collected as the controls. The patients
underwent a histopathological examination to establish the clinical
profile. Patients who had received pre-operative chemotherapy or
hormone therapy were excluded. This study was approved by the
Institutional Ethics Committee of GMC. All cases were reviewed by a
pathologist with haematoxylin and eosin (H&E)-stained slides.
The histological grade was assessed using the Nottingham grading
system. Clinicopathological parameters evaluated in each case
included patient age at initial diagnosis, tumour histological
type, clinical tumour staging, lymph node metastasis and
histopathological grade. The immune markers, ER, PR and HER-2, were
also evaluated. ER/PR was considered positive even when ≥1% tumour
cells exhibited nuclear staining. HER-2 staining was analysed
according to the American Society of Clinical Oncology
(ASCO)/College of American Pathologists (CAP) guidelines (14). HER-2 immunostaining was considered
positive when strong (3+) membranous staining in at least 30% of
the tumour cells was observed, whereas cases with 0 to 1+ were
regarded as negative.
DNA isolation and MASMP analysis
High-molecular-weight DNA was isolated from the
tissue samples of the breast cancer patients by proteinase-K
digestion and phenol/chloroform extraction [Fan and Gulley
(15)]. The extracted genomic DNA
was analysed by agarose gel electrophoresis using a UV illuminator
(Crystal-BioGlow).
PCR primers
The BRAF mutation primers (BRAF-mut) were
designed based on the BRAF sequence, with a mismatched nucleotide
at the 3' end, so that wild-type BRAF was not amplified by
these primers. The length of the PCR products was 126 bp. In
addition, thromboxane A synthase 1 (TBXAS1) was selected as
a reference gene as an indicator for successful PCR amplification.
Information concerning these primers is presented in Table I.
 | Table IMutation allele-specific multiplex PCR
primer sequences for the BRAF gene. |
Table I
Mutation allele-specific multiplex PCR
primer sequences for the BRAF gene.
| Gene | Sequence | Product size
(bp) |
|---|
| BRAF-mut | F:
5'-GGTGATTTTGGTCTAGCTACAGA-3' | 126 |
| | R:
5'-GGCCAAAAATTTAATCAGTGG-3' | |
| TBXAS1 | F:
5'-GCCCGACATTCTGCAAGTCC-3' | 100 |
| | R:
5'-GGTGTTGCCGGGAAGGGTT-3' | |
MASMP
BRAF-mut and TBXAS1 genes were
amplified in the same 25-µl amplification system, which included 30
ng DNA templates, 10 pmol BRAF-mut primers, 5 pmol
TBXAS1 primers, 0.1 mol/dNTPs and 1.5 units Taq enzyme. The
thermal cycling protocol for PCR (TAKARA-thermal cycler) involved
an initial denaturation step at 95˚C for 5 min followed by 30
cycles at 94˚C for 30 sec, 54˚C for 30 sec and 72˚C for 30 sec.
Statistical analysis
All statistical analyses were performed using S-PLUS
software (version 8.2, TIBCO software Inc). The Chi-square test was
performed to determine the associations between the presence of
mutations with various clinicopathological characteristics, such as
age, origin, menopausal status, provisional diagnosis, lymph node/s
involved, clinical tumour stage and histopathological grade of the
tumour. The association between the presence of BRAF mutation with
the ER, PR and HER-2 status was also evaluated. P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinical data
In total, 50 breast cancer patients with a mean age
at diagnosis of 50.2±12.3 years were recruited in the present
study; 36% (18/50) of the patients were <45 years of age and 64%
(32/50) were >45 years of age. Among the studied subjects, 36%
(18/50) were pre-menopausal and 64% (32/50) were post-menopausal
females. The majority of the patients (88%; 44/50) were diagnosed
with infiltrating ductal carcinomas (IDCs), 8% with inflammatory
breast carcinomas (IBCs) and 4% with Paget's disease. The majority
of cases were of clinical tumour stage II (a,b) 70% (35/50)
followed by stage III and IV 30% (15/50). The lymph node was
involved in 80% (40/50) of the cases. Histopathological examination
revealed that the majority of cases, 80% (40/50) were of the well
and moderately differentiated grade. The majority of the patients
under study were ER/PR-positive (70%) and HER-2 negative (94%).
