Introduction
Mechanical ventilation is the main respiratory
support provided for patients with lung injury or those under
general anesthesia. However, ventilator-induced lung injury (VILI),
involving inflammation, alveolar barrier disruption and hypertonic
pulmonary edema, is a common complication. Pro-inflammatory factors
(1-3) and macrophages (4) play a key role in VILI (5).
Alveolar macrophages (AMs) and alveolar epithelial
cells (ECs) are the most critical cells for maintaining lung
homeostasis (6,7). AMs are the major immune cells in
lung tissue (8), suppressing
inflammatory responses at homeostasis by releasing
anti-inflammatory mediators, and play a crucial role in the
inflammation progress in the lungs (9). Macrophages can be classified into
pro-inflammatory M1 macrophages and anti-inflammatory M2
macrophages according to environmental stimuli (10). Alveolar ECs represent physical
barrier that protects from external harmful substances. It has been
demonstrated that the maintenance of homeostasis in the lungs is
largely attributed to the interaction between ECs and macrophages
(11). AMs adhere to ECs through
CD200R, PD1 and their ligands CD200 and PDL1 on ECs. These protein
interactions suppress the activation of AMs. In a steady state, ECs
maintain macrophages in a quiescent state. In addition, ECs
regulate the immune response and AM functions by secreting multiple
mediators. Moreover, ECs participate in the recruitment and
regulation of macrophages (12,13). ECs are also involved in
mechanotransduction and the inflammatory response during mechanical
ventilation (14,15). Thus, the interactions between AMs
and ECs may serve as a novel therapeutic strategy for
mechanical-induced lung injury.
The interactions between ECs and AMs include gap
junctions (16), the secretion
of molecules (17) and exosomes
(7). Exosomes are small (20-200
nm in diameter) single-membrane vesicles, which have been
considered as a novel communication mechanism of intercellular due
to transfer microRNAs (miRNAs/miRs), DNAs, lipids and proteins, and
have various biological functions via autocrine and paracrine
mechanisms (18). miRNAs are
small non-coding RNAs that can induce the degradation or
transcriptional repression of target genes (19). miRNAs play crucial roles in cell
differentiation, proliferation, migration and apoptosis. Numerous
studies have demonstrated that miRNAs play an essential role in
lung diseases. miRNAs, such as miR-155, miR-26a, miR-21a-5p,
miR-223, etc., have been shown to be involved in lung homeostasis
and development (20). The
inhibition of miR-221 has been found to ameliorate
lipopolysaccharide (LPS)-induced lung injury (21), and miR-127 has been shown to be
involved in VILI through the NF-κB and p38MAPK signaling pathways
(22). Notably, miR-21a-5p, as
one of the most crucial miRNAs, has been reported to be involved in
the occurrence and development of various lung diseases (23,24), including asthma, chronic
obstructive pulmonary disease (COPD) and acute lung injury (ALI).
It has been demonstrated that miR-21a-5p negatively regulates the
inflammatory response by interacting with immune cells in
LPS-induced ALI (25). In
addition, miR-21a-5p has been defined as a biomarker and
therapeutic target (26).
Exosomal miRNAs can mediate inter-cellular communications between
several types of cells, which have been reported to influence
biological pathways and further alter cellular functions (27). Exosomal miR-30d-5p-derived from
polymorphonuclear neutrophil (PMN)-induced M1 macrophage
polarization to promote sepsis-related ALI (28). Alveolar EC-derived exosomal
miR-92a-3p has also been shown to activate macrophages by
regulating the NF-kB signaling pathway and suppressing PTEN
(29). However, the role of
exosomes in the epithelial-macrophage interaction in mechanical
ventilation has not yet been fully elucidated.
Considering the functions of exosomes, the present
study, aimed to determine whether exosomes-derived from ECs
subjected to cyclic stretching (CS) can regulate macrophage
polarization. In addition, miRNA intervention was used to reveal
the specific underlying mechanisms. The findings presented herein
provide novel information on the effects of EC-derived exosomal
miRNAs on the polarization of macrophages, which may be used as a
potential therapeutic approach for lung injury during mechanical
ventilation.
Materials and methods
Ethics approval
All procedures for the animal experiments were
approved (approval no. 2406) by the Institutional Animal Care and
Use Committee at Tongji Medical College, Huazhong University of
Science and Technology (Wuhan, China).
Bone marrow-derived macrophage (BMDM)
isolation
Male C57BL/6 mice (7 weeks old) were obtained from
Vital River Laboratory Animal Technology. BMDMs were isolated from
7 male C57BL/6 mice (7 weeks old; weight, 21±2 g). In brief, mice
were sacrificed using an overdose of 2% sodium pentobarbital (100
mg/kg) administred by intraperitoneal injection. Following
sacrifice, the muscle was trimmed and the bone marrow cells were
flushed from the bone shafts with Roswell Park Memorial Institute
(RPMI)-1640 medium (cat. no. 11875093; Gibco; Thermo Fisher
Scientific, Inc.) and centrifuged (500 × g, 4°C, 5 min).
Subsequently, 5 ml erythrocyte lysis buffer (cat. no. BL503A,
Biosharp) were added, followed by 10 ml RPMI-1640 medium. The
mixture was then centrifuged (500 × g, 4°C, 5 min). The cells were
filtered through a 70-µm strainer and incubated at 37°C for
7 days in RPMI-1640 medium containing 10% fetal bovine serum (FBS,
cat. no. 10100147; Gibco; Thermo Fisher Scientific, Inc.). The
macrophages were incubated with macrophage colony-stimulating
factor (10 ng/ml, cat. no. 416-ML-050, R&D Systems, Inc.) to
activate the M0 macrophages. Half the medium was replaced every 2
days.
Cells and cell culture
Raw264.7 cells (mouse macrophages) were obtained
from the American Type Culture Collection (ATCC). MLE-12 cells
(mouse lung ECs) were obtained from the Department of Thoracic
Surgery, Nanjing Medical University Affiliated Cancer Hospital,
Cancer Institute of Jiangsu Province. The Raw264.7 cells were
cultured in DMEM (cat. no. 11965092; Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS and the MLE-12 were cultured in
DMEM/F12 (cat. no. 11330032; Gibco; Thermo Fisher Scientific, Inc.)
with 10% FBS. THP1 cells (human monocytes) were purchased from ATCC
and cultured in RPMI-1640 with 10% FBS. All three types of cells
were maintained at 37°C in a humidified incubator containing 5%
CO2.
CS of cells
MLE-12 cells were seeded onto fibronectin-coated (10
µg/cm2) flexible silastic membranes
(5.0×105 cells/well) with 10% exosome-depleted FBS (cat.
no. AB-FBS-ED-0500, ABW GmbH), and divided into the following
groups: i) The non-CS group (non-CS); ii) and the CS group. In the
CS group, the cells were subjected to CS for 6 h, using the
Flexercell Tension Plus TM FX-5000T system (Flexcell International)
set at 20% elongation with 30 cycles/min as previously described
(30).
Treatment of macrophages
BMDM or Raw264.7 macrophages were seeded in 6-well
plates (5.0×105 cells/well), grown to 70-80% confluence,
and divided into the following groups: i) The non-CS-exo (incubated
with exosomes, which were isolated from non-CS group medium); and
ii) the CS-exo group (incubated with exosomes isolated from CS
group medium). The cells were treated with non-CS-exo or CS-exo
(100 µg/ml), as previously described (28) for 12 h.
Cell transfection
Raw264.7 macrophages were transfected with
miR-21a-5p mimic or negative control (NC) (100 nM, synthesized by
Guangzhou RiboBio Co., Ltd.) using HiPerFect transfection reagent
(cat. no. 301707; Qiagen GmbH). The miR-21a-5p mimic/NC sequences
were: miR-21a-5p mimic sense, 5′-UAG CUU AUC AGA CUG AUG UUG A-3′
and anti-sense, 5′-UCA ACA UCA GUC UGA UAA GCU A-3′; miR-21a-5p NC
sense, 5′-UUU GUA CUA CAC AAA AGU ACU G-3′ and anti-sense, 5′-CAG
UAC UUU UGU GUA GUA CAA A-3′. Raw264.7 macrophages were also
transfected with SOCS1 siRNA or NC (100 nM, synthesized by
Guangzhou RiboBio Co., Ltd.) for 21 h and then treated with
Jagged-1 (JAG; 20 ng/ml, cat. no. ab109346, Abcam) for 3 h. The
SOCS1 siRNA/NC target sequences were: SOCS1 siRNA, 5′-CTACCT GAG
TTC CTT CCC C-3′; SOCS1 NC, 5′-TTC TCC GAA CGT GTC ACG T-3′.
Raw264.7 macrophages were transfected with SOCS1 siRNA or NC at
37°C for 24 h. The results of the cell transfection efficiency with
miR-21a-5p mimic and SOCS1 siRNA are presented in Fig. S1A and B.
Raw264.7 were transfected with miR-21a-5p inhibitor
or NC (100 nM, Guangzhou RiboBio Co., Ltd.) using HiPerFect
transfection reagent (cat. no. 301707, Qiagen GmbH) 24 h before
co-culturing with exosomes derived from ECs subjected to CS.
Raw264.7 cells were transfected with miR-21a-5p inhibitor or NC at
37°C for 24 h. The miR-21a-5p inhibitor/NC sequences were:
miR-21a-5p inhibitor, 5′-UCA ACA UCA GUC UGA UAA GCU A-3′;
miR-21a-5p NC, 5′-CAG UAC UUU UGU GUA GUA CAA A-3′. The cell
transfection efficiency with miR-21a-5p inhibitor is presented in
Fig. S1C.
MLE12 cells were seeded onto fibronectin-coated (10
µg/cm2) flexible silastic membranes
(5.0×105 cells/well) and transfected with fluorescein
amidite (FAM)-miR-21a-5p mimic (100 nM, Guangzhou RiboBio Co.,
Ltd.) at 37°C for 24 h. The cell transfection efficiency with
FAM-miR-21a-5p mimic is presented in Fig. S2A. The cells were subjected to
CS for 6 h as described above. The exosomes which contained
FAM-miR-21a-5p mimic were isolated from the medium of MLE12 cells
subjected to CS.
THP1 cells were seeded in 6-well plates
(5.0×105 cells/well), and were differentiated from
monocytes following treatment with phorbol 12-myristate 13-acetate
(PMA, 100 nM, cat. no. P8139, MilliporeSigma) for 48 h. The THP1
cells were then transfected with miR-21a-5p mimic or NC (100 nM,
Guangzhou RiboBio Co., Ltd.) using HiPerFect transfection reagent
(cat. no. 301707; Qiagen GmbH) at 37°C for 48 h. The cell
transfection efficiency with miR-21a-5p mimic is presented in
Fig. S2B.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA, including miRNA, was extracted from
cultured cells, exosomes and lung tissues using RNAiso Plus (cat.
no. 9109, Takara Bio, Inc.) according to the manufacturer's
recommendations. The concentration of RNA was measured using a
NanoDrop Lite UV-Vis spectrometer (ND-LITE, Thermo Fisher
Scientific, Inc.). The PrimeScript™ RT kit (cat. no. RR037A, Takara
Bio, Inc.) was used for miRNA and mRNA reverse transcription. A
total of 500 ng RNA was used to synthesize the cDNA. miRNA and mRNA
qPCR analyses were performed using TB Green® Premix Ex
Taq™ II (cat. no. RR820A, Takara Bio, Inc.) and TB
Green® Premix Ex Taq™ (cat. no. RR420A, Takara Bio,
Inc.), respectively. The reactions were performed in triplicate at
95°C for 30 sec, followed by 40 cycles at 95°C for 15 sec, 60°C for
30 sec; and 95°C for 15 sec, 60°C for 1 min, 95°C for 15 sec on an
ABI StepOne Real-Time PCR System, with U6 and glyceraldehyde
3-phosphate dehydrogenase (GAPDH) or β-actin used as the reference
genes for miRNAs and mRNAs, respectively. GAPDH was used as a human
reference gene and β-actin was used as a mouse reference gene. The
mRNA primers (Tables SI and
SII) were synthesized by TSINGKE Biological Technology, Co.,
Ltd. and miRNA primers (Table
SI) were synthesized by Guangzhou RiboBio Co., Ltd. The
relative expression levels were calculated using the
2−ΔΔCq method (31).
