Introduction
Coronavirus disease 2019 (COVID-19) is caused by
severe acute respiratory coronavirus 2 (SARS-CoV-2) (1). SARS-CoV-2 has led to a serious
pandemic worldwide and has become a burden borne by health care
systems since its first outbreak in Wuhan, China, in 2019(2).
With the rapid spread of COVID-19 worldwide,
researchers are working on the development of vaccines and
effective therapeutics; however, the number of new cases and deaths
continues to increase at unfavorable rates (3,4). In
view of the high mortality rates of severe cases, rapid reliable
risk stratification tools and sensitive indicators of prognosis are
urgently required for timely disease monitoring and efficient
interventions targeting the reduction of morbidity and mortality
(5).
The initial clinical picture of several patients
with COVID-19 appears to be non-specific. Patients may present with
minimal symptoms and no radiological abnormalities. In certain
cases, rapid disease progression may occur, leading to acute
respiratory distress syndrome (ARDS), multiple organ failure and
even death (4,5). A cytokine storm has been identified in
several studies as a key factor causing COVID-19 exacerbation or
mortality (6). Several inflammatory
factors, coagulation parameters and cytokines have been proposed as
potential biomarkers of disease progression, occurrence of cytokine
storm and severity (7).
Serum amyloid A (SAA) is an important potential
biomarker. SAA is a highly conserved acute-phase protein primarily
produced by the liver in response to inflammatory cytokines such as
IL-6, IL-1, TNF, IFN-γ and TGF-β. SAA levels are associated with
the severity of inflammation (8).
In addition, SAA is the precursor of the amyloid A protein, which
is a fibrillar, insoluble product that is deposited in major
organs, thus contributing to organ failure and death during the
course of secondary amyloidosis (9).
Another important potential biomarker is
carcinoembryonic antigen (CEA); a glycoprotein formed in the
respiratory and colonic epithelium during embryogenesis, and it has
been widely utilized as a tumor marker to monitor tumor progression
(10). CEA is related to
respiratory or digestive cancers and infectious diseases, such as
gonorrhea, or chronic inflammatory diseases such as interstitial
lung diseases (ILD) (11).
Immunohistochemical staining of lung specimens from patients with
pulmonary fibrosis demonstrated strong expression of CEA in the
metaplastic bronchiolar and type II alveolar epithelia (12). Significant hyperplasia of type II
alveolar epithelial cells and interstitial fibrosis have been
described in several reports of COVID-19 autopsies and biopsies,
similar to the pathological changes observed in ILD (13).
In the present study, SAA and CEA were evaluated as
potential prognostic biomarkers in comparison to other commonly
used inflammatory predictors, including C-reactive protein (CRP)
and ferritin, and their association with the severity of COVID-19
and CT scan findings, and whether they may be a beneficial tool for
patient stratification was assessed.
Materials and methods
Patients
This cross-sectional study included 124 patients
diagnosed with COVID-19 enrolled from the Ain Shams University
Isolation Hospital (Cairo, Egypt). In this study cohort, the median
age was 48 years and the 25-75th IQR was 40-56 years (age range,
25-87 years). A total of 10 patients were >65 years old,
constituting 8% of the entire cohort. Of the 124 cases, 98 (79%)
were men and 26 were women (21%). The median male age was 48.5
years (IQR, 40-54) and the median female age was 44.5 years (IQR,
35-57). Data were collected from hospitalized patients between
September 2020 and February 2021. Patients were diagnosed and
categorized into mild and severe groups according to the World
Health Organization interim guidelines (14).
A definite COVID-19 case was identified as a
positive result using sequencing or reverse
transcription-quantitative PCR (RT-qPCR) of nasopharyngeal
swabs.
The patients were classified into 2 groups: i) Mild
group, had clinical symptoms of fever, fatigue, cough, anorexia,
malaise, muscle pain, sore throat, dyspnea, nasal congestion and/or
a headache; and ii) Severe group, had respiratory distress, a
respiratory rate of ≥30 breaths/min at resting state, a mean oxygen
saturation of ≤93% and an arterial blood oxygen partial pressure
(PaO2)/oxygen concentration (FiO2) of ≤300 mmHg.
The exclusion criteria were as follows: i) Patients
infected with other viruses or bacteria; ii) patients diagnosed
with autoimmune disorders; iii) patients diagnosed with arthritic
diseases and iv) patients with cancer and/or any chronic disease
related to elevated CEA and/or SAA levels, such as chronic kidney
disease (15).
Ethical considerations
This study was conducted in accordance with the
principles outlined in the Declaration of Helsinki of the World
Medical Association (16). The
study was approved by the Institutional Ethics Committee of Ain
Shams University. Informed consent was obtained from all enrolled
participants after receiving an explanation of the study's aim and
procedures.
Methods
The recruited cohort consisted of 124 patients
diagnosed with COVID-19 at the Ain Shams University Hospital. A
detailed history was obtained for all patients, with a particular
emphasis on age, sex, duration of disease and clinical symptoms.
Data from routine investigations were retrieved, including liver
enzyme levels [alanine transaminase (ALT) and aspartate
transaminase (AST)], kidney function tests [blood urea nitrogen
(BUN), creatinine], inflammatory parameters (CRP, ferritin) and
D-Dimer levels, in addition to complete blood count (CBC),
hemoglobin, white blood count (WBC), neutrophil count, lymphocyte
count and platelet (PLT) count (15).
Detection of viral RNA was performed using the
CerTest ViasureVR SARS-CoV-2 RT-qPCR Detection kit (CerTest,
Biotec) according to the manufacturer's instructions. The detection
was performed in a one-step real-time reverse-transcription format,
where the reverse transcription and subsequent amplification of a
specific target sequence occurred in the same reaction well. The
isolated RNA target was transcribed to generate cDNA using the
included reverse transcriptase, followed by the amplification of a
conserved region of the open reading frames (ORF) 1 ab and N genes
for SARS-CoV-2 using specific primers and a fluorescent-labelled
probe, all of which were included in the kit. The assay has a 97.5%
sensitivity and >99.9% specificity. The average estimated limit
of detection for SARS-CoV-2 was 18 copies/ml.
Blood samples from PCR-positive patients were
obtained, left to clot completely, and centrifuged at 3,000 x g for
20 min at 4˚C, and stored at -80˚C until required. SAA level
analysis was performed using the Invitrogen human SAA ELISA kit
(cat. no. EHSAA1; Invitrogen; Thermo Fisher Scientific, Inc.). The
detection limit of this assay is 0.004 mg/l. CEA serum
concentrations were assayed using a Cobas e411 immunoassay
autoanalyzer (Roche Diagnostics GmbH).
Computed tomography (CT)
All patients underwent non-contrast-enhanced chest
CT in the Radiology Department of Ain Shams University, which was
performed by expert radiologists using a Siemens 16-channel scope
(CTAWP92544; Siemens Healthineers). The following CT parameters
were evaluated: COVID-19 Reporting and Data System (CO-RADS) score
based on CT findings (17): i) The
CO-RADS score represents the level of suspicion of COVID-19.
CO-RADS 1, COVID-19 is highly doubtful, CT is normal, or findings
representing a non-infectious disease; CO-RADS 2, low level of
suspicion of COVID-19 infection, CT findings consistent with other
infections; CO-RADS 3, COVID-19 infection is indeterminate and
unsure whether CT abnormalities are caused by COVID-19; CO-RADS 4,
high suspicion level, and most CT findings are not extremely
typical; CO-RADS 5, high level of suspicion with typical CT
findings. ii) Semi quantitative scoring system: A quantitative
estimate of pulmonary involvement based on abnormalities in the
areas involved. The CT-severity score (CT-SS) is based on the
extent of lobar involvement. Each of the five lung lobes was
visually scored from 0-5 as follows: 0, no involvement; 1, <5%
involvement; 2, 5-25% involvement; 3, 26-49% involvement; 4, 50-75%
involvement; and 5, ≥75% involvement. The total CT score was the
sum of individual lobar scores and ranged from 0 (no involvement)
to 25 (maximum) (18).