MASMP for BRAFV600E mutational
detection
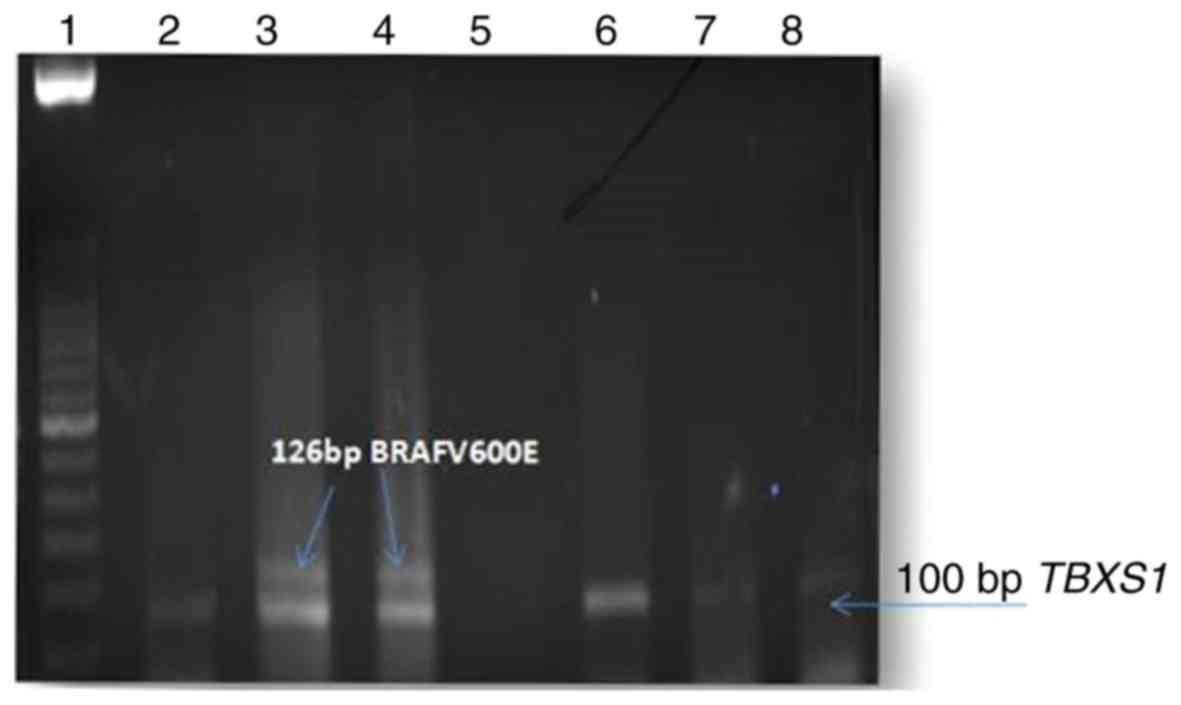
We assessed BRAFV600E/BRAFc.1799T>A
mutation in 50 breast carcinoma tissues and in an equal number of
adjacent normal tissues as controls by MASMP (Fig. 1). MASMP was repeated thrice for
each sample. The thick band of the 126 bp product was repeatedly
found in the same samples only and not in the controls or
presumably BRAFV600E-negative samples in the presence of the 100 bp
TBXAS1 reference gene. The authenticity of MASMP in picking up the
right product was previously confirmed by sequencing the product
using a reverse primer and the sensitivity of the MASMP method was
previously investigated and compared with direct DNA sequencing
[13]. The association of the BRAFV600E mutation status in the
breast cancer patients with the clinicopathological features and
immune markers, such as: ER/PR and HER-2 is summarised in Table II. The data regarding the ER/PR
and HER-2 status of these patients were collected from pathology
department and after reviewing the data in light of the ER/PR and
HER-2 status, it appeared that BRAFV600E/BRAFc.1799T>A
mutation was significantly present in ER/PR-negative
(χ2=4.36, P=0.03) and mostly in triple-negative breast
cancers (TNBCs) (χ2=2.5, P=0.11). In addition, although
not significant, BRAFV600E-positive breast cancers were mostly
found in older-aged (χ2=1.10, P=0.29) and in
post-menopausal women (χ2=1.10, P=0.29). No significant
association was found between BRAFV600E-mutated breast cancers and
traditional poor prognostic factors, such as clinical tumour stages
III and IV (χ2 0.036, P=0.84) and a poorly
differentiated (PD) histopathological grade (χ2 0.04,
P=0.82). However, there is no information regarding KI67, and thus
we could not classify BRAF-mutated ER-positive samples into luminal
A or luminal B molecular subtypes.