Exosome isolation, characterization and
treatment
Exosomes were isolated from the medium of ECs in the
CS or non-CS groups. To remove the cells, the medium was first
centrifuged at 300 × g for 10 min, then centrifuged at 12,000 × g
for 30 min to remove platelets and filtered using a 0.22 µm
polyvinylidene fluoride (PVDF) filter (MilliporeSigma). The
supernatant was then centrifuged at 120,000 × g with a Type 70 Ti
(Beckman Coulter, Inc.) for 70 min to pellet the exosomes. All
procedures for centrifugation were performed at 4°C. The final
pellet containing exosomes were resuspended in 200 µl
phosphate-buffered saline (PBS, cat. no. BL302A, Biosharp) and
stored at −80°C for use in further experiments. The size
distribution of exosomes was assessed using Nanosight tracking
analysis (NTA).
Transmission electron microscopy
(TEM)
TEM was performed as previously described (32). Briefly, exosome pellets from
medium from cells in the CS group were fixed with 2.5%
glutaraldehyde at 4°C for 30 min. A drop of exosome sample was
placed on a carbon-coated copper grid for 10 min, followed by
removal of excess exosomes with absorbent paper, immersion in 1%
phosphotungstic acid solution (pH 7.0) for 30 sec, drying and
examination using a JEOL JEM-1200EX (JEOL Ltd.) transmission
electron microscope at an acceleration voltage of 80 kV.
Macrophage uptake of labelled
exosomes
Exosomes or exosomes which contain FAM-miR-21a-5p
mimic were labeled with PKH26 (cat. no. MINI26, MilliporeSigma).
Subsequently, 100 µl exosome suspension were mixed with 500
µl Diluent C and 2 µl PKH26 to label the membranes.
Following incubation for 5 min at room temperature, 1 ml of 1%
bovine serum albumin (BSA, cat. no. BS114-100g, Biosharp) was added
to terminate the labeling reaction. Thereafter, PKH-26-labeled
exosomes (10 µg) were added to the Raw264.7 cells and
incubated at 37°C for 12 h. Nuclei were stained with
4′,6-diamidino-2-phenylindole (DAPI, cat. no. F6057,
MilliporeSigma) at room temperature for 5 min and viewed under a
confocal microscope (Olympus Corporation).
Enzyme-linked immunosorbent assay (ELISA)
of cytokines
The TNF-α, IL-1β, IL-6 and IL-10 levels in cell
culture supernatants were assessed using TNF-α (cat. no.
EMC102a.96), IL-1β (cat. no. EMC001b.96), IL-6 (cat. no. EMC004.96)
and IL-10 (cat. no. EMC005.96) (all from Neobioscience Technology
Co., Ltd.) ELISA kits following the manufacturer's protocols. The
activity of lactate dehydrogenase (LDH) was detected using an LDH
Activity Assay kit (cat. no. E-BC-K046-M, Elabscience
Biotechnology, Inc.).
Cells in bronchoalveolar lavage fluid
(BALF)
A total of 279 male C57BL/6 mice (8 weeks old) were
obtained from Vital River Laboratory Animal Technology. They were
raised in a specific pathogen-free environment at 23±2°C with a
relative humidity of 40-60%, 12-h light/dark cycle. The mice were
provided with free access to food and water.
A total of 63 male C57BL/6 mice (weight, 21±2 g)
were randomly divided into three groups as follows: The control
(ctrl), low-tidal-volume ventilation (LtVt, Vt=8 ml/kg) for 2 h,
and high-tidal-volume ventilation (HtVt, Vt=30 ml/kg) for 2 h. At
the end of ventilation, the mice were sacrificed using an overdose
of 2% sodium pentobarbital (100 mg/kg). The lungs were exposed and
BALF was collected with 1.0 ml pre-cooled PBS (cat. no. BL302A,
Biosharpa) through a tracheal cannula and repeated three times.
BALF from 3 mice was collected into a 50 ml centrifuge tube and
centrifuged at 1,000 × g for 5 min at 4°C to collect cells and
analyzed by flow cytometry. The lung tissues were collected for
further analysis.
Exosome administration in vivo
To explore the function of exosome in vivo, a
total of 51 male C57BL/6 mice were randomly divided into three
groups as follows: The control (ctrl), non-CS-exo and CS-exo. The
mice were administrated with non-CS-exo or CS-exo (300
µg/mouse) (28) through a
tracheal cannula. After 24 h, the animals were sacrificed with an
overdose of 2% sodium pentobarbital, and the lung tissues were then
collected. The cells in BALF were collected according to the
methods described above and analyzed using a LSRFortessa flow
cytometer (BD Biosciences).
A total of 60 male C57BL/6 mice were randomly
divided into two groups as follows: The CS-exo + NC and CS-exo +
anti-miR-21a-5p groups. For miR-21a-5p inhibition, the miR-21a-5p
antagomir (Guangzhou RiboBio Co., Ltd.) were used according to the
manufacturer's instructions. Briefly, the mice were administered
miR-21a-5p antagomir or NC (2.5 nM/mouse) 24 h prior to the
challenged with CS-exo (300 µg/mouse) through a tracheal
cannula.
Animal treatment
A total of 45 male C57BL/6 mice were randomly
divided into five groups as follows: The ctrl + NC, LtVt + NC, LtVt
+ anti-miR-21a-5p, HtVt + NC and HtVt + anti-miR-21a-5p groups. The
mice were administered miR-21a-5p antagomir or NC (2.5 nM/mouse) 24
h prior to the administration of LtVt or HtVt. Following
ventilation for 2 h, the mice were sacrificed using an overdose of
2% sodium pentobarbital (100 mg/kg) and the cells in BALF were
collected according to the methods described above.
A total of 60 male C57BL/6 mice were randomly
divided into four groups as follows: The PBS + NC, JAG + NC, PBS +
miR-21a-5p agomir and JAG + miR-21a-5p agomir groups. The
miR-21a-5p agomir or NC (Guangzhou RiboBio Corporation, China) were
used according to the manufacture's instruction. Mice were
administrated with miR-21a-5p agomir or NC (2.5 nM/mouse) 24 h and
sacrificed with an overdose of 2% sodium pentobarbital, then
collected the lung tissues and the cells in BALF according to the
methods described above. The miR-21a-5p agomir or NC (Guangzhou
RiboBio Co., Ltd.) and JAG were used according to the
manufacturer's instructions. The mice were administered miR-21a-5p
agomir or NC (2.5 nM/mouse) 23 h prior to the challenge with JAG
(0.5 mg/kg) or PBS through a tracheal cannula. Following the
challenge with JAG for 1 h, the mice were sacrificed as described
above, and the BALF and lung tissues were collected for further
analysis.
Dual-luciferase reporter assays
TargetScan (http://www.targetscan.org/vert_71/) assays were used
to identify potential miR-21a-5p targets. In this website, the
species 'mouse' was selected, the microRNA name 'miR-21-5p' was
entered and the 'submit' button was then clicked. Target genes were
then screened according to 'Cumulative weighted context + + score'
and 'Total context + + score'. 293T cells (obtained from ATCC) at
70-80% confluence was co-transfected with wild-type or mutant
Notch2 3′-UTR (i.e., with a mutated binding site) luciferase
plasmids (50 ng, pmiR-RB-m-Notch2-WT or pmiR-RB-m-Notch2-MUT,
respectively; Guangzhou RiboBio Co., Ltd.) and miR-21a-5p mimic or
NC (50 nM, Guangzhou RiboBio Co., Ltd.) using HiPerFect
transfection reagent. The Firefly luciferase activity was
employed as the reference. Following incubation for 48 h at 37°C,
the cells were assessed using a Dual-Luciferase Reporter Assay
System (Promega Corporation) with a Glomax96 spectrophotometer
(Promega Corporation). The cell transfection efficiency with
miR-21a-5p mimic is presented in Fig. S2C.
Western blot analysis
Proteins were extracted from lung tissues or
cultured cells using radioimmunoprecipitation assay (RIPA) buffer
(cat. no. P0013B, Beyotime Institute of Biotechnology) with
protease inhibitors (1:50, cat. no. 04693159001, Roche Diagnostics)
and phosphatase inhibitors (1:50, cat. no. 04906837001, Roche
Diagnostics). Protein concentrations were measured with a
bicinchoninic acid assay kit (cat. no. 23227, Thermo Fisher
Scientific, Inc.). Subsequently, 20 µg proteins were
subjected to gel electrophoresis using 10% SDS-PAGE and
electrophoretically transferred onto a PVDF membrane, then blocked
with 5% BSA (cat. no. BS114-100g, Biosharp) for 1 h at room
temperature. The membranes were then incubated with primary
antibodies overnight at 4°C: Notch2 (1:1,000, cat. no. 5732, Cell
Signaling Technology, Inc.), SOCS1 (1:500, cat. no. sc-518028,
Santa Cruz Biotechnology, Inc.), GAPDH (1:10,000, cat. no. AC002,
ABclonal Biotech Co., Ltd.), CD9 (1:400, cat. no. sc-13118, Santa
Cruz Biotechnology, Inc.), CD63 (1:500, cat. no. A19023, ABclonal
Biotech Co., Ltd.), CD81 (1:400; cat. no. sc-166029, Santa Cruz
Biotechnology, Inc.), inducible nitric oxide synthase (iNOS;
1:1,000, cat. no. A0312, ABclonal Biotech Co., Ltd.), TNF-α
(1:2,000, cat. no. 60291-1-lg, ProteinTech Group, Inc.), IL-6
(1:1,000, cat. no. 21865-1-AP, ProteinTech Group, Inc.), IL-1β
(1:1,000, cat. no. 66737-1-lg, ProteinTech Group, Inc.), arginase 1
(Arg1; 1:5,000, cat. no. 16001-1-AP, ProteinTech Group, Inc.),
CD206 (1:1,000, cat. no. 18704-1-AP, ProteinTech Group, Inc.),
IL-10 (1:500, cat. no. 60291-1-lg, ProteinTech Group, Inc.), or
β-actin (1:10,000, cat. no. AC026, ABclonal Biotech Co., Ltd.) and
then incubated with goat anti-rabbit IgG HRP (1:5,000, cat. no.
BL010A, Biosharp) or goat anti-mouse IgG HRP (1:5,000, cat. no.
BL008A, Biosharp) antibodies for 1 h at room temperature. Images
were captured using the UVP EC3 imaging system (Analytik Jena US
LLC Co., Ltd.) and analyzed using ImageJ v1.52v software (National
Institutes of Health).
Co-immunoprecipitation (Co-IP)
After the BMDMs were treated with non-CS-exo or
CS-exo, Co-IP was performed. Briefly, cells were lysed in RIPA
lysis buffer (cat. no. P0013B, Beyotime Institute of
Biotechnology), with protease inhibitors (1:50, cat. no.
04693159001, Roche Diagnostics) and phosphatase inhibitors (1:50,
cat. no. 04906837001, Roche Diagnostics) for 30 min on ice.
Following centrifugation at 12,000 × g for 15 min 4°C, the 1/10
volume of protein sample was isolated as input, and half of the
protein sample was incubated with 20 µl/ml protein A/G-beads
(cat. no. 1614813, Bio-Rad Laboratories, Inc.) at 4°C for 30 min.