Statistical analysis
Statistical analysis was performed using SPSS
version 25.0 (IBM Corp.). Continuous variables are presented as the
median and interquartile range (IQR), whereas categorical variables
are presented as the number (n) and percentage (%) of patients. A
Wilcoxon rank sum test was used to evaluate differences between
groups. A χ2 was used for analysis of sets of
categorical data, and a Spearman's rank correlation test was used
to measure the degree of correlation between the hierarchically
ordered variables in this study. A multivariate regression analysis
was performed. The selection of independent variables as potential
predictors of COVID-19 severity and CT-SS included age, duration of
disease, CBC (WBC, neutrophil, lymphocyte, PLT, monocyte,
eosinophil count and hemoglobin), creatinine, AST, ALT, BUN, CRP,
ferritin, D-Dimer, CEA and SAA. Stepwise multivariate-regression
analysis was performed including for all studied items as
independent variables (Model-1). The regression was re-run using
only the most significant items with exclusion of the
non-significant items iteratively. The least sensitive predictors
for the model had the highest F-ratio and the lowest P-value.
P<0.05 was considered to indicate a statistically significant
difference. Receiver operating characteristic (ROC) curves were
used for predictive analysis by calculating the area under the
curve (AUC), sensitivity and specificity.
Results
A total of 124 patients whose RT-qPCR tests for
COVID-19 were positive were recruited, their baseline demographics,
clinical characteristics, laboratory, radiological CO-RADS score
and CT-SS parameters are summarized in Table I, including age, duration of
disease, symptoms, laboratory parameters including CBC, liver
enzymes (AST and ALT), kidney function tests (BUN and creatinine),
inflammatory parameters (CRP, ferritin, and D-Dimer), in addition
to SAA and CEA levels. In this study cohort, the median age was 48
years and the 25-75th IQR was 40-56 years (age range, 25-87 years).
A total of 10 patients were >65 years old, constituting 8% of
the entire cohort. Of the 124 cases, 98 (79%) were men and 26 were
women (21%). The median male age was 48.5 years (IQR, 40-54) and
the median female age was 44.5 years (IQR, 35-57) (P>0.05).
 | Table IDemographics and baseline clinical,
laboratory and radiological parameters of the COVID-19
patients. |
Table I
Demographics and baseline clinical,
laboratory and radiological parameters of the COVID-19
patients.
| | Interquartile
range |
|---|
| Parameters | Value | Range | 25th
percentile | 75th
percentile |
|---|
| Age, years | 48 | 25-87 | 40 | 56 |
| Sex, n (%) | | - | - | - |
|
Male | 98(79) | | | |
|
Female | 26(21) | | | |
| Clinical
parameters | | | | |
|
Duration of
disease, days | 7 | 1-14 | 4 | 10 |
|
Fever, n
(%) | 118 (95.2) | - | - | - |
|
Cough, n
(%) | 108 (87.1) | - | - | - |
|
Loss of
smell and taste, n (%) | 14 (11.3) | - | - | - |
|
Dyspnea, n
(%) | 74 (59.7) | - | - | - |
|
Respiratory
distress, n (%) | 39 (31.5) | - | - | - |
|
Diarrhea, n
(%) | 8 (6.6) | - | - | - |
| Laboratory
parameters | | | | |
|
White blood
cell count, 109/l | 6.9 | 2.6-17 | 4.3 | 10.3 |
|
Neutrophil
count, 109/l | 4.5 | 1.5-13.6 | 2.82 | 7.2 |
|
Lymphocyte
count, 109/l | 1.22 | 0.46-4.38 | 0.841 | 1.6 |
|
Platelet
count, 109/l | 232 | 50-649 | 191 | 295 |
|
Hemoglobin,
g/dl | 14.2 | 8.0-17.6 | 13 | 15.1 |
|
Blood urea
nitrogen, mg/dl | 14 | 7.5-46 | 11.7 | 19.2 |
|
Creatinine,
mg/dl | 0.8 | 0.47-7.5 | 0.7 | 1.04 |
|
Aspartate
transaminase, U/l | 26 | 12-116 | 17 | 43 |
|
Alanine
transaminase, U/l | 32 | 11-346 | 16 | 54 |
|
C-reactive
protein, mg/l | 43 | 1.8-444.0 | 18 | 161.3 |
|
Ferritin,
ng/ml | 361 | 24-2,450 | 203.75 | 839.75 |
|
D-Dimer,
ng/ml | 500 | 90-6088 | 300 | 1200 |
|
Serum
amyloid A, mg/l | 22.5 | 3-90 | 10 | 49 |
|
Carcinoembryonic
antigen, ng/ml | 6 | 3-18 | 5 | 9 |
| Radiological
parameters | | | | |
|
CO-RADS
score | 3 | 0-5 | 1 | 4 |
|
Computed
tomography-severity score | 8.5 | 0-25 | 3 | 16 |
All patients were divided into two groups as shown
in Table II: Mild or severe
COVID-19 infection. Of the 124 patients, 58 patients (46.7%) were
classified as having mild COVID-19, with a median age of 43 years
(IQR, 35-52.5) and 66 patients (53.3%) were classified as having
severe COVID-19, with a median age of 50 years (IQR, 43-56); this
showed a highly significant statistical difference between both
groups with median ages higher in the severe group (P<0.001). In
the severe COVID-19 group, 7 out of 66 (10.6%) patients were >65
years old compared to three cases out of 58 (5.2%) in the mild
group.
 | Table IIComparison between mild and severe
COVID-19 patients. |
Table II
Comparison between mild and severe
COVID-19 patients.
| Parameter | Mild, n=58 | Severe, n=66 | Z | χ2 | P-value | Significance |
|---|
| Age, years
(IQR) | 43 (35-52.5) | 50 (43-56) | -3.24 | 2.88 | <0.001 | HS |
| Sex, n (%) | | | | | >0.05 | NS |
|
Male | 42/58 (72.4) | 56/66 (84.8) | | | | |
|
Female | 16/58 (27.6) | 10/66 (15.2) | | | | |
| Clinical
parameters | | | | | | |
|
Duration of
disease, days (IQR) | 5 (4-9) | 8 (6-11) | -3.7 | | <0.001 | HS |
|
Fever, n
(%) | 52 (89.7) | 66(100) | | 7.175 | <0.05 | S |
|
Cough, n
(%) | 48 (82.7) | 60 (90.9) | | 1.825 | >0.05 | NS |
|
Loss of
smell, n (%) | 4 (6.8) | 10 (15.1) | | 2.1 | >0.05 | NS |
|
Dyspnea, n
(%) | 26 (44.8) | 48 (72.7) | | 9.986 | <0.05 | S |
|
Respiratory
distress, n (%) | 1 (1.7) | 38 (57.6) | | 41.39 | <0.001 | HS |
|
Diarrhea, n
(%) | 4 (6.8) | 4 (6.1) | | 0.036 | >0.05 | NS |
| Laboratory
parameters | | | | | | |
|
White blood
cell count, 109/l, median (IQR) | 5.5 (4.3-10.5) | 7.8 (4.8-10.4) | -1.02 | | >0.05 | NS |
|
Neutrophil
count, 109/l, median (IQR) | 4 (2.7-6.9) | 5.5 (2.9-8.3) | -1.66 | | >0.05 | NS |
|
Lymphocyte
count, 109/l, median (IQR) | 1.24 (0.9-1.7) | 1.1 (0.8-1.5) | -0.62 | | >0.05 | NS |
|
Platelet
count, 109/l, median (IQR) | 229 (194-259) | 246 (168-330) | -0.62 | | >0.05 | NS |
|
Hemoglobin,
g/dl, median (IQR) | 14.2
(11.9-15.4) | 14 (13.2-15) | -0.58 | | >0.05 | NS |
|
Blood urea
nitrogen, mg/dl, median (IQR) | 14 (11.5-16.9) | 15.9 (11.7-22) | -2.16 | | <0.05 | S |
|
Creatinine,
mg/dl, median (IQR) | 0.8 (0.7-1.03) | 0.9 (0.7-1.05) | -0.54 | | >0.05 | NS |
|
Aspartate
transaminase, U/l, median (IQR) | 23
(15.5-37.25) | 34 (18-43.25) | -1.85 | | >0.05 | NS |
|
Alanine
transaminase, U/l, median (IQR) | 27 (15-51) | 34 (17-54) | -0.96 | | >0.05 | NS |
|
C-reactive
protein, mg/l, median (IQR) | 22 (11.8-48.5) | 83 (29-222) | -4.51 | | <0.001 | HS |
|
Ferritin,
ng/ml, median (IQR) | 227 (127-290) | 778
(455-1,225) | -7.86 | | <0.001 | HS |
|
D-Dimer,
ng/ml, median (IQR) | 336 (230-451) | 1200
(569-1,451) | -7.09 | | <0.001 | HS |
|
Serum
amyloid A, mg/l, median (IQR) | 10 (7-12) | 48.5 (34-59) | -9.28 | | <0.001 | HS |
|
Carcinoembryonic
antigen, ng/ml, median (IQR) | 5 (5-7) | 9 (6-13) | -6.04 | | <0.001 | HS |
| Radiological
parameters | | | | | | |
|
CO-RADS
score, median (IQR) | 1 (0.2) | 4 (3-5) | -8.1 | | <0.001 | HS |
|
Computed
tomography-severity score | 3 (2-8) | 15 (8.7-20) | -6.82 | | <0.001 | HS |
All studied parameters were compared in both groups
and are presented in Table II.