 | Table IIAssociation of BRAF gene mutation with
clinicopathological features and immune markers of sporadic breast
cancer patients from Kashmir, India (n=50). |
Table II
Association of BRAF gene mutation with
clinicopathological features and immune markers of sporadic breast
cancer patients from Kashmir, India (n=50).
| | | BRAF mutation | | |
|---|
| Feature | Cases | Positive | Negative | χ2 | P-value |
|---|
| Age | | | | | |
|
<45
years | 18 | 1 | 17 | 1.10 | 0.29 |
|
<45
years | 32 | 5 | 27 | | |
| Living
environment | | | | | |
|
Rural | 30 | 3 | 27 | 0.28 | 0.5 |
|
Urban | 20 | 3 | 17 | | |
| Menopausal
status | | | | | |
|
Pre-menopausal | 18 | 1 | 17 | 1.10 | 0.29 |
|
Post-menopausal | 32 | 5 | 27 | | |
| Provisional
diagnosisa | | | | | |
|
IDC | 44 | 5 | 39 | 0.14 | 0.70 |
|
IBC + | 4 | 1 | 3 | | |
|
Paget's
disease | 2 | 0 | 2 | | |
| Lymph node
involvement | | | | | |
|
Yes | 40 | 4 | 36 | 0.75 | 0.38 |
|
No | 10 | 2 | 8 | | |
| Clinical tumour
stagingb | | | | | |
|
II
(a,b) | 35 | 4 | 31 | 0.036 | 0.84 |
|
III +
IV | 15 | 2 | 13 | | |
| Histopathological
gradingc | | | | | |
|
WD + MD | 40 | 5 | 35 | 0.04 | 0.82 |
|
PD | 10 | 1 | 9 | | |
| ER/PR
statusd | | | | | |
|
Strong +
weak positive | 35 | 2 | 33 | 4.36 | 0.03 |
|
Negative | 15 | 4 | 11 | | |
| HER-2
statusd | | | | | |
|
Positive | 3 | 1 | 2 | 1.37 | 0.24 |
|
Negative | 47 | 5 | 42 | | |
|
TNBC
(ER-, PR-, HER-) | | | | | |
|
Yes | 21 | 3 | 9 | 2.5 | 0.11 |
|
No | 38 | 3 | 35 | | |
Discussion
A wealth of data now suggest that molecular
aberrations may be shared across multiple histologies. As an
example, BRAF is mutated in approximately 15% of all cancers,
either in solid tumours, haematological malignancies and related
disease types (7,16). However, the frequencies vary; in
some, it is more frequently present in cancers such as melanoma
(40-60%) and hairy cell leukaemia (100%) patients (12,17). In others types of cancer, such as
breast cancer, the BRAFV600E mutation is uncommon (0-13%) although
BRAF amplification has been reported in 30% of basal-like
carcinomas (10,11). In the present study, BRAFV600E
mutation has been found in 12% of breast cancer patients, which is
in conformity with that of Korea, but relatively higher than that
reported worldwide (18).
Although certain studies have indicated geographical and
histological subtype classification factors that may account for
these differences in frequencies, the reliability of the detection
methods used must also be taken into consideration (19,20). The gold standard method for the
detection of BRAF mutation is Sanger sequencing and other PCR-based
methods, such as single-strand conformation polymorphism,
restriction fragment length polymorphism (RFLP), mass-array
spectrometry, pyrosequencing. However, these methods require
expensive equipment, high technical skills and are associated with
other problems, such as tissue heterogeneity and sampling error.