Following centrifugation at 12,000 × g for 15 min 4°C, 1 µg
Notch2 antibody or rabbit control IgG (cat. no. AC005, ABclonal
Biotech Co., Ltd.) was co-incubated with the remaining sample at
4°C for 1 h with gentle rotation. Subsequently, 20 µl beads
were added and incubated overnight at 4°C and the beads were then
extracted and washed three times using RIPA buffer, and the bound
proteins were boiled in 2X Laemmli buffer (cat. no. 1610737,
Bio-Rad Laboratories, Inc.). Finally, the purified proteins were
examined using western blot analysis.
Immunofluorescence
Raw264.7 macrophages were treated with non-CS-exo or
CS-exo, washed twice with pre-cooled PBS, and fixed with 4%
paraformaldehyde at 4°C for 30 min. After blocking with 5% BSA
(cat. no. BS114-100g, Biosharp) at room temperature for 1 h, the
cells were incubated with SOCS1 (1:100, cat. no. sc-518028, Santa
Cruz Biotechnology, Inc.) and Notch2 (1:100, cat. no. 5732, Cell
Signaling Technology, Inc.) antibodies overnight at 4°C, followed
by incubation with Dylight 549, goat anti-rabbit IgG (1:200, cat.
no. A23320, Abbkine Scientific Co., Ltd.) and DyLight 488, goat
anti-mouse IgG (1:200, cat. no. A23210, Abbkine Scientific Co.,
Ltd.) at room temperature for 1 h on a horizontal shaker. Nuclei
were stained with DAPI at room temperature for 5 min and images
were captured using a fluorescence microscope (magnification, ×400,
Olympus Corporation).
mRNA library construction and
sequencing
The MGISEQ2000 platform (BGI-Shenzhen) was applied
to sequence the final ligation PCR products. i) RNA extraction: RNA
was extracted from BMDMs treated with non-CS-exo or CS-exo using an
RNeasy Mini kit (cat. no. 74104, Qiagen GmbH) according to the
manufacturer's protocols, and 1.3 µg/sample total RNA was
used for the construction of the sequencing libraries.
Subsequently, total RNA was qualified and quantified using a Nano
Drop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, Inc.).
ii) mRNA library construction: mRNA molecules were purified with
oligo (dT)-attached magnetic beads. The resulting mRNAs were cut
into small sections with fragmentation reagent. First-strand cDNA
was generated using random hexamer-primed reverse transcription,
followed by a second-strand cDNA synthesis. The synthesized cDNA
was subjected to end-repair and then was 3′ adenylated. Adapters
were ligated to the ends of these 3′ adenylated cDNA fragments, and
the cDNA fragments were then amplified and products were purified
with Ampure XP Beads (Agencourt, cat. no. A63882, Beckman Coulter,
Inc.), and dissolved in EB solution. For quality control, the
library was validated on an Agilent Technologies 2100 bioanalyzer.
Following heat denaturation, the double-stranded PCR products were
circularized with the splint oligo sequence. The single-strand
circle DNA (ssCir DNA) were formatted as the final library. The
library was amplified with phi29 to yield DNA nanoball (DNB) with a
molecular copy number >300. The DNBs were load into the
patterned nanoarray, and the method of combinatorial Probe-Anchor
Synthesis (cPAS) was used to generated single end 50 (pair end
100/150) bases reads. iii) Bioinformatics workflow: Bioinformatics
was completed at BGI-Shenzhen. The detailed protocols were provided
and demonstrated at the BGI website: http://www.bgitechsolutions.com/). The sequencing data
were filtered with SOAPnuke (v1.5.2) as follows: a) Reads
containing sequencing adapter were removed; b) reads with
low-quality base ratio (base quality ≤5) >20% were removed; c)
reads with an unknown base ('N' base) ratio >5% were removed,
and clean reads were then obtained and stored in FASTQ format. The
clean reads were mapped to the reference genome using HISAT2
(v2.0.4) and aligned to the reference coding gene set using Bowtie2
(v2.2.5), and RSEM (v1.2.12) was then used to calculate the
expression levels of genes. Essentially, differential expression
analysis was performed using the DESeq2 (v1.4.5) with a Q value
≤0.05. Pheatmap (v1.0.8) was used to draw the heatmap according to
the expression of gene in different samples. The obtained novel
genes have been deposited in the NCBI Gene Expression Omnibus
Archive (accession no. GSE200932). Kyoto Encyclopedia of Genes and
Genomes (KEGG) (https://www.kegg.jp/) enrichment
analysis of annotated differentially expressed genes was
performed.
Flow cytometry
Cells in BALF, Raw264.7 or BMDMs were passed through
a 100-µm cell strainer, resuspended in
fluorescence-activated cell sorting (FACS) buffer (0.5% FBS and 1
ml of 2 mM EDTA in PBS), followed by Fc Block (0.5 µg/ml,
cat. no. 553141, BD Biosciences) at room temperature for 10 min.
The cells were then stained with the following antibodies for 30
min: Fixable Viability Stain 510 (1:200, cat. no. 564406, BD
Biosciences), APC-Cy7 CD45 (1:200, cat. no. 561037, BD
Biosciences), V450 anti-CD11b (1:200, cat. no. 560456, BD
Biosciences), phycoerythrin (PE) anti-F4/80 (1:400, cat. no.
565410, BD Biosciences), BB700 anti-CD11c (1:200, cat. no. 566505,
BD Biosciences) and Alexa Fluor® 647 anti-CD206 (1:100,
cat. no. 565250, BD Biosciences). The cells were incubated with
antibodies at 4°C in the dark and then analyzed using a BD
LSRFortessa flow cytometer. Data were analyzed using FlowJo
software (v10, TreeStar, Inc.).
Statistical analysis
Data are presented as the mean ± standard error of
the mean (SEM). Means were compared between two groups using a
two-tailed unpaired Student's t-test. One-way analysis of variance
(ANOVA) was used among multiple groups followed by Tukey's post hoc
test. The expression of mRNAs or miRNAs was log-transformed.
P<0.05 was considered to indicate a statistically significant
difference. All calculations were performed using GraphPad Prism
7.0 software (GraphPad Software, Inc.).
Results
Exosomes-derived from CS-treated ECs
transferred miR-21a-5p to macrophages
Alveolar ECs are involved in mechanotransduction
during mechanical ventilation (15). Thus, the present study subjected
MLE-12 cells to CS for 0 and 6 h. No marked differences were
observed in the levels of TNF-α, IL-6, IL-1β and LDH (Fig. S3). However, CS increased
miR-21a-5p expression in MLE-12 cells (Fig. S4). Thus, CS did not result in
significant changes in ECs, apart from the increase in miR-21a-5p
expression in ECs.
As inflammatory cytokine levels were not
significantly altered by subjecting the MLE-12 cells to CS, the
medium of MLE-12 cells was collected after CS, and exosomes were
isolated (Fig. 1A-C) and then
labeled with PKH26. The labeled exosomes were engulfed by
macrophages following incubation with macrophages for 12 h
(Fig. 1D). Exosomal miR-21a-5p
expression (contained within the exosomes) was also increased in
the exosomes derived from CS-exo compared with non-CS group cells
(Fig. 1E). miR-21a-5p was
particularly upregulated compared to other miRNAs related to lung
injury, such as miR-155-5p (33), miR-181a-5p (34), miR-200c-3p (35), miR-221-3p (21), miR-429-3p (36) (Fig. 1E). In addition, the levels of
miR-21a-5p in macrophages was increased in the CS-exo group
compared with the non-CS-exo group (Fig. 1F).
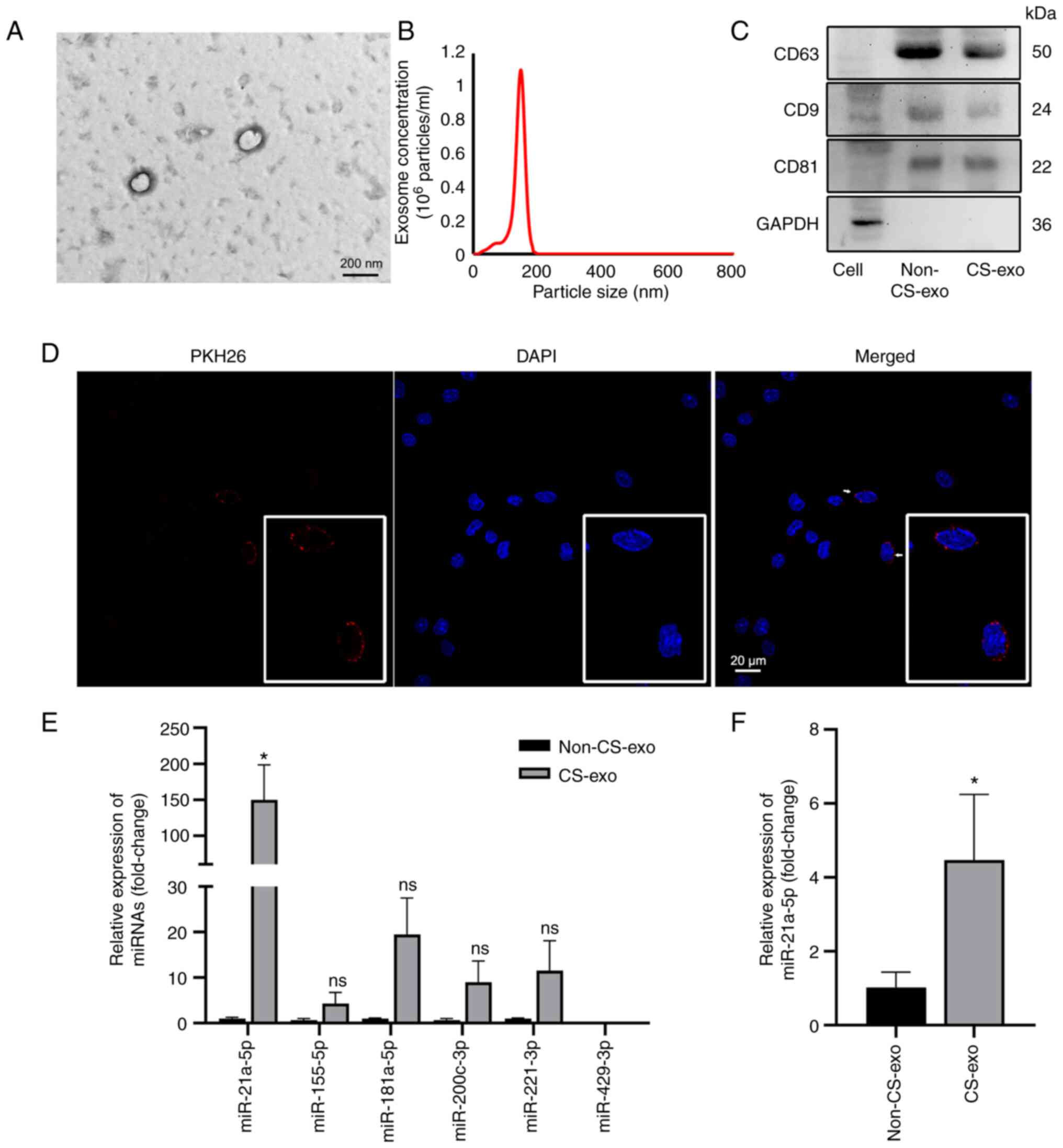 | Figure 1Exosomes from MLE-12 cells subjected
to CS transfer miR-21a-5p to macrophages. (A) Electron microscopy
images of exosomes isolated from medium of cells subjected to CS.
Scale bar, 200 nm. (B) Particle size distribution, assessed using
dynamic light scattering. (C) The levels of the exosomal markers,
CD9, CD63 and CD81, and the internal reference, GAPDH, were
assessed using western blot analysis. (D) Immunofluorescence images
of macrophages incubated with PKH26-labeled exosomes (red) for 12
h. Nuclei was counterstained with DAPI (blue). Scale bar, 20
µm. (E) The expression levels of miRNAs in exosomes were
assessed using reverse transcription-quantitative PCR (n=3). (F)
The levels of miR-21a-5p in macrophages following treatment with
exosomes (n=4). Data are expressed as the mean ± SEM.