Patients with severe COVID-19 (n=66) showed a statistically
significant higher median disease duration of 8 days (IQR, 6-11),
while the remaining 58 mild cases had a median duration of disease
of 5 days (IQR, 4-9) (P<0.001). Regarding laboratory parameters,
such as total WBC, neutrophil, lymphocyte counts, PLT, creatinine,
ALT and AST, there were no significant differences between the
groups (P>0.05; Table II). In
the entire cohort, it was noticed that high serum levels of
inflammatory parameters, including CRP, ferritin and D-Dimer, as
well as SAA and CEA, were predictors of disease severity (Table I); their levels in severe COVID-19
group were significantly higher than that in the mild group
(P<0.001). Patients with mild COVID-19 showed median SAA levels
of 10 ng/ml (IQR, 7-12), while in the severe COVID-19 group, the
median SAA was 48.5 (IQR, 34-59), which was significantly higher
than that in the mild group (P<0.001). The median CEA level was
5 ng/ml (IQR,5-7) while in the severe group, the median CEA level
was 9 (IQR, 6-13), which was significantly higher than that in the
mild group (P<0.001) Table II.
The median CT-SS was significantly higher in the severe group
(median, 15; IQR, 8.75-20) than in the mild COVID-19 group (median,
3; IQR, 2-8) (P<0.001).
Using Spearman's rank correlation analysis, SAA
showed a highly significant positive correlation with age (r=0.349,
P<0.001) and disease duration (r=0.337, P <0.001). SAA showed
a significant positive correlation with neutrophil count (r=0.179,
P<0.05) and a highly significant positive correlation with all
inflammatory indices, including CRP (r=0.422, P<0.001), ferritin
(r=0.574, P<0.001), D-Dimer (r=0.613, P<0.001) and CEA
(r=0.514, P<0.001). In addition, SAA showed a highly significant
positive correlation with the CO-RADS score (r=0.812, P<0.001)
and CT-SS (r=0.691, P<0.001) (Table III).
 | Table IIICorrelation between SAA and CEA
concentration with all the studied parameters. |
Table III
Correlation between SAA and CEA
concentration with all the studied parameters.
| | SAA | CEA |
|---|
| Parameter | R | P-value | Significance | R | P-value | Significance |
|---|
| Age | 0.349 | <0.001 | HS | 0.328 | <0.001 | HS |
| Duration of
disease | 0.337 | <0.001 | HS | 0.212 | <0.05 | S |
| White blood cell
count | 0.12 | >0.05 | NS | -0.026 | >0.05 | NS |
| Neutrophil
count | 0.179 | <0.05 | S | 0.072 | >0.05 | NS |
| Lymphocyte
count | -0.06 | >0.05 | NS | -0.155 | >0.05 | NS |
| Platelet count | 0.07 | >0.05 | NS | 0.007 | >0.05 | NS |
| Hemoglobin | -0.011 | >0.05 | NS | 0.023 | >0.05 | NS |
| Blood urea
nitrogen | 0.345 | <0.001 | HS | 0.296 | <0.001 | HS |
| Creatinine | 0.118 | >0.05 | NS | 0.15 | >0.05 | NS |
| Aspartate
transaminase | 0.149 | >0.05 | NS | 0.149 | >0.05 | NS |
| Alanine
transaminase | 0.092 | >0.05 | NS | 0.052 | >0.05 | NS |
| C- reactive
protein | 0.422 | <0.001 | HS | 0.441 | <0.001 | HS |
| Ferritin | 0.574 | <0.001 | HS | 0.349 | <0.001 | HS |
| D-Dimer | 0.613 | <0.001 | HS | 0.309 | <0.001 | HS |
| Carcinoembryonic
antigen | 0.514 | <0.001 | HS | | | |
| CO-RADS score | 0.812 | <0.001 | HS | 0.434 | <0.001 | HS |
| Computed tomography
severity score | 0.691 | <0.001 | HS | 0.374 | <0.001 | HS |
CEA showed a highly significant positive correlation
with age (r=0.328, P<0.001), a significant positive correlation
with disease duration (r=0.212, P<0.05) and a highly significant
positive correlation with all inflammatory indices, including CRP
(r=0.441, P<0.001), ferritin (r=0.349, P<0.001) and D-Dimer
(r=0.309, P<0.001). CEA showed a highly significant positive
correlation with the CO-RADS score (r=0.434, P<0.001) and CT-SS
(r=0.374, P<0.001) (Table
III).
In the multivariate regression model in Table IV, including age, disease duration,
CBC, SAA, ferritin, CEA, CRP, D-Dimer, BUN, creatinine, AST and
ALT, both SAA and ferritin were the best independent predictors of
COVID-19 severity (F=170.861, P<0.001). Furthermore, in another
multivariate regression model for predictors of CT-SS and pulmonary
involvement (Table V), SAA and PLT
count were independent predictors of CT-SS and lung involvement
(F=58.014, P<0.001).
 | Table IVRegression model outcomes of
significant predictors of COVID-19 severity. |
Table IV
Regression model outcomes of
significant predictors of COVID-19 severity.