These drawbacks limit their general use in the clinical field
(20). To overcome the
above-mentioned limitations, in this study, we used previously
developed MASMP for BRAFV600E mutational detection in breast
cancers (13). It has been
previously demonstrated by us and others that the detection
sensitivity of MASMP technique is higher than that of direct DNA
sequencing (12,13). Our frequency (12%) of breast
cancers harbouring the BRAFV600E mutation by MASMP was higher than
that reported worldwide by various methodologies, but comparable to
the immunohistochemistry (IHC) method of detection (13%) using
BRAFV600E mutation-specific antibody (18). As previously demonstrated, while
direct sequencing picks up altered alleles from the sample only
when the tumour purity is >60%, MASMP picks up variations even
if the tumour purity is <1% which may be the reason of the high
frequency of mutations detected in the present study (12). These results, therefore, suggest
that MASMP is a reliable method of mutation detection in clinical
samples.
Association analysis revealed that BRAFV600E-mutated
breast cancers were characterized by unique
clinical-epidemiological and pathological characteristics. Although
the absolute number in this study was small, several characteristic
features of BRAFV600E-mutated breast cancers have become apparent.
A casual association between BRAFV600E-positive breast cancers and
an older age (>45 years; χ2=1.10, P=0.29) and the
post-menopausal status of women (χ2=1.10, P=0.29) was
observed in the present study (Table
II). In addition, a casual association between
BRAFV600E-mutated breast cancer and infiltrating ductal carcinoma
and lymph node involvement was evident from the present study
(Table II), which is in
conformity with various other studies (18). No significant association was
found between BRAFV600E-mutated breast cancer was found and
traditional poor prognostic factors, such as clinical tumour stages
III and IV (χ2=0.036 P=0.84) and the PD
histopathological grade (χ2=0.04, P=0.82). In previous
studies, BRAF mutation in different cancer types has been reported
to be associated with several clinicopathologic features, such as
lymph node metastasis and distant metastasis in papillary thyroid
cancer (PTC); younger age and tumour occurrence from intermittently
sun-exposed skin in malignant melanoma; poor prognosis in colon
cancer which are different from the result from our study regarding
breast cancer (21,22). In non-small cell lung cancer, the
BRAF mutation status has not been associated with a clinical
outcome, such as the result of this study with breast cancer
(22). Further association
analysis of immune markers revealed that BRAFV600E-positive breast
cancers were significantly observed in ER/PR-negative
(χ2=4.36, P=0.03) and in the TNBC (χ2=2.5,
P=0.11) subtypes, which is in conformity, to a certain extent, to
worldwide reports, which have found that BRAFV600E mutation
predominantly in TNBC molecular subtype (11,18).
The clinical implication of this study is the
potential use of the BRAF mutation status for the application of
BRAF targeted therapy in breast carcinomas. Drugs targeting BRAF
mutation, such as vemurafenib and dabrafenib, have been approved
for BRAF-mutant melanoma based on the results from the phase III
BRIM-3 study (23) and the phase
III BREAK-3 study (24),
respectively. Since the approval of vemurafenib for
BRAFV600E-mutated melanoma, accumulating evidence presented in
published reports supports the hypothesis that what works for
BRAFV600E-mutated melanoma is often also effective for other
cancers characterized by the BRAFV600E aberration. For example,
dabrafenib for BRAFV600E mutation-positive metastatic non-small
cell lung cancer (with a response rate of 54%) [non-small cell lung
cancer (25)]; BRAFV600E-positive
papillary thyroid cancer (26)];
BRAFV600E-positive gastrointestinal stromal tumours (27). Complete regression with
vemurafenib has been attained in a child with glioblastoma
harbouring the BRAFV600E mutation (28), in BRAFV600E-mutated glioma
patients (29), in patients with
BRAFV600E-mutated hairy cell leukaemia (9,30),
and in BRAFV600E mutated Erdheim-Chester disease (31). These studies confirm that multiple
histological types of cancer with BRAF mutations respond to BRAF
inhibitors, although the precise response rates may differ. Based
on the results from the present study and worldwide reports,
ER/PR-negative and TNBC molecular subtypes harbour comparatively
BRAF mutations or amplifications at a higher frequency. As these
subtypes of breast cancer cannot respond to traditional endocrine
therapy, BRAF targeted therapy can thus be considered as a possible
treatment for these. Thus, screening patients for BRAF mutations
may aid in the selection of the initial therapy mode and in the
follow-up of these molecular subtypes of breast cancer.