*P<0.05, vs. non-CS-exo group. CS, cyclic stretching;
non-CS-exo, incubated with exosomes isolated from medium of cells
not subjected to CS; CS-exo, incubated with exosomes isolated from
medium of cells subjected to CS. |
To verify whether exosomal miR-21a-5p could be taken
up by macrophages, macrophages were cultured with PKH26-labeled
exosomes-derived from ECs transfected with FAM-miR-21a-5p mimic
prior to CS, and FAM-miR-21a-5p signals (green) were detected in
the macrophages' cytoplasm, colocalizing with PKH26 (red) (Fig. S5). These results demonstrated
that ECs treated with CS could secrete exosomes carrying high
levels of miR-21a-5p, and transfer this to macrophages.
Exosomes from CS-treated ECs promote M2
polarization
M1 macrophages have been implicated in several
inflammatory conditions (37),
while M2 macrophages exhibit anti-inflammatory or reparative
functions (38,39). Distinct macrophage phenotypes
contribute to lung inflammation or resolution (40). In the present study, CS did not
induce notable changes in MLE-12 cells apart from the increase in
miR-21a-5p expression. Exosomes were derived from CS-treated ECs
(CS-exo) containing high levels miR-21a-5p, and the present study
then determined whether 12 h of incubation with CS-exo induced
macrophage inflammation in BMDMs. No marked differences were
observed in the levels of IL-6 or TNF-α in the exosome-treated
BMDMs (Fig. 2A); however, the
levels of IL-10 increased in the CS-exo group compared with the
non-CS-exo group (Fig. 2A). Flow
cytometry revealed that the percentage of M2 macrophages
(CD206+CD11c-) increased, while that of M1 macrophages
(CD11c+CD206-) decreased in the CS-exo group (Fig. 2B and C). The levels of M1 and M2
macrophage markers detected using RT-qPCR (Fig. 2D and E) were basically consistent
with the flow cytometry results. In addition, the protein
expression levels of M1 macrophage-related markers (iNOS, TNF-α,
IL-6 and IL-1β) and M2 macrophage-related markers (Arg1, CD206 and
IL-10) in BMDMs in the non-CS-exo and CS-exo groups were examined
using western blot analysis. The protein expression levels of iNOS,
TNF-α, IL-6 and IL-1β were decreased in the CS-exo group compared
with the non-CS-group, while the expression levels of Arg1, CD206
and IL-10 were increased in the CS-exo group (Fig. 2F-H). CS-exo also increased the M2
macrophage polarization of Raw264.7 cells compared to non-CS-exo
(Fig. 2I and J). However, the
percentage of M2 macrophages decreased following the inhibition of
miR-21a-5p prior to co-culturing with exosomes, while the
percentage of M1 macrophages increased (Fig. 2K and L). These data indicated
that CS-treated EC-derived exosomal miR-21a-5p promoted M2
macrophage polarization.
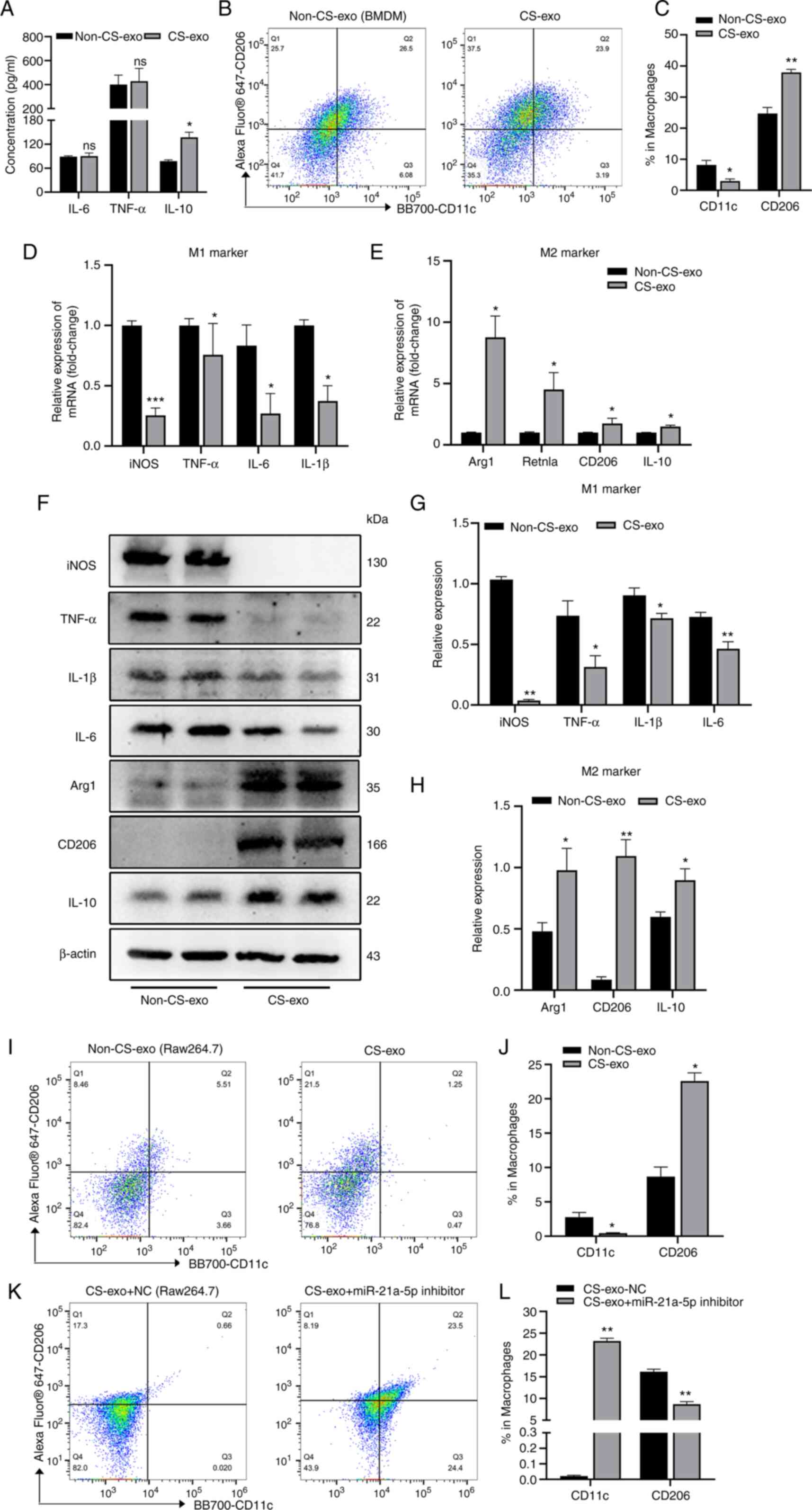 | Figure 2CS-exo promotes the M2 polarization
of macrophages. Exosomes derived from MLE-12 cells subjected to CS
for 6 h were added to BMDMs or Raw264.7 macrophages for 12 h. (A)
IL-6, TNF-α and IL-10 levels in supernatant of BMDMs, assessed
using ELISA (n=4). (B and C) Measurement of the expression of CD11c
and CD206 on BMDMs, assessed using flow cytometry (n=4). (D and E)
Macrophage markers, assessed using reverse
transcription-quantitative PCR (n=7). (F-H) Protein expression
levels of iNOS, TNF-α, IL-1β, IL-6, Arg1, CD206 and IL-10 in BMDMs
following treatment with exosomes (n=4). (I and J) Measurement of
the expression of CD11c and CD206 in Raw264.7 cells (n=3). (K and
L) Inhibition of miR-21a-5p (using miR-21a-5p inhibitor) prior to
CS-exo. Flow cytometry was used to detect CD11c and CD206
expression (n=3). Data are expressed as the mean ± SEM.
*P<0.05, **P<0.001 and
***P<0.001, vs. non-CS-exo group. ns, not
significant; CS, cyclic stretching; non-CS-exo, incubated with
exosomes isolated from medium of cells not subjected to CS; CS-exo,
incubated with exosomes isolated from medium of cells subjected to
CS; BMDM, bone marrow-derived macrophage; iNOS, inducible nitric
oxide synthase; Arg1, arginase 1. |
Exosomal miR-21a-5p from CS-treated ECs
targets Notch2 to regulate macrophage polarization
To determine the exact mechanisms by which
miR-21a-5p induced macrophage polarization, TargetScan (http://www.targetscan.org/vert_71/) assays were
used to identify potential miR-21a-5p targets, which led to the
identification of Notch2. As the dual-luciferase reporter assays
revealed, when wild-type Notch2 3′-UTR was used with the plasmid,
miR-21a-5p overexpression decreased the relative luciferase
activity (Fig. 3A); however, no
significant change was observed when the miR-21a-5p binding site in
the Notch2 3′-UTR was mutated. Moreover, miR-21a-5p mimic
downregulated the expression of Notch2 in Raw264.7 macrophages at
24 h following transfection (Fig. 3B
and C). Furthermore, exosomes from the medium of CS-treated ECs
also downregulated the expression of Notch2 mRNA (Fig. 3D) and protein (Fig. 3E and F) in macrophages. However,
miR-21a-5p inhibition prior to co-culture with exosomes increased
the protein expression of Notch2 (Fig. 3G and H).