| A, Model 1 |
|---|
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
|---|
| Constant | 0.555 | 1.996 | <0.05 | S | | | |
| Age, years | 0.001 | 0.527 | >0.05 | NS | | | |
| Duration of
disease, days | 0.004 | 0.496 | >0.05 | NS | | | |
| White blood cell
count, 109/l | -0.12 | -1.18 | >0.05 | NS | | | |
| Neutrophil count,
109/l | 0.123 | 1.238 | >0.05 | NS | | | |
| Lymphocyte count,
109/l | 0.164 | 1.557 | >0.05 | NS | | | |
| Platelet count,
109/l | 0 | 0.446 | >0.05 | NS | | | |
| C-reactive protein,
mg/l | -0.0000972 | -0.284 | >0.05 | NS | | | |
| Ferritin,
ng/ml | 0 | 4.36 | <0.001 | HS | | | |
| D-Dimer, ng/ml | 5.795E-05 | 1.795 | >0.05 | NS | | | |
| Carcinoembryonic
antigen, ng/ml | 0.026 | 3.045 | <0.05 | S | | | |
| Serum amyloid A,
mg/l | 0.01 | 5.297 | <0.001 | HS | | | |
| Monocyte count,
109/l | 0.07 | 0.522 | >0.05 | NS | | | |
| Eosinophil count,
109/l | 0.04 | 0.236 | >0.05 | NS | | | |
| Hemoglobin,
g/dl | 0.017 | 1.283 | >0.05 | NS | | | |
| Blood urea
nitrogen, mg/dl | -0.007 | -1.742 | >0.05 | NS | | | |
| Creatinine,
mg/dl | -0.07 | -1.828 | >0.05 | NS | | | |
| Aspartate
transaminase, U/l | 0.003 | 1.688 | >0.05 | NS | | | |
| Alanine
transaminase, U/l | -0.002 | -2.606 | <0.05 | S | | | |
| CO-RADS score | 0.043 | 1.218 | >0.05 | NS | | | |
| CT Severity
score | -0.001 | -0.18 | >0.05 | NS | | | |
| Overall | | | | | 22.574 | <0.001 | HS |
| B, Model 2 |
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
| Constant | 0.854 | 14.114 | <0.001 | HS | | | |
| Ferritin,
ng/ml | 0 | 6.903 | <0.001 | HS | | | |
| D-Dimer, ng/ml | 0.0000732 | 2.723 | <0.05 | S | | | |
| Carcinoembryonic
antigen, ng/ml | 0.027 | 3.577 | <0.05 | S | | | |
| Serum amyloid A,
mg/l | 0.012 | 10.453 | <0.001 | HS | | | |
| Blood urea
nitrogen, mg/dl | -0.004 | -1.237 | >0.05 | NS | | | |
| Creatinine,
mg/dl | -0.105 | -3.245 | <0.05 | S | | | |
| Aspartate
transaminase, U/l | 0.003 | 2.317 | <0.05 | S | | | |
| Alanine
transaminase, U/l | -0.002 | -2.827 | <0.05 | S | | | |
| Overall | | | | | 60.933 | <0.001 | HS |
| C, Model 3 |
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
| Constant | 0.871 | 16.09 | <0.001 | HS | | | |
| Ferritin,
ng/ml | 0 | 6.748 | <0.001 | HS | | | |
| D-Dimer, ng/ml | 8.154E-05 | 3.007 | <0.05 | S | | | |
| Carcinoembryonic
antigen, ng/ml | 0.026 | 3.398 | <0.05 | S | | | |
| Serum amyloid A,
mg/l | 0.012 | 10.381 | <0.001 | HS | | | |
| Creatinine,
mg/dl | -0.116 | -4.421 | <0.001 | HS | | | |
| Alanine
transaminase, U/l | -0.001 | -1.91 | 0.059 | NS | | | |
| Overall | | | | | 77.565 | <0.001 | HS |
| D, Model 4 |
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
| Constant | 0.856 | 15.983 | <0.001 | HS | | | |
| Ferritin,
ng/ml | 0 | 6.979 | <0.001 | HS | | | |
| Carcinoembryonic
antigen, ng/ml | 0.026 | 3.329 | <0.05 | S | | | |
| Serum amyloid A,
mg/l | 0.013 | 11.272 | <0.001 | HS | | | |
| Creatinine,
mg/dl | -0.116 | -4.263 | <0.001 | HS | | | |
| Overall | | | | | 105.489 | <0.001 | HS |
| E, Model 5 |
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
| Constant | 0.97 | 22.718 | <0.001 | HS | | | |
| Ferritin,
ng/ml | 0 | 6.923 | <0.001 | HS | | | |
| Serum amyloid A,
mg/l | 0.015 | 13.539 | <0.001 | HS | | | |
| Creatinine,
mg/dl | -0.087 | -3.239 | <0.05 | S | | | |
| Overall | | | | | 126.339 | <0.001 | HS |
| F, Model 6 |
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
| Constant | 0.915 | 22.492 | <0.001 | HS | | | |
| Ferritin,
ng/ml | 0 | 6.007 | <0.001 | HS | | | |
| Serum amyloid A,
mg/l | 0.015 | 12.889 | <0.001 | HS | | | |
| Overall | | | | | 170.861 | <0.001 | HS |
 | Table VMultivariate regression models of
significant predictors of lung CT severity score. |
Table V
Multivariate regression models of
significant predictors of lung CT severity score.
| A, Model 1 |
|---|
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
|---|
| Constant | 2.865 | 0.448 | >0.05 | NS | | | |
| Age, years | -0.035 | -0.647 | >0.05 | NS | | | |
| Duration of
disease, days | 0.245 | 1.387 | >0.05 | NS | | | |
| White blood cell
count, 109/l | -1.92 | -0.832 | >0.05 | NS | | | |
| Neutrophil count,
109/l | 1.765 | 0.781 | >0.05 | NS | | | |
| Lymphocyte count,
109/l | 0.877 | 0.368 | >0.05 | NS | | | |
| Platelet count,
109/l | -0.013 | -2.12 | <0.05 | S | | | |
| C-reactive protein,
mg/l | -0.005 | -0.686 | >0.05 | NS | | | |
| Ferritin,
ng/ml | 0.001 | 0.993 | >0.05 | NS | | | |
| D-Dimer, ng/ml | 0.000 | 0.149 | >0.05 | NS | | | |
| Carcinoembryonic
antigen, ng/ml | -0.097 | -0.488 | >0.05 | NS | | | |
| Serum amyloid A,
mg/l | 0.213 | 6.475 | <0.001 | HS | | | |
| Monocyte count,
109/l | 3.773 | 1.217 | >0.05 | NS | | | |
| Eosinophil count,
109/l | 0.101 | 0.027 | >0.05 | NS | | | |
| Hemoglobin,
g/dl | 0.316 | 1.009 | >0.05 | NS | | | |
| Blood urea
nitrogen, mg/dl | 0.214 | 2.29 | <0.05 | S | | | |
| Creatinine,
mg/dl | -1.302 | -1.479 | >0.05 | NS | | | |
| Aspartate
transaminase, U/l | -0.007 | -0.182 | >0.05 | NS | | | |
| Alanine
transaminase, U/l | 0.006 | 0.375 | >0.05 | NS | | | |
| Overall | | | | | 7.991 | <0.001 | HS |
| B, Model 2 |
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
| Constant | 5.446 | 3.519 | < 0.001 | HS | | | |
| Platelet count,
109/l | -0.016 | -3.345 | < 0.001 | HS | | | |
| Serum amyloid A,
mg/l | 0.219 | 9.552 | <0.001 | HS | | | |
| Blood urea
nitrogen, mg/dl | 0.125 | 1.937 | >0.05 | NS | | | |
| Overall | | | | | 40.805 | <0.001 | HS |
| C, Model 3 |
| Item | Regression
coefficient | t | P-value | Significance | F-Ratio | P-value | Significance |
| Constant | 6.817 | 4.898 | <0.001 | HS | | | |
| Platelet count,
109/l | -0.015 | -3.108 | <0.001 | HS | | | |
| Serum amyloid A,
mg/l | 0.234 | 10.642 | <0.001 | HS | | | |
| Overall | | | | | 58.014 | <0.001 | HS |
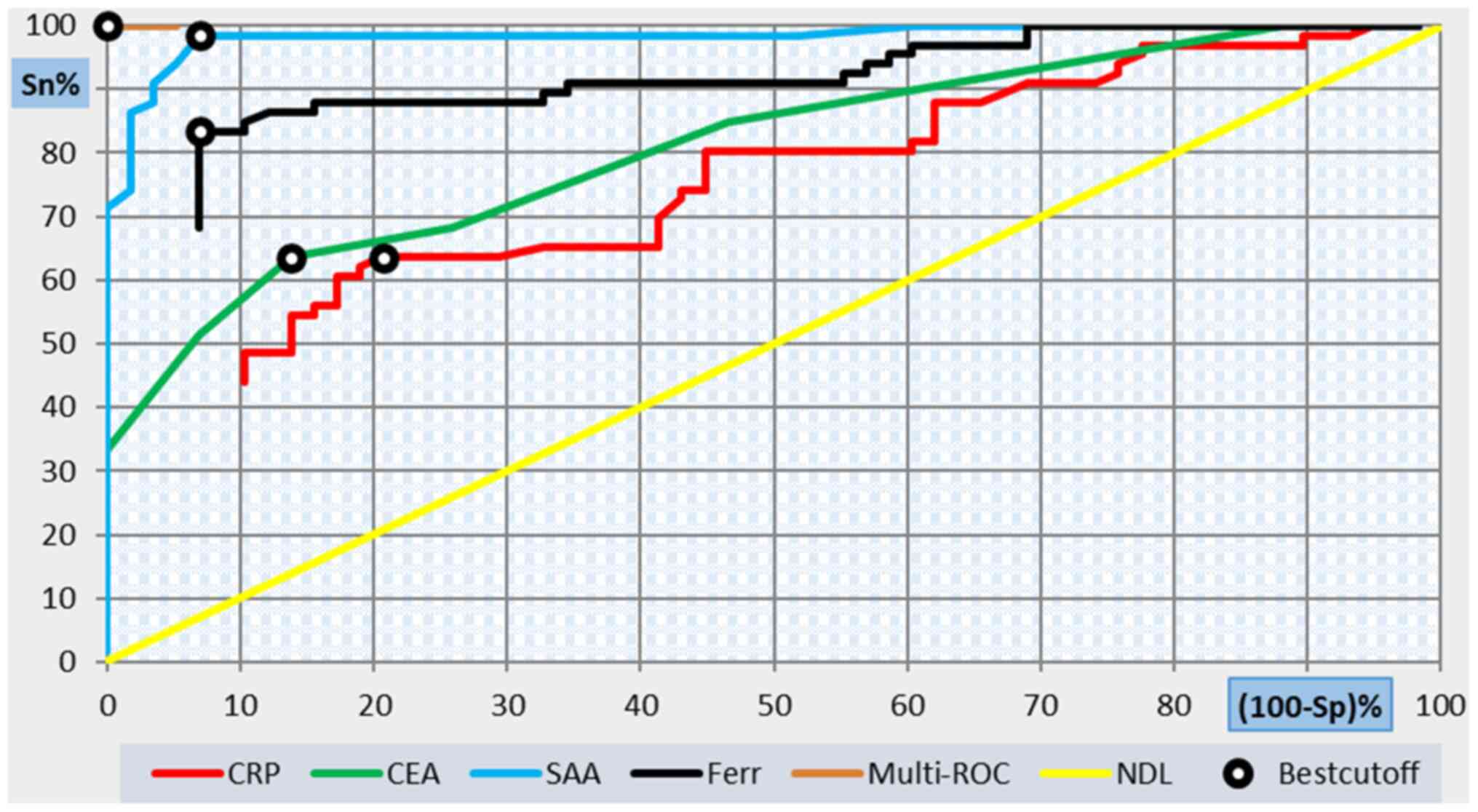
ROC curves showing the performance of SAA, CEA,
ferritin and CRP levels in predicting severity in COVID-19
patients, cutoff value and performance characteristics are
presented in Table VI and Fig. 1. An SAA value of 16 was predictive
of a poor prognosis with a specificity of 93.1%, sensitivity of
98.5, positive predictive value (PPV) of 94.2, negative predictive
value (NPV) of 98.2, efficiency of 96, and an AUC of 0.928. In a
multi-ROC curve showing the performance of SAA in addition to
ferritin. Specificity, sensitivity, PPV, NPV and efficiency were
100% and AUC was equal to 1.000.