In conclusion, in this study, the positivity for
BRAFV600E was noted in a fraction of elderly post-menopausal women,
predominantly of the ER/PR-negative and TNBC molecular subtype.
MASMP turned out to be a simple and sensitive for the rapid
identification of BRAFV600E-mutated breast cancers for which BRAF
targeted therapy, in place of endocrine therapy, can be considered
a possible targeted treatment in the future. The present study,
however, is a preliminary report and studies using larger patient
populations are warranted to substantiate the results of this
study.
Acknowledgements
The authors gratefully acknowledge the Pathology
Department and Multidisciplinary Research Unit (MRU) of GMC
(Government Medical College) Srinagar for providing the equipment
for conducting the experiments.
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
SF performed the research work. RE designed the
research study, analysed the data and wrote the manuscript. RR
assisted in the selection of histopathologically confirmed breast
cancer samples along with their ER/PR/HER-2 status, and in data
analysis. MUR, TR and NAN performed the research work. SA and AM
analysed the data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional Ethics
Committee of Government Medical College and all patients provided
informed consent prior to enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics 2018. CA Cancer J Clin. 68:7–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wani MA, Jan FA, Khan NA, Pandita KK,
Khurshid R and Khan SH: Cancer trends in Kashmir; common types,
site incidence and demographic profiles: National Cancer Registry
2000-2012. Indian J Cancer. 51:133–137. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Qureshi MA, Khan SM, Masoodi MA, Qureshi
U, Ain Q, Jan Y, Haq I and Sheikh ZA: Epidemiology of cancers in
Kashmir, India: An analysis of hospital data. Adv Prev Med.
2016(1896761)2016. View Article : Google Scholar
|
|
4
|
Malvia S, Bagadi SA, Dubey US and Saxena
S: Epidemiology of breast cancer in Indian women. Asia Pac J Clin
Oncol. 13:289–295. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature 490: 61-70,
2012.
|
|
6
|
Garnett MJ and Marais R: Guilty as
charged: B-RAF is a human oncogene. Cancer Cell. 6:313–319.
2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hall RD and Kudchadkar RR: BRAF mutations:
Signalling, epidemiology, and clinical experience in multiple
malignancies. Cancer Control. 21:221–230. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hollestelle A, Elstrodt F, Nagel JH,
Kallemeijn WW and Schutte M: Phosphatidylinositol-3-OH kinase or
RAS pathway mutations in human breast cancer cell lines. Mol Cancer
Res. 5:195–201. 2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tilch E, Seidens T, Cocciardi S, Reid LE,
Byrne D, Simpson PT, Vargas AC, Cummings MC, Fox SB, Lakhani SR and
Chenevix Trench G: Mutations in EGFR, BRAF and RAS are rare in
triple-negative and basal-like breast cancers from Caucasian women.
Breast Cancer Res Treat. 143:385–392. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Santarpia L, Qi Y, Stemke-Hale K, Wang B,
Young EJ, Booser DJ, Holmes FA, O'Shaughnessy J, Hellerstedt B,
Pippen J, et al: Mutation profiling identifies numerous rare drug
targets and distinct mutation patterns in different clinical
subtypes of breast cancers. Breast Cancer Res Treat. 134:333–343.
2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sapio MR, Posca D, Troncone G, Pettinato
G, Palombini L, Rossi G, Fenzi G and Vitale M: Detection of BRAF
mutation in thyroid papillary carcinomas by mutant allele-specific
PCR amplification (MASA). Eur J Endocrinol. 154:341–348.
2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Eachkoti R, Farooq S, Syeed SI, Wani HA,
Majid S and Pampori MR: Prevalence and prognostic relevance of
BrafV600E mutation in colorectal carcinomas from Kashmir (North
India) valley. Mutagenesis. 33:225–230. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bui MM, Riben MW, Allison KH, Chlipala E,
Colasacco C, Kahn AG, Lacchetti C, Madabhushi A, Pantanowitz L,
Salama ME, et al: Quantitative image analysis of human epidermal
growth factor receptor 2 immunohistochemistry for breast cancer:
Guideline from the college of american pathologists. Arch Pathol
Lab Med. Jan 15. 2019.(Epub ahead of print). PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fan H and Gulley ML: DNA extraction from
fresh or frozen tissues. Methods Mol Med. 49:5–10. 2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
El-Osta H, Falchook G, Tsimberidou A, Hong
D, Naing A, Kim K, Wen S, Janku F and Kurzrock R: BRAF mutations in
advanced cancers: Clinical characteristics and outcomes. PLoS One.