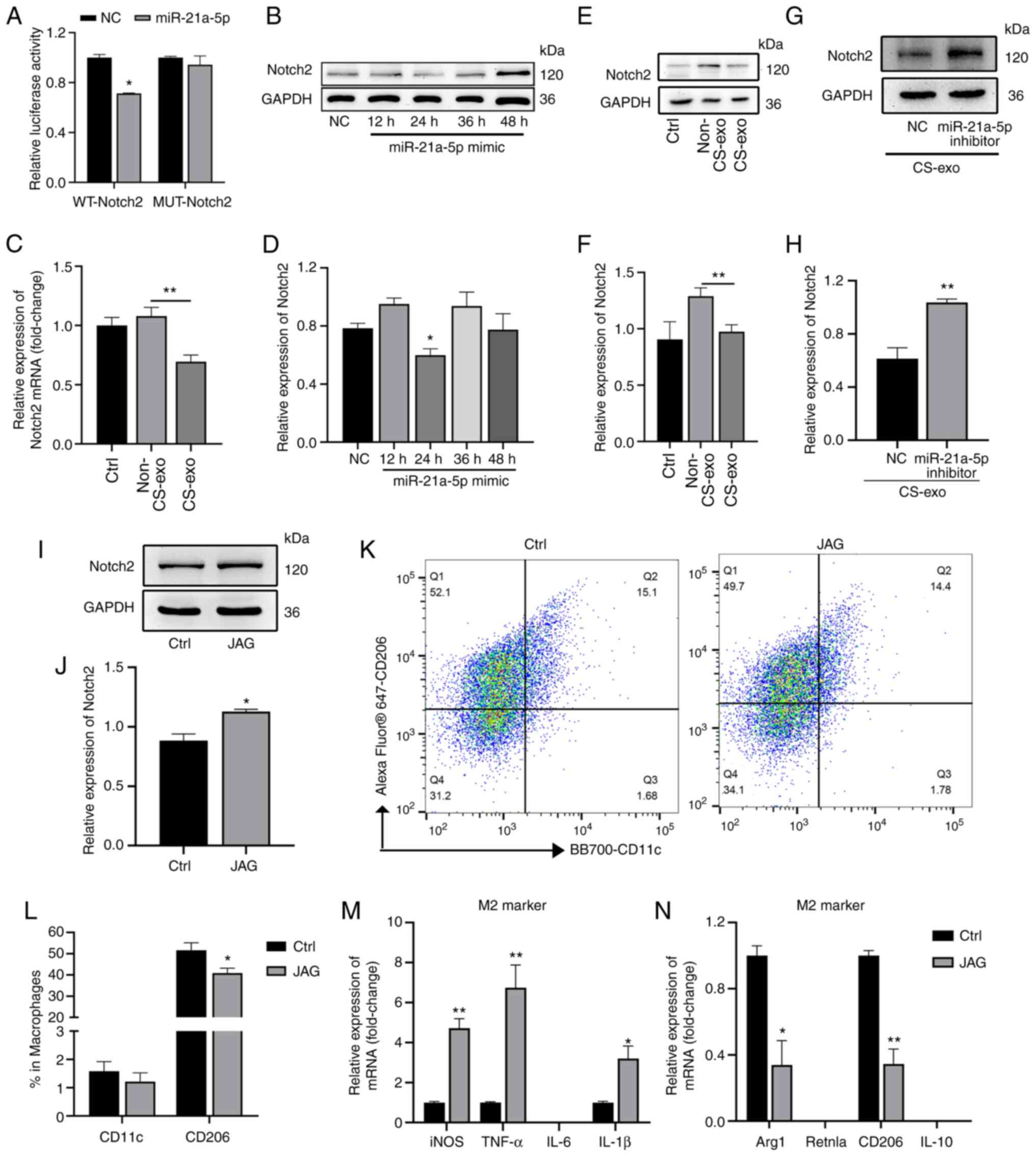 | Figure 3Exosomal miR-21a-5p derived from
epithelial cells subjected to CS targets Notch2 in macrophages. (A)
Luciferase reporter assay following co-transfection with miR-21a-5p
mimic or NC and wild-type or mutant Notch2 3′-UTR luciferase
plasmid (50 ng) (n=3). (B and C) Notch2 expression in Raw264.7
macrophages treated with miR-21a-5p mimic for 12, 24, 36 and 48 h,
assessed using western blot analysis (n=3). (D) Raw264.7 cells were
treated with exosomes and Notch2 mRNA expression was assessed using
RT-qPCR (n=5). (E and F) Raw264.7 cells were treated with exosomes
and Notch2 protein expression was assessed using western blot
analysis (n=6). (G and H) Raw264.7 cells were treated with
miR-21a-5p inhibitor prior to CS-exo treatment. Western blot
analysis was used to detect the expression of Notch2 (n=3). (I and
J) Notch2 protein expression in Raw264.7 macrophages was assessed
using western blot analysis following treatment with JAG for 3 h
(n=3). (K and L) Proportions of M1 and M2 macrophages (determined
using anti-CD11c and anti-CD206 antibodies, respectively) following
treatment with 20 ng/ml JAG for 3 h, assessed using flow cytometry
(n=7). (M and N) Macrophage markers following treatment with 20
ng/ml JAG for 3 h, assessed using RT-qPCR (n=3). Data are expressed
as the mean ± SEM. *P<0.05 and
**P<0.001, vs. the respective control. ctrl, control;
NC, negative control; JAG, Jagged-1; WT, wild-type; MUT, mutant;
RT-qPCR, reverse transcription-quantitative PCR; CS, cyclic
stretching; non-CS-exo, incubated with exosomes isolated from
medium of cells not subjected to CS; CS-exo, incubated with
exosomes isolated from medium of cells subjected to CS; iNOS,
inducible nitric oxide synthase; Arg1, arginase 1; Retnla, resistin
like alpha. |
The present study then activated Notch2 using its
ligand, JAG (20 ng/ml), for 3 h (Fig. 3I and J). Flow cytometry revealed
that the proportion of M2 macrophages was decreased in the Notch2
activation (JAG treatment) group (Fig. 3K and L). RT-qPCR was used to
detect M1 and M2 markers; Arg1 and CD206 (M2 macrophage markers)
mRNA expression was decreased, while iNOS, TNF-α, and IL-1β mRNA
expression was increased (Fig. 3M
and N). Thus, Notch2 was shown to be involved in macrophage
polarization.
Notch2/SOCS1 axis regulates macrophage
polarization
The Notch2 intracellular domain is transported to
the nucleus, and then regulates the transcription of target genes
(41). In the present study, to
explore the downstream mechanisms of Notch2 during mechanical
ventilation, the BMDMs in the non-CS-exo and CS-exo groups were
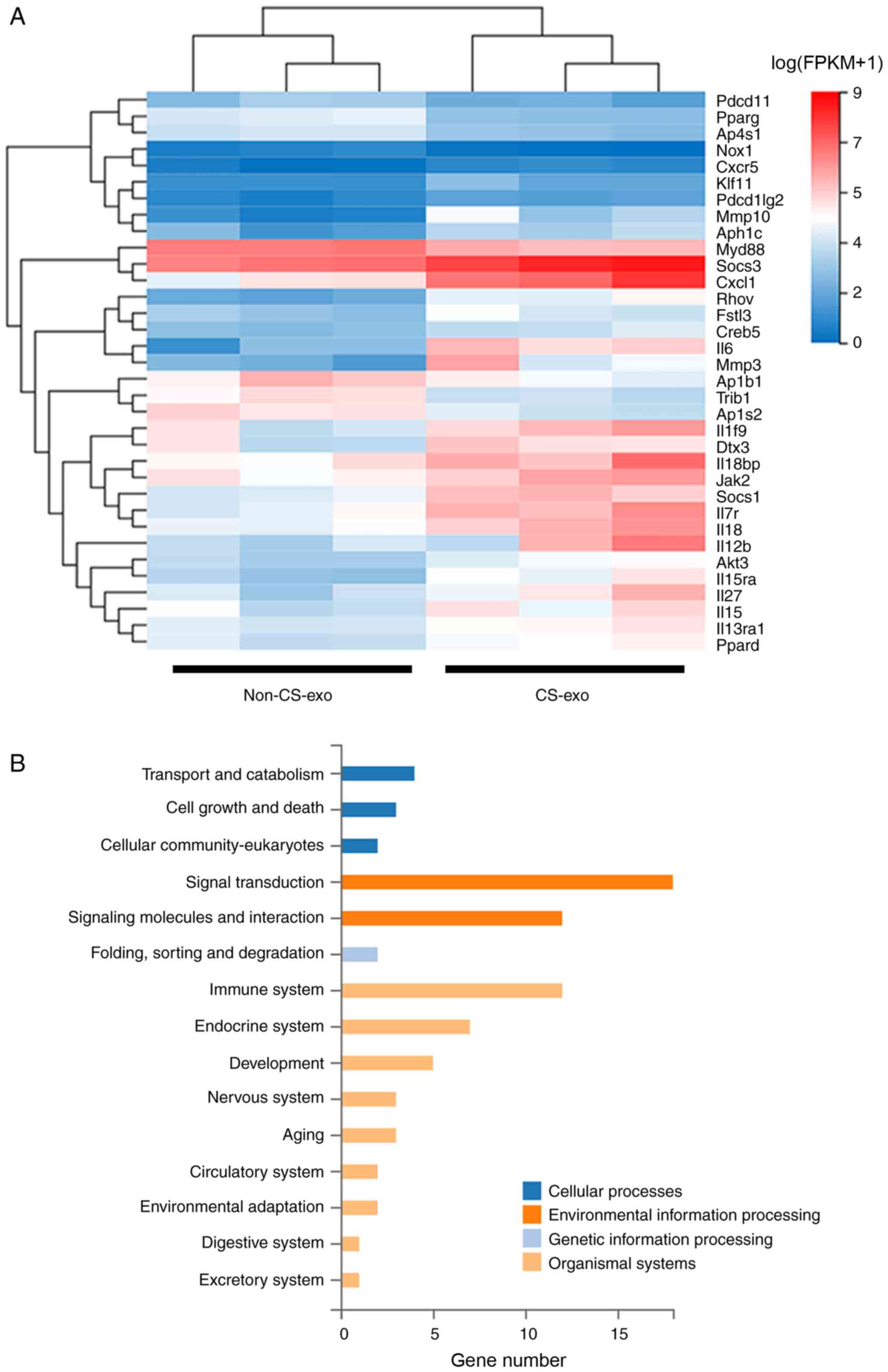
subjected to RNA-seq. The genes associated with M2 macrophage
polarization are presented in Fig.
4A. KEGG analysis revealed that these genes were enriched in
signal transduction (Fig. 4B).
Co-IP assays revealed that Notch2 interacted with SOCS1 in
macrophages (Fig. 5A). Following
treatment with non-CS-exo or CS-exo, SOCS1 mRNA expression
(assessed using RT-qPCR) in macrophages was increased (Fig. 5B) in the CS-exo group compared
with the non-CS-exo group, as in the RNA-seq analysis. SOCS1
protein expression (assessed using western blot analysis) was
downregulated in macrophages (Fig.
5C and D). There are numerous complex and varied
post-transcriptional mechanisms involved in turning mRNA into
protein that are not yet sufficiently well-defined to be able to
compute protein concentrations from mRNA (42); this may be the reason for the
poor concordance between the level of SOCS1 mRNA and protein. The
inhibition of miR-21a-5p also increased SOCS1 expression that was
downregulated by CS-exo (Fig. 5E and
F). The results of immunofluorescence staining revealed that
CS-exo also reduced the fluorescence intensity of Notch2 and SOCS1
(Fig. 5G). As Notch2 activation
(using JAG) decreased the percentage of M2 macrophages compared to
the control group (Fig. 3K and
L), the present study then verified the regulation of
macrophage polarization by Notch2/SOCS1. Raw264.7 macrophages were
transfected with SOCS1 siRNA or NC prior to JAG treatment. The
percentage of M2 macrophages was increased in the siRNA group
compared to the NC group (Fig. 5H
and I). Thus, the Notch2/SOCS1 axis is involved in the M2
polarization of macrophages.
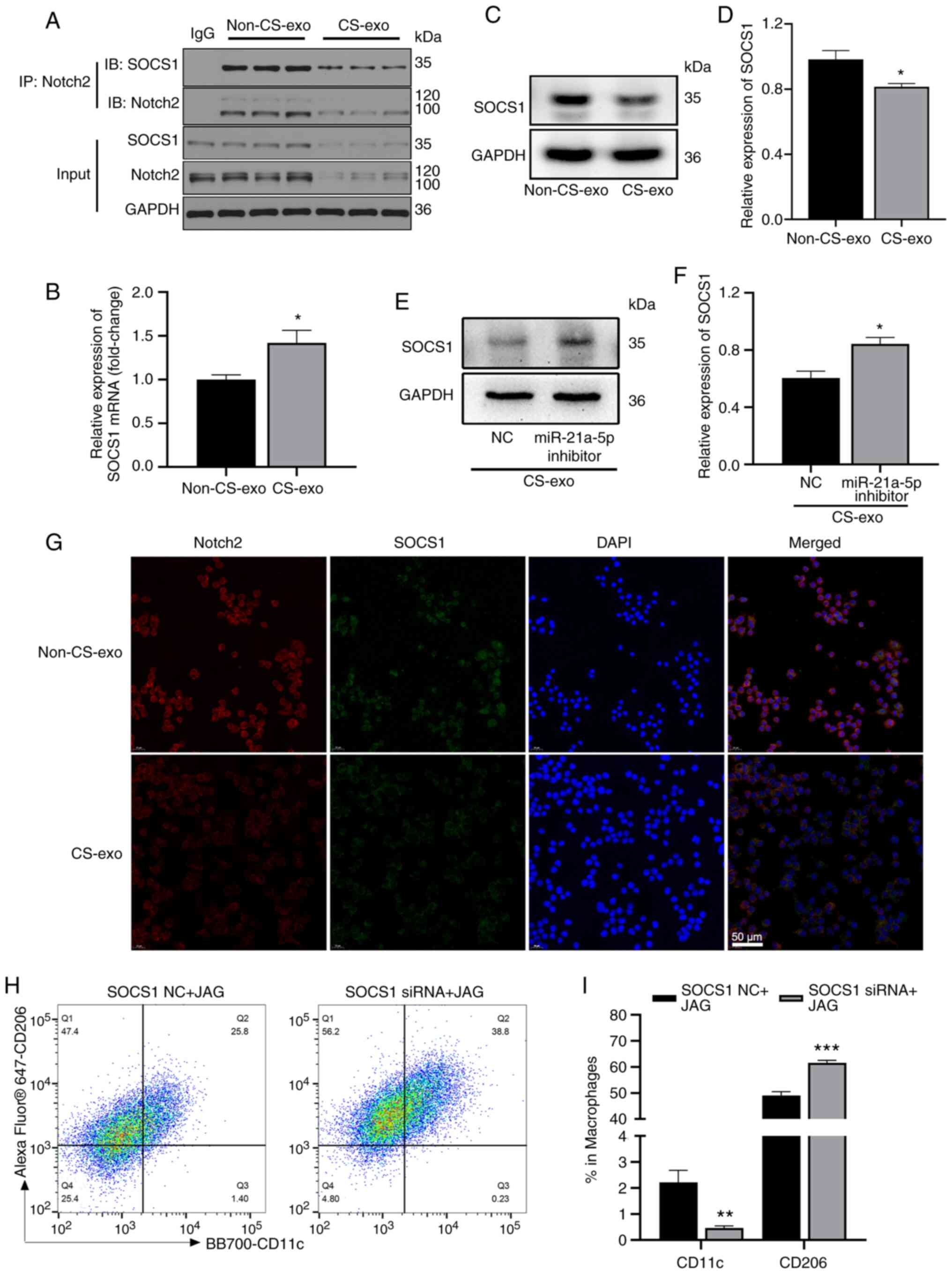 | Figure 5The Notch2/SOCS1 axis regulates
macrophage polarization. (A) Bone marrow-derived macrophages in the
non-CS-exo and CS-exo groups were immunoprecipitated with
anti-Notch2 antibody, or rabbit control IgG, and the precipitates
were examined using western blot analysis [immunoblotting (IB)]
with anti-Notch2 or anti-SOCS1 antibodies (n=3). (B) SOCS1 mRNA
expression in macrophages following treatment with exosomes (n=6).