 | Table VICutoff and performance
characteristics of SAA, CEA, ferritin and CRP in predicting
severity in COVID-19 patients. |
Table VI
Cutoff and performance
characteristics of SAA, CEA, ferritin and CRP in predicting
severity in COVID-19 patients.
| Parameter | Cutoff | Sensitivity, % | Specificity, % | Positive predictive
value | Negative predictive
value | Efficiency | Area under the
curve |
|---|
| SAA, mg/l | 16 | 98.5 | 93.1 | 94.2 | 98.2 | 96 | 0.928 |
| CEA, ng/ml | 7 | 63.6 | 86.2 | 84 | 67.6 | 74.2 | 0.78 |
| Ferritin,
ng/ml | 397 | 83.3 | 93.1 | 93.2 | 83.1 | 87.9 | 0.86 |
| CRP, mg/l | 54 | 63.6 | 79.3 | 77.8 | 65.7 | 71 | 0.616 |
| SAA + ferritin | | 100 | 100 | 100 | 100 | 100 | 1.000 |
Discussion
COVID-19 is an acute infectious disease caused by
SARS-CoV-2. The initial clinical picture of several patients may be
non-specific. Patients may present with minimal symptoms, including
mild to moderate fever, chills, malaise, respiratory distress
and/or gastroenterological disorders such as nausea and vomiting
(19). In several cases, rapid
disease progression can occur within a few days with ARDS,
uncompensated acidosis, septic shock and coagulation dysfunction,
followed by multiple organ failure and death (4,5,20).
The severity and prognosis of COVID-19 are
complicated by the diversity of symptoms, imaging manifestations
and the degree of disease progression (21,22).
Consequently, rapid reliable risk stratification tools and
sensitive indicators are required for timely disease monitoring to
develop efficient interventions targeting the reduction of
morbidity and mortality in patients with COVID-19.
RT-qPCR is the primary technique used for accurate
COVID-19 diagnosis. Although PCR detection can determine whether a
patient is infected with COVID-19(23), this technique cannot detect disease
progression and severity. Several modalities are used to aid the
diagnosis and provide evidence of disease progression, including
chest radiography, CT scans and biomarkers.
Imaging methods, primarily CT scans, can identify
chest infections and provide a reference for pathogen type
determination. However, the risks related to patient transportation
and examination, in addition to the extra protection required for
healthcare personnel during the examination, can be a disadvantage
(21,24). Several studies have focused on the
role of inflammatory markers as predictors of severity of COVID-19
infection, including lymphocyte and PLT counts, and CRP, D-Dimer
and ferritin levels (7,25,26).
Considering the above, the current study aimed to
evaluate potential biomarkers, including SAA and CEA, in addition
to other commonly used inflammatory predictors, such as CRP and
ferritin, and their association with COVID-19 severity and CT scan
findings, and to determine their role as beneficial tools for
patient stratification.
Based on COVID-19 severity, the recruited cohort was
divided into two groups: Patients with mild and patients with
severe COVID-19. The median age of the mild group was 43 years,
whereas that of the severe group was 50 years, and the difference
was highly significant, suggesting an increased probability of
severe disease in the elderly, as stated by several studies. These
results agree with a study by Zheng et al (27), where the median age of 161 patients
with COVID-19 admitted to Changsha Public Health Centre was 45
years, the median age of the severe group was 57 years, and the
non-severe group was 40 years, showing a statistical difference in
age between the two groups. Older age is a risk factor for a more
severe infectious course, and several clinical studies on influenza
virus pneumonia have shown that old age is a risk factor for severe
illness, especially when associated with at least one underlying
disease (27,28). The results of the present study also
agreed with the results of Liu et al (29), in which it was stated that age was
an important risk factor for severity in patients with COVID-19.
The median age of the non-severe group was 43 years, and that of
the severe group was 64 years (27,29).
In the current study, 10 patients were >65 years old,
constituting 8% of the entire cohort. In the severe COVID-19 group,
7 patients out of 66 (10.6%) compared to 3 cases out of 58 (5.2%)
in the mild group were >65 years taking into consideration that
the current life expectancy for the Egyptian population in 2021 is
74.3 years, and the number of individuals aged ≥60 years is 6.8
million, representing 6.7% of the total population according to
official data by the Central Agency for Public Mobilization and
Statistics (CAPMAS, 2021) (30).
A meta-analysis conducted by Peckham et al
(31) on 3,111,714 cases worldwide
reported no difference in the proportion of confirmed COVID-19
cases between men and women (31).
In the present study, several social, cultural and behavioral
differences between the sexes may have contributed to the high
overall male to female ratio (79 to 21%, respectively) seen in this
cohort, including habits more common in men, such as smoking
(32,33) and sex-based differences in hygiene.
Additionally, based on anecdotal evidence, men are likely to leave
their houses and enter crowded areas in Egypt. Furthermore,
differences in health-seeking behaviors, unequal access to
healthcare facilities and testing between sexes may have skewed the
data further towards a male bias (34,35).
However, a consistent feature of the COVID-19
pandemic is the male bias towards severe disease (36). This agrees with the results of the
present study, as 85% (56 out of 66) of the severe cases were men
compared with 15% of women. Male sex is associated with nearly a 3x
risk of requiring intensive care unit admission and a higher
probability of death compared to women (31).
Fundamental differences in the immune response
between males and females are likely to be the driving factor
behind the significant sex bias observed in severe COVID-19 cases.
Sex differences in innate and adaptive immunity have been reported,
which may account for the reduced risk of severe disease in
females. A robust antiviral innate IFN response and enhanced
adaptive immunity in females, higher numbers of CD4+ T
cells, robust CD8+ T cells and increased B cell
production of immunoglobulin compared to males, may lead to more
effective viral control in females, and a relatively lower risk of
developing severe disease (37,38).
The X chromosome encodes several immune-related genes, which can be
variably expressed on both alleles, increasing the diversity of the
immune response (39). Hormonal
differences also play a role; estradiol augments the immune
response in contrast to testosterone, which was found to suppress
the immune system (40).