6(e25806)2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tiacci E, Trifonov V, Schiavoni G, Holmes
A, Kern W, Martelli MF, Pucciarini A, Bigerna B, Pacini R, Wells
VA, et al: BRAF mutations in hairy-cell leukemia. N Engl J Med.
364:2305–2315. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jung YY, Jung WH and Koo JS: BRAF mutation
in breast cancer by BRAF V600E mutation-specific antibody. Int J
Clin Exp Pathol. 9:1545–1556. 2016.
|
|
19
|
Turski ML, Vidwans SJ, Janku F,
Garrido-Laguna I, Munoz J, Schwab R, Subbiah V, Rodon J and
Kurzrock R: Genomically driven tumors and actionability across
histologies: BRAF-Mutant cancers as a paradigm. Mol Cancer Ther.
15:533–547. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bösmüller H, Fischer A, Pham DL, Fehm T,
Capper D, von Deimling A, Bonzheim I, Staebler A and Fend F:
Detection of the BRAF V600E mutation in serous ovarian tumors: A
comparative analysis of immunohistochemistry with a
mutation-specific monoclonal antibody and allele-specific PCR. Hum
Pathol. 44:329–335. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
COSMIC: Catalogue of Somatic Mutations in
Cancer. Wellcome Trust Sanger Institute; http://cancer.sanger.ac.uk/cancer-genome/projects/cosmic.
Accessed January 2, 2014.
|
|
22
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6(pl1)2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et
al: Improved survival with vemurafenib in melanoma with BRAF V600E
mutation. N Engl J Med. 364:2507–2516. 2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hauschild A, Grob JJ, Demidov LV, Jouary
T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH Jr,
Kaempgen E, et al: Dabrafenib in BRAF-mutated metastatic melanoma:
A multicentre, open-label, phase 3 randomized controlled trial.
Lancet. 380:358–365. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tafinlar® receives FDA
Breakthrough Therapy designation for non-small cell lung cancer
with BRAF mutation. GlaxoSmithKline, London, 2013. http://us.gsk.com/en-us/media/press-releases/2014/tafinlar-receives-FDA-breakthrough-therapy-designation-for-non-small-cell-lung-cancer-with-braf-mutation/"ref-label" rowspan="1" colspan="1">
26
|
Kim KB, Cabanillas ME, Lazar AJ, Williams
MD, Sanders DL, Ilagan JL, Nolop K, Lee RJ and Sherman SI: Clinical
responses to vemurafenib in patients with metastatic papillary
thyroid cancer harboring BRAF(V600E) mutation. Thyroid.
23:1277–1283. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Falchook GS, Trent JC, Heinrich MC,
Beadling C, Patterson J, Bastida CC, Blackman SC and Kurzrock R:
BRAF mutant gastrointestinal stromal tumor: First report of
regression with BRAF inhibitor dabrafenib (GSK2118436) and whole
exomic sequencing for analysis of acquired resistance. Oncotarget.
4:310–315. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Robinson GW, Orr BA and Gajjar A: Complete
clinical regression of a BRAF V600E-mutant pediatric glioblastoma
multiforme after BRAF inhibitor therapy. BMC Cancer.
14(258)2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bautista F, Paci A, Minard-Colin V, Dufour
C, Grill J, Lacroix L, Varlet P, Valteau-Couanet D and Geoerger B:
Vemurafenib in pediatric patients with BRAFV600E mutated high-grade
gliomas. Pediatr Blood Cancer. 61:1101–1103. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Samuel J, Macip S and Dyer MJ: Efficacy of
vemurafenib in hairy-cell leukemia. N Engl J Med. 370:286–288.
2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Munoz J, Janku F, Cohen PR and Kurzrock R:
Erdheim-Chester disease: Characteristics and management. Mayo Clin
Proc. 89:985–996. 2014.PubMed/NCBI View Article : Google Scholar
|