(C and D) SOCS1 protein expression in macrophages following
treatment with exosomes (n=3). (E and F) Raw264.7 cells were
transfected with miR-21a-5p inhibitor or NC prior to CS-exo
treatment. SOCS1 expression in macrophages was detected using
western blot analysis (n=3). (G) Immunofluorescence co-staining of
SOCS1 and Notch2 in Raw264.7 macrophages following treatment with
exosomes (n=3). (H and I) Proportions of M1 and M2 macrophages
(determined using anti-CD11c and anti-CD206 antibodies,
respectively) following treatment with SOCS1 siRNA or NC prior to
JAG treatment, assessed using flow cytometry (n=6). Data are
expressed as the mean ± SEM. *P<0.05, vs. non-CS-exo
group; **P<0.001 and ***P<0.0001, vs.
NC. NC, negative control; JAG, Jagged-1; CS, cyclic stretching;
non-CS-exo, incubated with exosomes isolated from medium of cells
not subjected to CS; CS-exo, incubated with exosomes isolated from
medium of cells subjected to CS; SOCS1, suppressor of cytokine
signaling 1. |
THP1 cells were also transfected with miR-21a-5p
mimic to determine whether the miR-21a-5p/Notch2/SOCS1 axis
regulates macrophage polarization in human cells. The expression
levels of Notch2 and SOCS1 (Fig.
S6A-C) were suppressed following transfection with miR-21a-5p
mimic. The results of RT-qPCR also revealed that the levels of M1
macrophage markers were downregulated and those of M2 macrophage
markers were upregulated (Fig. S6D
and E). These results further confirmed that the
miR-21a-5p/Notch2/SOCS1 axis regulated macrophage polarization.
miR-21a-5p regulates mechanical
ventilation-induced M2 polarization through Notch2/SOCS1 in
vivo
The present study then verified the function of
epithelial-derived exosomal miR-21a-5p in vivo. The
administration of CS-exo significantly increased the expression of
miR-21a-5p in mouse lung tissues (Fig. 6A). Following treatment with
CS-exo, the iNOS, TNF-α and IL-1β mRNA (M1 macrophage markers)
expression levels were decreased (Fig. 6B), while those of Arg1, resistin
like alpha (Retnla), CD206 and IL-10 mRNA (M2 macrophage markers)
were increased (Fig. 6C). CS-exo
also increased the proportion of M2 macrophages, and decreased the
proportion of M1 macrophages in BALF (Fig. 6D and E). Moreover, CS-exo
decreased Notch2 expression in lung tissues (Fig. S7A and B). Subsequently,
miR-21a-5p antagomir (anti-miR-21a-5p) or NC was administered to
the mice prior to the challenge with CS-exo. miR-21a-5p inhibition
decreased the percentage of M2 macrophages in BALF (Fig. 6F and G), and increased Notch2 and
SOCS1 expression in lung tissues (Fig. S7C and D).
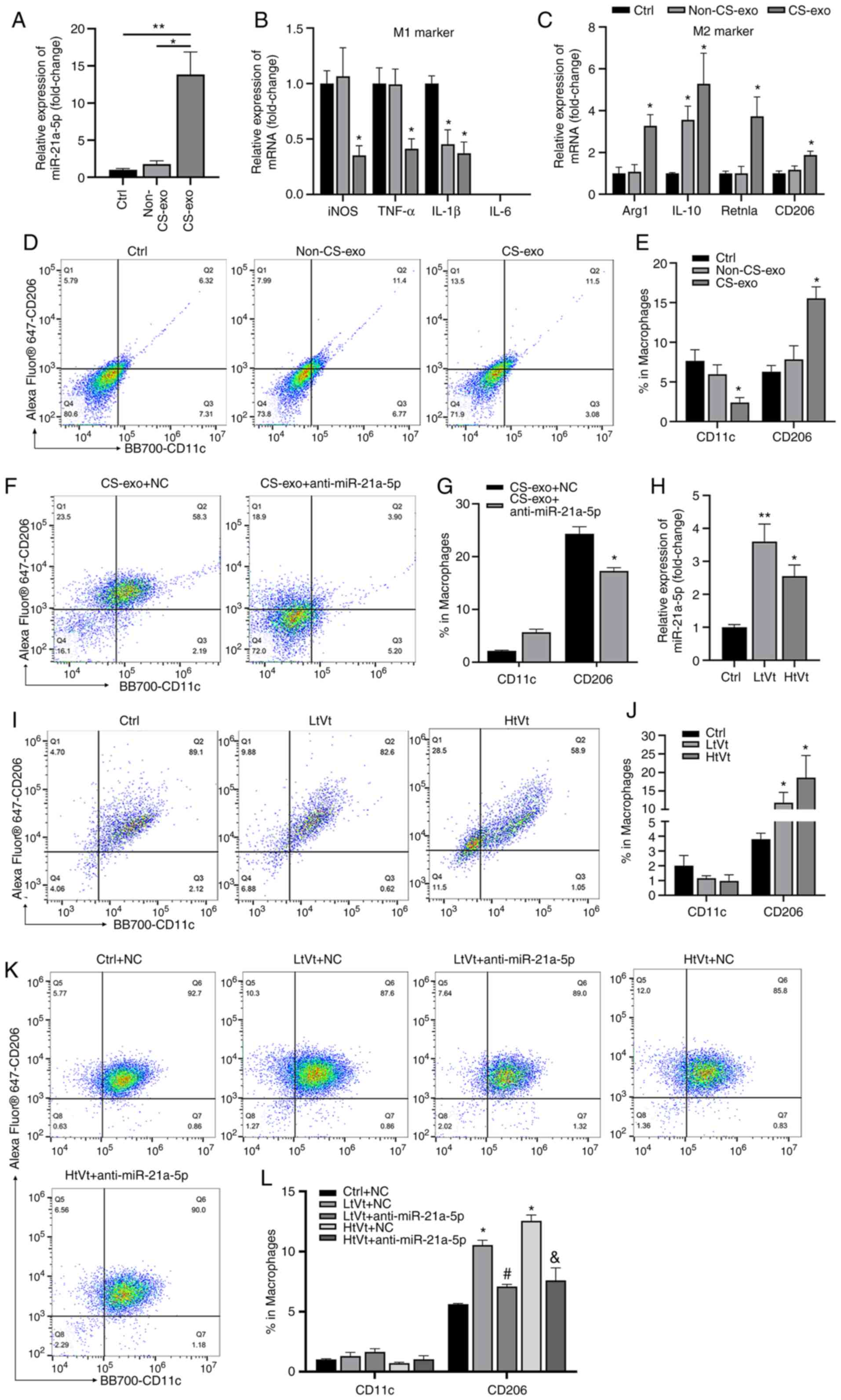 | Figure 6miR-21a-5p regulates M2 polarization
induced by mechanical ventilation in vivo. Mice were
challenged CS-exo through a tracheal cannula or subjected to
mechanical ventilation. Mice were administered miR-21a-5p antagomir
or NC 24 h prior to treatment with CS-exo or mechanical
ventilation. (A) Mice were treated with CS-exo, and the level of
miR-21a-5p in lung tissue was assessed using RT-qPCR (n=8). (B and
C) Macrophage markers, assessed using RT-qPCR (n=6). (D and E) The
expression of CD11c and CD206 in cells in BALF was measured using
flow cytometry (n=9). (F and G) Mice were administered miR-21a-5p
antagomir or NC 24 h prior to treatment with CS-exo. The expression
of CD11c and CD206 in cells in BALF was detected using flow
cytometry (n=9). (H) The expression of miR-21a-5p in lung tissues
of mice subjected to LtVt or HtVt or the ctrl procedure for 2 h was
assessed using RT-qPCR (n=8). (I and J) Macrophage phenotypes in
BALF of mice subjected to LtVt or HtVt or the ctrl procedure for 2
h were assessed using flow cytometry (n=12). (K and L) The
expression of CD11c and CD206 in cells in BALF was detected using
flow cytometry (n=9). Data are expressed as the mean ± SEM.
*P<0.05 and **P<0.01, vs. control;
#P<0.05, vs. LtVt + NC; &P<0.05,
vs. HtVt + NC. NC, negative control; CS, cyclic stretching;
non-CS-exo, incubated with exosomes isolated from medium of cells
not subjected to CS; CS-exo, incubated with exosomes isolated from
medium of cells subjected to CS; RT-qPCR, reverse
transcription-quantitative PCR; BALF, bronchoalveolar lavage fluid;
Arg1, arginase 1; Retnla, resistin like alpha; LtVt,
low-tidal-volume ventilation; HtVt, high-tidal-volume
ventilation. |
Furthermore, mice were treated with mechanical
ventilation to further validate the effects of miR-21a-5p in
vivo. C57BL/6 mice were ventilated with low (8 ml/kg) or high
(30 ml/kg) tidal volumes for 2 h. As shown in Fig. 6H, the expression of miR-21a-5p in
the mechanical ventilated model was significantly increased. The
percentage of M2 macrophages in BALF was upregulated in both the
HtVt and LtVt groups compared to the control group; however, there
was no difference between high- and low-tidal volume groups, while
the percentage of M1 macrophages was not significantly altered
(Fig. 6I and J). Mechanical
ventilation also decreased the expression of Notch2 and SOCS1 in
lung tissues (Fig. S7E and F).
Notably, as shown in Fig. 6K and
L, the proportion of M2 macrophages was significantly decreased
in mice that were treated with miR-21a-5p antagomir prior to
ventilation.
In addition, miR-21a-5p agomir was used to mimic the
high levels of miR-21a-5p in ventilated mice. The Notch2 and SOCS1
levels were decreased after the mice were administered miR-21a-5p
agomir (Fig. S7G and H), and
the proportion of M2 macrophages increased (Fig. S7I and J). However, the increase
in the expression of Notch2 using its ligand, JAG, reversed these
effects (Fig. S7G, H, K and L).