The median WBC, PNL and PLT counts were higher in
the severe group than in the mild group, whereas the lymphocyte
count was lower. However, the differences between both groups
regarding these parameters were not statistically significant. Of
the 124 COVID-19 positive cases, a high prevalence of increased
inflammatory marker levels was identified. These results showed
that in all COVID-19 cases, there was an overall increased level of
inflammatory parameters, including SAA, CEA, CRP and ferritin, in
addition to pro-coagulation markers. D-Dimer levels were similar to
the results of previous studies (29,41,42),
and they were significantly higher at admission in the severe
patient group than in the mild group, whereas total WBC, lymphocyte
and PLT counts were within the normal range, consistent with the
results of Li et al (42)
and Wang et al (43).
Huang et al (44) showed that patients with severe
COVID-19 had higher levels of IL-1β, IFN-γ, IP-10 and monocyte
chemoattractant protein compared with mild cases, causing Th1 cell
activation and stimulating the production of SAA, CRP,
procalcitonin and PLT. These inflammatory factors may be useful as
indicators reflecting the body's response to infection (45).
According to Li et al (42) and Wang et al (43) as the disease progressed from mild to
severe, SAA and CRP levels increased, while lymphocyte counts
gradually decreased. However, WBC and PLT were all within normal
ranges, suggesting that SAA and CRP are closely related to disease
classification, while WBC and PLT are of little significance.
SAA showed a highly significant positive correlation
with age, disease duration, neutrophil count and all inflammatory
indices, including CRP, ferritin, CEA and D-Dimer in the present
study. Similarly, CEA showed a significant positive correlation
with age, disease duration and all inflammatory indices.
Additionally, both SAA and CEA levels showed a highly significant
positive correlation with the CT-SS. These results agreed with the
results from various studies showing significantly higher SAA
levels in PCR positive COVID-19 cases, and significant associations
between SAA levels and the number of COVID-19 cases, severity and
mortality rate (29,41,42).
SAA is an acute phase protein produced by the liver
and is induced by cytokines, including IL-1β, IL-6 and TNF-α. SAA
promotes an inflammatory response by activating chemokines and
inducing chemotaxis, even at very low concentrations (46,47).
In current clinical practices, SAA is frequently used as an
indicator for monitoring of inflammation and estimation of
prognosis (48,49). SAA showed promising results when
used to monitor the effectiveness of antibiotics (cefotiam and
Augmentin) in early onset neonatal sepsis according to a study by
Liu et al (49).
Previous studies have shown that patients with
severe ARDS have significantly increased levels of SAA, suggesting
that SAA could be used as a biomarker to monitor the progression of
respiratory diseases (50,51). As stated by Cheng et al
(52), in a study involving 89
COVID-19 patients, dynamic changes in SAA could be used to predict
prognosis.
In the present study, serum CEA levels were
significantly higher in patients with severe COVID-19 than in those
with a mild infection. Moreover, higher serum CEA levels were
associated with a higher CT-SS. A limited number of studies
reported elevated serum CEA levels in COVID-19 patients with
significantly increased CEA levels compared with healthy controls,
and a potential association between CEA and CRP levels was reported
(53,54). However, its role in predicting
clinical outcomes or CT involvement in patients with COVID-19 is
still under investigation. The present study validated the previous
results that CEA levels were related to COVID-19 severity.
Moreover, the association between CEA levels and CT scores were
also validated, which are in agreement with the findings of Chen
et al (41).
CEA is a biomarker of adenocarcinoma in respiratory
or digestive system cancers, in addition to non-neoplastic lung
diseases (55,56). Increased CEA expression was detected
in type II pneumocytes in allergic bronchopulmonary aspergillosis
(57) and atypical epithelial
proliferation in idiopathic pulmonary fibrosis (13,58).
Bronchiolar and type II alveolar epithelial cells are the main
targets of SARS-CoV-2 in the lungs. SARS-CoV-2 infection-induces
massive type II alveolar epithelial cell death and aberrant
regeneration of type II pneumocytes along with the production of
CEA, such as that observed in the uncontrolled proliferation in
lung adenocarcinoma. Moreover, atypical epithelial and fibroblast
proliferation may also worsen the obstruction of bronchioles and
lung consolidation, causing refractory hypoxemia along with worse
CT scores (59,60).
Consequently, it is possible that serum CEA levels
may correlate with the severity and prognosis of COVID-19. Based on
the relationship between CEA and type II pneumocyte hyperplasia and
lung fibrosis, medications such as nintedanib, which target
atypical epithelial and fibrotic proliferation, may be a potential
therapeutic option to decrease mortality in patients with
COVID-19(41).
An elevated D-Dimer level represents microangiopathy
and a hypercoagulable state in COVID-19 patients (61). COVID-19 is implicated in aggressive
pro-inflammatory responses causing endothelial cell dysfunction and
excessive thrombin formation (62),
which can be responsible for oxygen desaturation and respiratory
distress seen in severe cases (63,64).
In the present study, D-Dimer levels were significantly higher in
patients with severe COVID-19, and there was a highly significant
positive correlation with SAA and CEA in addition to CRP and
ferritin levels. These results are also in agreement with a
retrospective study of 183 patients with COVID-19 performed by Tang
et al (65) which showed a
significant increase in D-Dimer levels and fibrin degradation
products, and this was indicative of a poor prognosis.
Similarly, CRP is also an acute phase protein, the
levels of which rise rapidly, and the rate of increase is
positively correlated with the severity of infection (7,29). A
higher CRP level is linked to an increased risk of severe COVID-19
and may contribute to pneumonia, ARDS and the rapid multiple organ
damage (7,66). According to a study by Liu et
al (66), elevated CRP and
decreased albumin levels were important factors affecting the
prognosis of COVID-19.
Ferritin is another crucial mediator of the immune
response that showed a statistically significant increase in severe
COVID-19 cases, and showed a highly significant positive
correlation with SAA levels in addition to CEA, CRP and D-Dimer
levels in the present study. Increased ferritin levels may be
implicated in the cytokine storm due to its direct
immunosuppressive and pro-inflammatory effects (25). Efstathiou et al (67) indicated that viral infections
increased ferritin levels. This finding may be attributed to the
fact that the inflammatory mediators induce an increase in ferritin
levels, in addition to denaturation and necrosis of cells to break
down cell membranes, causing a leakage of ferritin from damaged
cells (68).
The results of the present study showed that the
levels of SAA, CEA, CRP and ferritin increased as the disease
severity increased. CT imaging is an important clinical diagnostic
tool for evaluating COVID-19 infections. According to the
correlation analysis in this study, SAA, CEA, CRP and ferritin
levels at admission were highly correlated with the CT-SS,
suggesting a possible role in predicting disease progression.
ROC curve analysis was used to comprehensively and
accurately compare and evaluate the diagnostic performance of SAA,
CEA, CRP and ferritin levels on admission and to explore their
clinical and prognostic utility. The results of the current study
showed that the AUC was highest for SAA, followed by ferritin, CEA
and CRP. According to the multivariate regression analysis and ROC
curve results, the combined use of SAA and ferritin was more
sensitive than SAA or ferritin alone as predictors of severity, as
their combination had the highest predictive value for disease
severity, with an AUC of 1.000.
Accordingly, the combined detection of SAA and
ferritin may have guiding significance for assessing the severity,
disease progression and prognosis of COVID-19 cases. This may aid
effective intervention measures to be implemented in timely manner
and reduce the rates of severe illness and mortality, as we
continue to face upcoming waves of COVID-19.
The present study has some limitations. The
elevations in SAA and CEA may have been due to various other
conditions and comorbidities, which themselves may be associated
with a higher COVID-19 risk. Additionally, this study was a
single-center study with a relatively small cohort, which may have
limited the power of the statistical analyses.
In conclusion, the combined detection of SAA and
ferritin may have guiding significance for the severity of COVID-19
and may be correlated with CT-SS in patients with COVID-19. Serum
CEA levels were correlated with the severity of the CT scores and
the prognosis of COVID-19. Therefore, assessment and monitoring of
these laboratory markers at the earliest stage of the disease may
have a significant impact on halting disease progression and
decreasing mortality. Further prospective and multicenter studies
with validation cohorts should be performed in the future.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DAA and MAEMT designed the study, contributed to
data collection and interpretation, and wrote the manuscript. NMBE
assisted with sample collection, data interpretation and editing of
the manuscript. FΜB and MAEM contributed to sample collection, data
interpretation and drafting of the manuscript. FΜB and MAEMT
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
This study was conducted in accordance with the
principles outlined in the Declaration of Helsinki of the World
Medical Association. The study was approved by the Institutional
Ethics Committee of Ain Shams University. Informed consent was
obtained from all enrolled participants after receiving an
explanation of the study's aim and procedures.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gorbalenya AE, Baker SC, Baric RS, De
Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich
AM, et al: Coronaviridae Study Group of the International Committee
on Taxonomy of Viruses: The species Severe acute respiratory
syndrome-related coronavirus: Classifying 2019-nCoV and naming it
SARS-CoV-2. Nat Microbiol. 5:536–544. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z,
Xiang J, Wang Y, Song B, Gu X, et al: Clinical course and risk
factors for mortality of adult inpatients with COVID-19 in Wuhan,
China: A retrospective cohort study. Lancet. 395:1054–1062.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Calina D, Docea AO, Petrakis D, Egorov AM,
Ishmukhametov AA, Gabibov AG, Shtilman MI, Kostoff R, Carvalho F,
Vinceti M, et al: Towards effective COVID-19 vaccines: Updates,
perspectives and challenges (Review). Int J Mol Med. 46:3–16.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Khan M, Khan H, Khan S and Nawaz M:
Epidemiological and clinical characteristics of coronavirus disease
(COVID-19) cases at a screening clinic during the early outbreak
period: A single-centre study. J Med Microbiol. 69:1114–1123.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H,
Wang W, Song H, Huang B, Zhu N, et al: Genomic characterisation and
epidemiology of 2019 novel coronavirus: Implications for virus
origins and receptor binding. Lancet. 395:565–574. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ye Q, Wang B and Mao J: The pathogenesis
and treatment of the ‘Cytokine Storm’ in COVID-19. J Infect.
80:607–613. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tan C, Huang Y, Shi F, Tan K, Ma Q, Chen
Y, Jiang X and Li X: C-reactive protein correlates with computed
tomographic findings and predicts severe COVID-19 early. J Med
Virol. 92:856–862. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wilson PG, Thompson JC, Webb NR, de Beer
FC, King VL and Tannock LR: Serum amyloid A, but not C-reactive
protein, stimulates vascular proteoglycan synthesis in a
pro-atherogenic manner. Am J Pathol. 173:1902–1910. 2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vietri L, Fui A, Bergantini L,
d'Alessandro M, Cameli P, Sestini P, Rottoli P and Bargagli E:
Serum amyloid A: A potential biomarker of lung disorders. Respir
Investig. 58:21–27. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Goldenberg DM, Neville AM, Carter AC, Go
VL, Holyoke ED, Isselbacher KJ, Schein PS and Schwartz M: CEA
(carcinoembryonic antigen): Its role as a marker in the management
of cancer. J Cancer Res Clin Oncol. 101:239–242. 1981.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hao C, Zhang G and Zhang L: Serum CEA
levels in 49 different types of cancer and noncancer diseases. Prog
Mol Biol Transl Sci. 162:213–227. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu Q, Wang RS, Qu GQ, Wang YY, Liu P, Zhu
YZ, Fei G, Ren L, Zhou YW and Liu L: Gross examination report of a
COVID-19 death autopsy. Fa Yi Xue Za Zhi. 36:21–23. 2020.PubMed/NCBI View Article : Google Scholar : (In English,
Chinese).
|
|
13
|
Fahim A, Crooks MG, Wilmot R, Campbell AP,
Morice AH and Hart SP: Serum carcinoembryonic antigen correlates
with severity of idiopathic pulmonary fibrosis. Respirology.
17:1247–1252. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
WHO: World Health Organization.
Coronavirus Disease (COVID-19) Outbreak. World Health Organization,
Geneva, 2020.
|
|
15
|
Teama MA, Abdelhakam DA, Elmohamadi MA and
Badr FM: Vitamin D deficiency as a predictor of severity in
patients with COVID-19 infection. Sci Prog.
104(368504211036854)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
World Medical Association. World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194.
2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Prokop M, van Everdingen W, van Rees
Vellinga T, Quarles van Ufford H, Stöger L, Beenen L, Geurts B,
Gietema H, Krdzalic J, Schaefer-Prokop C, et al: COVID-19
Standardized Reporting Working Group of the Dutch Radiological
Society: CO-RADS: A categorical CT assessment scheme for patients
suspected of having COVID-19-definition and evaluation. Radiology.
296:E97–E104. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Francone M, Iafrate F, Masci GM, Coco S,
Cilia F, Manganaro L, Panebianco V, Andreoli C, Colaiacomo MC,
Zingaropoli MA, et al: Chest CT score in COVID-19 patients:
Correlation with disease severity and short-term prognosis. Eur
Radiol. 30:6808–6817. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, et al: A novel coronavirus from patients
with pneumonia in China. N Engl J Med. 382:727–733. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX,
Wang QZ, Sun C, Sylvia S, Rozelle S, Raat H, et al: Epidemiology,
causes, clinical manifestation and diagnosis, prevention and
control of coronavirus disease (COVID-19) during the early outbreak
period: A scoping review. Infect Dis Poverty. 9(29)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Xu XW, Wu XX, Jiang XG, Xu KJ, Ying LJ, Ma
CL, Li SB, Wang HY, Zhang S, Gao HN, et al: Clinical findings in a
group of patients infected with the 2019 novel coronavirus
(SARS-Cov2) outside of Wuhan, China: Retrospective case series.
BMJ: Feb 19, 2020 (Epub ahead of print).
|
|
22
|
Guan W, Ni Z, Hu Y, Liang W, Ou C, He J,
Liu L, Shan H, Lei C, Hui DS, et al: Clinical characteristics of
coronavirus disease 2019 in China. N Engl J Med. 382:1708–1720.
2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wu J, Liu J, Li S, Peng Z, Xiao Z, Wang X,
Yan R and Luo J: Detection and analysis of nucleic acid in various
biological samples of COVID-19 patients. Travel Med Infect Dis.
37(101673)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lin C, Ding Y, Xie B, Sun Z, Li X, Chen Z
and Niu M: Asymptomatic novel coronavirus pneumonia patient outside
Wuhan: The value of CT images in the course of the disease. Clin
Imaging. 63:7–9. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Vargas-Vargas M and Cortés-Rojo C:
Ferritin levels and COVID-19. Rev Panam Salud Publica.
44(e72)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yang Z, Shi J, He Z, Lu Y, Xu Q, Ye C,
Chen S, Tang B, Yin K, Lu Y, et al: Predictors for imaging
progression on chest CT from coronavirus disease 2019 (covid-19)
patients. Aging (Albany NY). 12:6037–6048. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zheng F, Tang W, Li H, Huang YX, Xie YL
and Zhou ZG: Clinical characteristics of 161 cases of corona virus
disease 2019 (COVID-19) in Changsha. Eur Rev Med Pharmacol Sci.
24:3404–3410. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ishiguro T, Kagiyama N, Uozumi R, Odashima
K, Takaku Y, Kurashima K, Morita S and Takayanagi N: Clinical
characteristics of influenza-associated pneumonia of adults:
Clinical features and factors contributing to severity and
mortality. Yale J Biol Med. 90:165–181. 2017.PubMed/NCBI
|
|
29
|
Liu SL, Wang SY, Sun YF, Jia QY, Yang CL,
Cai PJ, Li JY, Wang L and Chen Y: Expressions of SAA, CRP, and FERR
in different severities of COVID-19. Eur Rev Med Pharmacol Sci.
24:11386–11394. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
CAPMAS, Central Agency for Public
Mobilization and Statistics: 2021. Available from: https://www.capmas.gov.eg/.
|
|
31
|
Peckham H, De Gruijter NM, Raine C,
Radziszewska A, Ciurtin C, Wedderburn LR, Rosser EC, Webb K and
Deakin CT: Male sex identified by global COVID-19 meta-analysis as
a risk factor for death and ITU admission. Nat Commun. 9(11:
6317)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Giovino GA, Mirza SA, Samet JM, Gupta PC,
Jarvis MJ, Bhala N, Peto R, Zatonski W, Hsia J, Morton J, et al:
Tobacco use in 3 billion individuals from 16 countries: An analysis
of nationally representative cross-sectional household surveys.