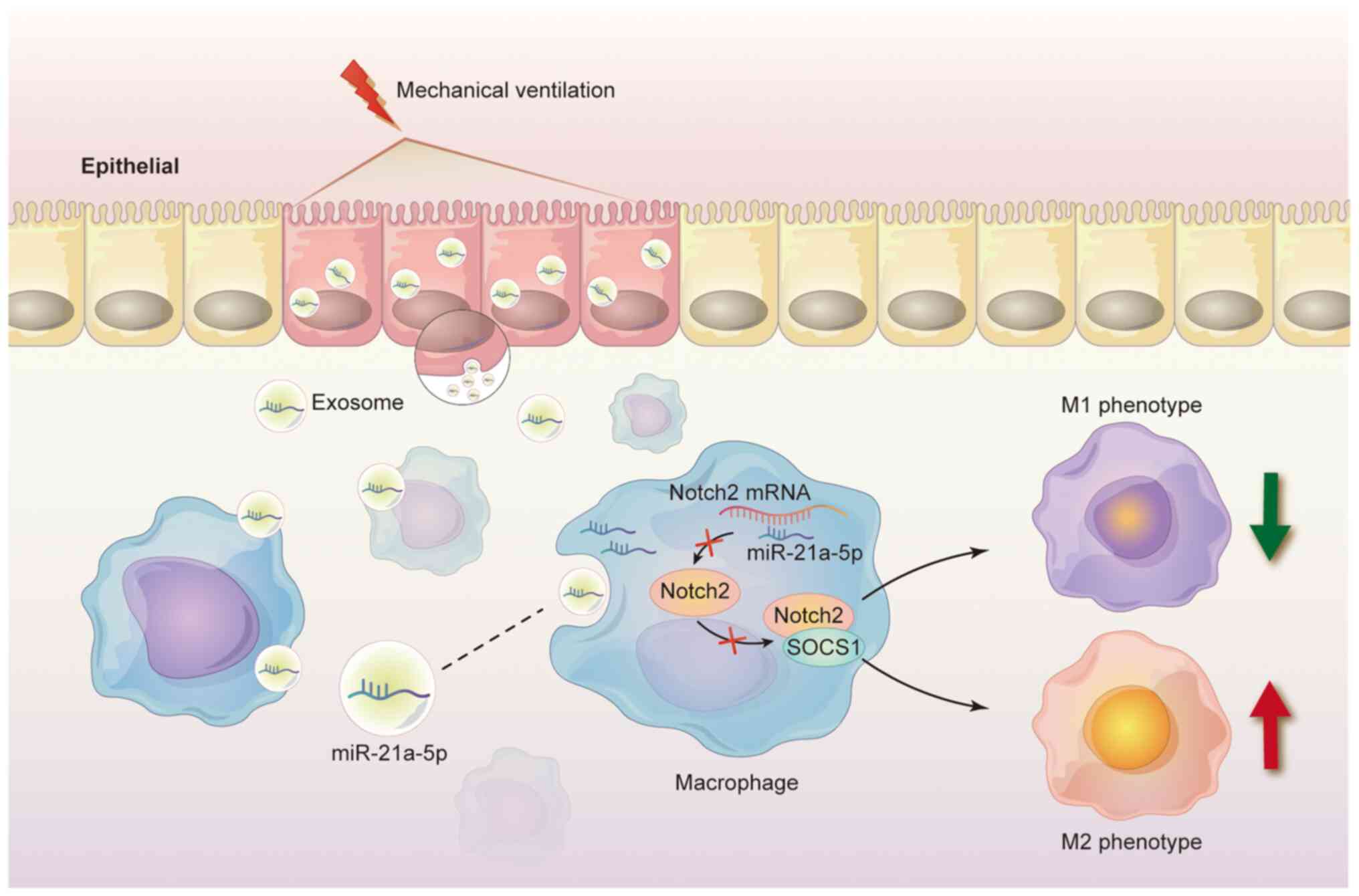
In summary, these data indicated that mechanical ventilation
induced the release of exosomal miR-21a-5p from epithelial cells,
which can be taken up by macrophages to promote M2 macrophages via
downregulating Notch2/SOCS1 (Fig.
7).
Discussion
The present study demonstrated that exosomes
released by CS-treated ECs were internalized by macrophages,
resulting in M2 macrophage polarization. Additionally, it was
demonstrated that EC-derived exosomes contained miR-21a-5p and
promoted macrophage M2 polarization via the downregulation of the
Notch2/SOCS1 signaling axis in the recipient macrophages in
mechanical ventilation (Fig.
7).
Intercellular communication between pulmonary ECs
and macrophages maintains pulmonary homeostasis under physiological
conditions (7). Macrophages are
activated when homeostasis is disturbed. Previous research has
emphasized the emerging role of exosomes in intercellular signal
transmission and exosomes, as extracellular vesicles, can transfer
RNAs, DNAs, lipids, and proteins via autocrine and paracrine
mechanisms (18); this provides
a medium between ECs and macrophages in lung tissue. In lung
tissue, exosomes can mediate macrophage polarization through
multiple pathways. Adipose-derived mesenchymal stem cell-derived
exosomes promote macrophage polarization by inhibiting TLR4 and
alleviating lung injury (43).
Previous studies have demonstrated that alveolar epithelial-derived
exosomes also mediate macrophage activation (29,44). These results indicate that
exosomes play a critical role in AM activation. The present study
demonstrated that exosomes derived from alveolar ECs treated with
CS were internalized by macrophages and promoted M2 polarization.
CS-exo also induced the release of IL-10 in macrophages, suggesting
that exosomes-derived from CS-treated ECs exerted an
anti-inflammatory effect.
miRNAs are the main contents transferred in
exosomes, which serve as novel regulators of cellular functions by
inhibiting translation or inducing mRNA degradation. Certain miRNAs
may be transferred into exosomes and delivered to target cells to
regulate cell functions (45).
Yao et al (46)
demonstrated that miR-21 in mesenchymal stem cell-derived exosomes
promoted M2 macrophage polarization in sepsis. In addition, Lee
et al (47) demonstrated
that miR-21 antagonism inhibited M2 macrophage polarization and
reduced airway hyperresponsiveness. These findings indicated that
miR-21 plays an essential role in the regulation of M2 macrophage
polarization. It was also suggested that CS-treated EC-derived
exosomal miR-21a-5p promoted M2 macrophage polarization, which may
function as a regulator of macrophage polarization.
The specific mechanisms by which miR-21a-5p induced
the polarization of macrophages remain to be further investigated.
TagertScan assays were used to predict the possible targets mRNA of
miR-21a-5p. When exploring miR-21a-5p protein interactions, Notch2,
a highly conserved cell surface receptor, attracted our interest,
as it is closely related to macrophage polarization (48-50). Notch2 is mainly expressed in stem
cells and primordial cells, and also widely exists on the surface
of macrophages, dendritic cells and B cells (41). Zhang et al (51) found that Notch2 signal
transduction negatively regulated inflammatory cytokines and signal
transduction in macrophages. Herein, a dual-luciferase reporter
assay confirmed that miR-21a-5p could bind to the predicted binding
site of Notch2. CS-exo containing miR-21a-5p decreased the
expression of Notch2 mRNA and protein in macrophages. miR-21a-5p
mimic transfection also decreased the protein expression of Notch2.
CS-exo containing miR-21a-5p also resulted in M2 macrophage
polarization. By contrast, the overexpression of Notch2 reversed
these results. However, the inhibition of miR-21a-5p prior to
CS-exo treatment decreased the M2 macrophage polarization which
induced by CS-exo.
Decreased Notch signaling promotes M2 macrophage
polarization (50), while SOCS3
expression is selectively associated with M1 macrophages and is
essential for maintaining their properties (52). In the present study, SOCS1
protein downregulation was strongly associated with M2 macrophage
polarization in the CS-exo group compared to the non-CS-exo group.
However, SOCS1 mRNA expression was increased in the CS-exo group.
There are several reasons for the poor concordance between the
level of SOCS1 mRNA and protein. First, the process of mRNA
translation into protein is complex and regulated by
post-transcriptional modification, which renders it impossible to
compute protein concentration from mRNA; second, protein half-life
may differ in vivo (53,54). These mechanisms are ubiquitous in
eukaryotic cells, which may lead to inconsistencies in the levels
of SOCS1 mRNA and protein. In the present study, the Co-IP results
confirmed the interaction between SOCS1 and Notch2 in macrophages.
Notch2 activation decreased the percentage of M2 macrophages, while
SOCS1 downregulation (using SOSC1 siRNA) prior to Notch2 activation
reversed this decrease. These findings suggested that the
Notch2/SOCS1 axis affected macrophage polarization.
Finally, the present study demonstrated the role of
exosomal miR-21a-5p in mechanical ventilation. CS-exo
administration increased the expression level of miR-21a-5p in lung
tissue and promoted M2 macrophage polarization in vivo,
while miR-21a-5p inhibition prior to the administration of CS-exo
markedly reduced M2 macrophage activation; this indicated that
miR-21a-5p contributed to M2 macrophage polarization induced by
CS-exo. Furthermore, mechanical ventilation for 2 h increased the
expression of miR-21a-5p and the proportion of M2 macrophages in
BALF. The inhibition of miR-21a-5p (using miR-21a-5p antagomir) led
to the inactivation of M2 macrophages in BALF during ventilation.
These results further suggested that epithelial-derived exosomes
regulated M2-type macrophage polarization during short-term
mechanical ventilation.
In conclusion, the presents study demonstrated that
exosomes derived from mechanically stretched ECs mediated the
polarization of M2 macrophages. Exosomal miR-21a-5p mediated the
crosstalk between ECs and macrophages, and promoted M2 macrophage
activation by inhibiting Notch2/SOCS1. The findings presented
herein provide novel information about the effects of exosomal
miR-21a-5p-derived from ECs on M2 macrophage polarization during
mechanical ventilation, and may serve as a potential treatment
strategy for VILI.
Supplementary Data
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI Gene Expression Omnibus
Archive, with the accession no. GSE200932. The other raw data
supporting the conclusions of the current study are available from
the corresponding author on reasonable request.
Authors' contributions
YW, WX and QW designed and conceived the study. YW,
YH, XL and JL performed the cell experiments and acquired the data.
YX, LC, GL and YF performed the animal experiments. ZX and YW
analyzed and interpreted the results. QW provided the reagents and
expertise. YW and WX wrote the manuscript. YW and QW confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All procedures for the animal experiments were
approved (approval no. 2406) by the Institutional Animal Care and
Use Committee at Tongji Medical College, Huazhong University of
Science and Technology (Wuhan, China).
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Key Research and
Development Program of China (grant no. 2018YFC2001900), the
National Natural Science Foundation of China (grant no. 81873952)
and the National Natural Science Foundation of China (grant no.
81901948).
Abbreviations:
|
ALI
|
acute lung injury
|
|
AMs
|
alveolar macrophages
|
|
BALF
|
bronchoalveolar lavage fluid
|
|
BMDM
|
bone marrow-derived macrophage
|
|
BSA
|
bovine serum albumin
|
|
COPD
|
chronic obstructive pulmonary
disease
|
|
CS
|
cyclic stretching
|
|
Co-IP
|
co-immunoprecipitation
|
|
DAPI
|
4′,6-diamidino-2-phenylindole
|
|
ECs
|
epithelial cells
|
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
FACS
|
fluorescence-activated cell
sorting
|
|
LDH
|
lactate dehydrogenase
|
|
LPS
|
lipopolysaccharide
|
|
PBS
|
phosphate-buffered saline
|
|
PMNs
|
polymorphonuclear neutrophils
|
|
RIPA
|
radioimmunoprecipitation assay
|
|
SOCS1
|
suppressor of cytokine signaling
1
|
|
VILI
|
ventilator-induced lung injury
|
References
|
1
|
Tremblay L, Valenza F, Ribeiro SP, Li J
and Slutsky AS: Injurious ventilatory strategies increase cytokines
and c-fos m-RNA expression in an isolated rat lung model. J Clin
Invest. 99:944–952. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ranieri VM, Suter PM, Tortorella C, De
Tullio R, Dayer JM, Brienza A, Bruno F and Slutsky AS: Effect of
mechanical ventilation on inflammatory mediators in patients with
acute respiratory distress syndrome. JAMA. 282:54–61. 1999.