Lancet. 380:668–679. 2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Loffredo CA, Radwan GN, Eltahlawy EM,
El-Setouhy M, Magder L and Hussein MH: Estimates of the prevalence
of tobacco smoking in Egypt. Open J Epidemiol. 5:129–135. 2015.
|
|
34
|
Spagnolo PA, Manson JAE and Joffe H: Sex
and gender differences in health: What the COVID-19 pandemic can
teach us. Ann Intern Med. 173:385–386. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Regitz-Zagrosek V: Sex and gender
differences in health. Science and society series on sex and
science. EMBO Rep. 13:596–603. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Singh S, Chowdhry M, Chatterjee A and Khan
A: Gender-based disparities in COVID-19: Clinical characteristics
and propensity matched analysis of outcomes. MedRxiv: May 12, 2020
(Epub ahead of print).
|
|
37
|
Abdullah M, Chai PS, Chong MY, Tohit ERM,
Ramasamy R, Pei CP and Vidyadaran S: Gender effect on in vitro
lymphocyte subset levels of healthy individuals. Cell Immunol.
272:214–219. 2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hewagama A, Patel D, Yarlagadda S,
Strickland FM and Richardson BC: Stronger inflammatory/cytotoxic
T-cell response in women identified by microarray analysis. Genes
Immun. 10:509–516. 2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gal-Oz ST, Maier B, Yoshida H, Seddu K,
Elbaz N, Czysz C, Zuk O, Stranger BE, Ner-Gaon H and Shay T: ImmGen
report: Sexual dimorphism in the immune system transcriptome. Nat
Commun. 10(4295)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Klein SL and Flanagan KL: Sex differences
in immune responses. Nat Rev Immunol. 16:626–638. 2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chen Q, Kong H, Qi X, Ding W, Ji N, Wu C,
Huang C, Wu W, Huang M, Wu W, et al: Carcinoembryonic antigen: A
potential biomarker to evaluate the severity and prognosis of
COVID-19. Front Med (Lausanne). 7(579543)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li H, Xiang X, Ren H, Xu L, Zhao L, Chen
X, Long H, Wang Q and Wu Q: Serum amyloid A is a biomarker of
severe coronavirus disease and poor prognosis. J Infect.
80:646–655. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J,
Wang B, Xiang H, Cheng Z, Xiong Y, et al: Clinical characteristics
of 138 hospitalized patients with 2019 novel coronavirus-infected
pneumonia in Wuhan, China. JAMA. 323:1061–1069. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu
Y, Zhang L, Fan G, Xu J, Gu X, et al: Clinical features of patients
infected with 2019 novel coronavirus in Wuhan, China. Lancet.
395:497–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Behrens K and Alexander WS: Cytokine
control of megakaryopoiesis. Growth Factors. 36:89–103.
2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Connolly M, Rooney PR, Mcgarry T, Maratha
AX, Mccormick J, Miggin SM, Veale DJ and Fearon U: Acute serum
amyloid A is an endogenous TLR2 ligand that mediates inflammatory
and angiogenic mechanisms. Ann Rheum Dis. 75:1392–1398.
2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Sack GH: Serum amyloid A - A review. Mol
Med. 24(46)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wakai M, Hayashi R, Tanaka S, Naito T,
Kumada J, Nomura M, Takigawa H, Oka S, Ueno Y, Ito M, et al: Serum
amyloid A is a better predictive biomarker of mucosal healing than
C-reactive protein in ulcerative colitis in clinical remission. BMC
Gastroenterol. 20(85)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Liu C, Zhang Y, Shang Y, Fang C, He Q and
Xie L: Clinical values of common biomarkers for efficacy monitoring
of antibiotics in early onset neonatal sepsis. Transl Pediatr.
9:669–676. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Yip TT, Chan JW, Cho WC, Yip TT, Wang Z,
Kwan TL, Law SC, Tsang D, Chan JK, Lee KC, et al: Protein chip
array profiling analysis in patients with severe acute respiratory
syndrome identified serum amyloid A protein as a biomarker
potentially useful in monitoring the extent of pneumonia. Clin
Chem. 51:47–55. 2005.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zinellu A, Paliogiannis P, Carru C and
Mangoni AA: Serum amyloid A concentrations, COVID-19 severity and
mortality: An updated systematic review and meta-analysis. Int J
Infect Dis. 105:668–674. 2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Cheng L, Yang JZ, Bai WH, Li ZY, Sun LF,
Yan JJ, Zhou CL and Tang BP: Prognostic value of serum amyloid A in
patients with COVID-19. Infection. 48:715–722. 2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wei X, Su J, Yang K, Wei J, Wan H, Cao X,
Tan W and Wang H: Elevations of serum cancer biomarkers correlate
with severity of COVID-19. J Med Virol. 92:2036–2041.
2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Yang C, Wang J, Liu J, Huang S and Xiong
B: Elevated carcinoembryonic antigen in patients with COVID-19
pneumonia. J Cancer Res Clin Oncol. 146:3385–3388. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
He G, Jiang Z, Xue S, Sun X and Wang W:
Expression of LDH and CEA in serum in the process of targeted
therapy of lung adenocarcinoma and the association between them and
prognosis. Oncol Lett. 17:4550–4556. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Mohr AM, Gould JJ, Kubik JL, Talmon GA,
Casey CA, Thomas P, Tuma DJ and McVicker BL: Enhanced colorectal
cancer metastases in the alcohol-injured liver. Clin Exp
Metastasis. 34:171–184. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Noguchi T, Yamamoto K, Moriyama G, Saito
Y, Kyoyama H, Mikami S, Ono R, Kobayashi T, Yamana K and Uematsu K:
Evaluation of serum levels of carcinoembryonic antigen in allergic
bronchopulmonary aspergillosis. J Nippon Med School. 80:404–409.
2013.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Abbona GC, Papotti M, Gugliotta P, Pecchio
F and Rapellino M: Immunohistochemical detection of
carcinoembryonic antigen (CEA) in non-neoplastic lung disease. Int
J Biol Markers. 8:240–243. 1993.PubMed/NCBI
|
|
59
|
Yao XH, Li TY, He ZC, Ping YF, Liu HW, Yu
SC, Mou HM, Wang LH, Zhang HR, et al: A pathological report of
three COVID-19 cases by minimally invasive autopsies. Zhonghua Bing
Li Xue Za Zhi:. 49:411–417. 2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
60
|
Fox SE, Akmatbekov A, Harbert JL, Li G,
Brown JQ and Heide RS: Pulmonary and cardiac pathology in African
American patients with COVID-19: An autopsy series from New
Orleans. Lancet Respir Med. 8:681–686. 2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Zhang L, Yan X, Fan Q, Liu H, Liu X, Liu Z
and Zhang Z: D-Dimer levels on admission to predict in-hospital
mortality in patients with Covid-19. J Thromb Haemost.
18:1324–1329. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Levi M and Van der Poll T: Coagulation and
sepsis. Thromb Res. 149:38–44. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Gattinoni L, Coppola S, Cressoni M, Busana
M, Rossi S and Chiumello D: COVID-19 does not lead to a ‘typical’
acute respiratory distress syndrome. Am J Respir Crit Care Med.
201:1299–1300. 2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Oudkerk M, Buller HR, Kuijpers D, van Es
N, Oudkerk SF, McLoud T, Gommers D, Dissel JV, Cate HT and van Beek
EJ: Diagnosis, prevention, and treatment of thromboembolic
complications in COVID-19: Report of the national institute for
public health of the Netherlands. Radiology. 297:E216–E222.
2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Tang N, Li D, Wang X and Sun Z: Abnormal
coagulation parameters are associated with poor prognosis in
patients with novel coronavirus pneumonia. J Thromb Haemost.
18:844–847. 2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Liu W, Tao ZW, Wang L, Yuan ML, Liu K,
Zhou L, Wei S, Deng Y, Liu J, Liu HG, et al: Analysis of factors
associated with disease outcomes in hospitalized patients with 2019
novel coronavirus disease. Chin Med J (Engl). 133:1032–1038.
2020.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Efstathiou SP, Pefanis AV, Tsiakou AG,
Skeva II, Tsioulos DI, Achimastos AD and Mountokalakis TD: Fever of
unknown origin: Discrimination between infectious and
non-infectious causes. Eur J Intern Med. 21:137–143.
2010.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Kell DB and Pretorius E: Serum ferritin is
an important inflammatory disease marker, as it is mainly a leakage
product from damaged cells. Metallomics. 6:748–773. 2014.PubMed/NCBI View Article : Google Scholar
|