View Article : Google Scholar
|
|
3
|
Imai Y, Parodo J, Kajikawa O, de Perrot M,
Fischer S, Edwards V, Cutz E, Liu M, Keshavjee S, Martin TR, et al:
Injurious mechanical ventilation and end-organ epithelial cell
apoptosis and organ dysfunction in an experimental model of acute
respiratory distress syndrome. JAMA. 289:2104–2112. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Silva PL, Negrini D and Macêdo Rocco PR:
Mechanisms of ventilator-induced lung injury in healthy lungs. Best
Pract Res Clin Anaesthesiol. 29:301–313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Imanaka H, Shimaoka M, Matsuura N,
Nishimura M, Ohta N and Kiyono H: Ventilator-induced lung injury is
associated with neutrophil infiltration, macrophage activation, and
TGF-beta 1 mRNA upregulation in rat lungs. Anesth Analg.
92:428–436. 2001.PubMed/NCBI
|
|
6
|
Bissonnette EY, Lauzon-Joset JF, Debley JS
and Ziegler SF: Cross-talk between alveolar macrophages and lung
epithelial cells is essential to maintain lung homeostasis. Front
Immunol. 11:5830422020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guillot L, Nathan N, Tabary O, Thouvenin
G, Le Rouzic P, Corvol H, Amselem S and Clement A: Alveolar
epithelial cells: Master regulators of lung homeostasis. Int J
Biochem Cell Biol. 45:2568–2573. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Opitz B, van Laak V, Eitel J and Suttorp
N: Innate immune recognition in infectious and noninfectious
diseases of the lung. Am J Resp Crit Care. 181:1294–1309. 2010.
View Article : Google Scholar
|
|
9
|
Cheng P, Li S and Chen H: Macrophages in
lung injury, repair, and fibrosis. Cells. 10:4362021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Laskin DL, Malaviya R and Laskin JD: Role
of macrophages in acute lung injury and chronic fibrosis induced by
pulmonary toxicants. Toxicol Sci. 168:287–301. 2019. View Article : Google Scholar :
|
|
11
|
Bhattacharya J and Westphalen K:
Macrophage-epithelial interactions in pulmonary alveoli. Semin
Immunopathol. 38:461–469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Corvol H, Flamein F, Epaud R, Clement A
and Guillot L: Lung alveolar epithelium and interstitial lung
disease. Int J Biochem Cell Biol. 41:1643–1651. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Skerrett SJ, Liggitt HD, Hajjar AM, Ernst
RK, Miller SI and Wilson CB: Respiratory epithelial cells regulate
lung inflammation in response to inhaled endotoxin. Am J Physiol
Lung Cell Mol Physiol. 287:L143–L152. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cohen TS, Cavanaugh KJ and Margulies SS:
Frequency and peak stretch magnitude affect alveolar epithelial
permeability. Eur Respir J. 32:854–861. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heise RL, Stober V, Cheluvaraju C,
Hollingsworth JW and Garantziotis S: Mechanical stretch induces
epithelial-mesenchymal transition in alveolar epithelia via
hyaluronan activation of innate immunity. J Biol Chem.
286:17435–17444. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Beckmann A, Grissmer A, Meier C and
Tschernig T: Intercellular communication between alveolar
epithelial cells and macrophages. Ann Anat. 227:151417. 2020.
View Article : Google Scholar
|
|
17
|
Mayer AK, Bartz H, Fey F, Schmidt LM and
Dalpke AH: Airway epithelial cells modify immune responses by
inducing an anti-inflammatory microenvironment. Eur J Immunol.
38:1689–1699. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iraci N, Leonardi T, Gessler F, Vega B and
Pluchino S: Focus on extracellular vesicles: Physiological role and
signalling properties of extracellular membrane vesicles. Int J Mol
Sci. 17:1712016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tomankova T, Petrek M and Kriegova E:
Involvement of microRNAs in physiological and pathological
processes in the lung. Respir Res. 11:1592010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang T, Jiang L, Wei X, Dong Z, Liu B,
Zhao J, Wang L, Xie P, Wang Y and Zhou S: Inhibition of miR-221
alleviates LPS-induced acute lung injury via inactivation of
SOCS1/NF-κB signaling pathway. Cell Cycle. 18:1893–1907. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Q, Ge YL, Li M, Fang XZ, Yuan YP, Liang
L and Huang SQ: miR-127 contributes to ventilator-induced lung
injury. Mol Med Rep. 16:4119–4126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu ZL, Wang H, Liu J and Wang ZX:
MicroRNA-21 (miR-21) expression promotes growth, metastasis, and
chemo- or radioresistance in non-small cell lung cancer cells by
targeting PTEN. Mol Cell Biochem. 372:35–45. 2013. View Article : Google Scholar
|
|
24
|
da Costa Martins PA and De Windt LJ:
miR-21: A miRaculous socratic paradox. Cardiovasc Res. 87:397–400.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu WD, Xu J, Zhang M, Zhu TM, Zhang YH
and Sun K: MicroRNA-21 inhibits lipopolysaccharide-induced acute
lung injury by targeting nuclear factor-κB. Exp Ther Med.
16:4616–4622. 2018.PubMed/NCBI
|
|
26
|
Ding Y, Hou Y, Liu Y, Xie X, Cui Y and Nie
H: Prospects for miR-21 as a target in the treatment of lung
diseases. Curr Pharm Des. 27:415–422. 2021. View Article : Google Scholar
|
|
27
|
Alipoor SD, Mortaz E, Garssen J,
Movassaghi M, Mirsaeidi M and Adcock IM: Exosomes and exosomal
miRNA in respiratory diseases. Mediat Inflamm. 216:56284042016.
|
|
28
|
Jiao Y, Zhang T, Zhang C, Ji H, Tong X,
Xia R, Wang W, Ma Z and Shi X: Exosomal miR-30d-5p of neutrophils
induces M1 macrophage polarization and primes macrophage pyroptosis
in sepsis-related acute lung injury. Crit Care. 25:3562021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu F, Peng W, Chen J, Xu Z, Jiang R, Shao
Q, Zhao N and Qian K: Exosomes derived from alveolar epithelial
cells promote alveolar macrophage activation mediated by miR-92a-3p
in sepsis-induced acute lung injury. Front Cell Infect Microbiol.
11:6465462021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Q, Xie W, Wang Y, Chen S, Han J, Wang
L, Gui P and Wu Q: JAK2/STAT1-mediated HMGB1 translocation
increases inflammation and cell death in a ventilator-induced lung
injury model. Lab Invest. 99:1810–1821. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
32
|
Xu H, Ling M, Xue J, Dai X, Sun Q, Chen C,
Liu Y, Zhou L, Liu J, Luo F, et al: Exosomal microRNA-21 derived
from bronchial epithelial cells is involved in aberrant
epithelium-fibroblast cross-talk in COPD induced by cigarette
smoking. Theranostics. 8:5419–5433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang K, Yang J, Guo S, Zhao G, Wu H and
Deng G: Peripheral circulating exosome-mediated delivery of miR-155
as a novel mechanism for acute lung inflammation. Mol Ther.
27:1758–1771. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li W, Qiu X, Jiang H, Han Y, Wei D and Liu
J: Downregulation of miR-181a protects mice from LPS-induced acute
lung injury by targeting Bcl-2. Biomed Pharmacother. 84:1375–1382.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Q, Du J, Yu X, Xu J, Huang F, Li X,
Zhang C, Li X, Chang J, Shang D, et al: miRNA-200c-3p is crucial in
acute respiratory distress syndrome. Cell Discov. 3:170212017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiao J, Tang J, Chen Q, Tang D, Liu M, Luo
M, Wang Y, Wang J, Zhao Z, Tang C, et al: miR-429 regulates
alveolar macrophage inflammatory cytokine production and is
involved in LPS-induced acute lung injury. Biochem J. 471:281–291.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wynn TA, Chawla A and Pollard JW:
Macrophage biology in development, homeostasis and disease. Nature.
496:445–455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Murray PJ, Allen JE, Biswas SK, Fisher EA,
Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence
T, et al: Macrophage activation and polarization: Nomenclature and
experimental guidelines. Immunity. 41:14–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ivashkiv LB: Epigenetic regulation of
macrophage polarization and function. Trends Immunol. 34:216–223.
2013. View Article : Google Scholar :
|
|
40
|
Hu G and Christman JW: Editorial: Alveolar
macrophages in lung inflammation and resolution. Front Immunol.
10:22752019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fortini ME: Notch signaling: The core
pathway and its posttranslational regulation. Dev Cell. 16:633–647.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Greenbaum D, Colangelo C, Williams K and
Gerstein M: Comparing protein abundance and mRNA expression levels
on a genomic scale. Genome Biol. 4:1172003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tian J, Cui X, Sun J and Zhang J: Exosomal
microRNA-16-5p from adipose mesenchymal stem cells promotes
TLR4-mediated M2 macrophage polarization in septic lung injury. Int
Immunopharmacol. 98:1078352021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen Z, Wu H, Shi R, Fan W, Zhang J, Su W,
Wang Y and Li P: miRNAomics analysis reveals the promoting effects
of cigarette smoke extract-treated Beas-2B-derived exosomes on
macrophage polarization. Biochem Bioph Res Co. 572:157–163. 2021.
View Article : Google Scholar
|
|
45
|
He C, Zheng S, Luo Y and Wang B: Exosome
theranostics: Biology and translational medicine. Theranostics.
8:237–255. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yao M, Cui B, Zhang W, Ma W, Zhao G and
Xing L: Exosomal miR-21 secreted by IL-1β-primed-mesenchymal stem
cells induces macrophage M2 polarization and ameliorates sepsis.
Life Sci. 264:1186582021. View Article : Google Scholar
|
|
47
|
Lee HY, Hur J, Kang JY, Rhee CK and Lee
SY: MicroRNA-21 inhibition suppresses Alveolar M2 macrophages in an
ovalbumin-induced allergic asthma mice model. Allergy Asthma
Immunol Res. 13:312–329. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kovall RA: Structures of CSL, notch and
mastermind proteins: Piecing together an active transcription
complex. Curr Opin Struct Biol. 17:117–127. 2007. View Article : Google Scholar
|
|
49
|
Fiúza UM and Arias AM: Cell and molecular
biology of notch. J Endocrinol. 194:459–474. 2007. View Article : Google Scholar
|
|
50
|
Wang Y, He F, Feng F, Liu XW, Dong GY, Qin
HY, Hu XB, Zheng MH, Liang L, Feng L, et al: Notch signaling
determines the M1 versus M2 polarization of macrophages in
antitumor immune responses. Cancer Res. 70:4840–4849. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Q, Wang C, Liu Z, Liu X, Han C, Cao
X and Li N: Notch signal suppresses toll-like receptor-triggered
inflammatory responses in macrophages by inhibiting extracellular
signal-regulated kinase 1/2-mediated nuclear factor κB activation.
J Biol Chem. 287:6208–6217. 2012. View Article : Google Scholar
|
|
52
|
Liu Y, Stewart KN, Bishop E, Marek CJ,
Kluth DC, Rees AJ and Wilson HM: Unique expression of suppressor of
cytokine signaling 3 is essential for classical macrophage
activation in rodents in vitro and in vivo. J Immunol.
180:6270–6278. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pascal LE, True LD, Campbell DS, Deutsch
EW, Risk M, Coleman IM, Eichner LJ, Nelson PS and Liu AY:
Correlation of mRNA and protein levels: Cell type-specific gene
expression of cluster designation antigens in the prostate. BMC
Genomics. 9:2462008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gygi SP, Rochon Y, Franza BR and Aebersold
R: Correlation between protein and mRNA abundance in yeast. Mol
Cell Biol. 19:1720–1730. 1999. View Article : Google Scholar : PubMed/NCBI
|