Introduction
Prostate cancer (PCa) is the most common malignant
tumor of the male genitourinary system and it is characterized by a
high recurrence rate (1,2). Despite novel technologies and
diagnostic methods applied in clinical practice, a large percentage
of patients have no obvious symptoms or present with an advanced
stage at the time of diagnosis (3). In particular, the incidence of PCa in
the Asia-Pacific region is increasing (4). Therefore, precise treatment
strategies for PCa and the underlying mechanisms of tumor
initiation need to be urgently addressed.
Long non-coding RNAs (lncRNAs) are a specific form
of RNA that consist of a transcript >200 nucleotides in length
that does not code for protein (5). The mechanisms associated with the
effects of lncRNAs on malignant processes in tumors have been
reported extensively and some lncRNAs have been demonstrated to
serve a critical role in the functionality of pathways associated
with PCa. For example, lncRNA PCAT6 has been shown to promote bone
metastasis in PCa by interacting with IGF2BP2 to stabilize IGF1R
(6). Furthermore, lncRNA SNHG17
can aggravate PCa progression in a SNORA71B-dependent manner
(7). LncRNA NXTAR has been
reported to serve as a tumor suppressor that downregulates AR/AR-V7
expression and augments enzalutamide resistance in PCa (8).
Current evidence has suggested that lncRNA cancer
susceptibility candidate 11 (CASC11) is upregulated in numerous
types of cancer and promotes tumorigenesis (9). As well as having value in cancer
diagnosis and prognostic prediction, CASC11 may also have potential
as a target treatment in lung and liver cancer. Zhang et al
(10) reported that c-Myc enhanced
promoter histone acetylation to increase CASC11 expression. Song
et al (11) demonstrated
that CASC11 promoted the progression of hepatocellular carcinoma by
means of EIF4A3-mediated E2F1 upregulation. CASC11 is highly
expressed and acts as an oncogene in various human malignancies,
and can also function as a competitive endogenous RNA (ceRNA) to
exert influence on tumor-related genes by adhering to microRNA
response elements or interacting with proteins (12). Furthermore, abnormal expression of
CASC11 has been reported to be associated with overall survival of
patients with tumors, and may be considered an important indicator
of diagnosis and prognostic assessment (13). Previous studies have illustrated
that CASC11 is associated with PCa proliferation through a ceRNA
mechanism (14). However, the
specific function of CASC11 has not been well investigated in the
context of PCa development and the underlying mechanisms require
elucidation.
The present study aimed to investigate the role of
CASC11 in PCa and its potential oncogenic properties. The present
findings supported the evidence regarding the biological functions
of CASC11 and its underlying role in the Y-box binding protein 1
(YBX1)/p53 axis, which could be used as a promising therapeutic
target for clinical treatment of PCa.
Materials and methods
Patients and tissue samples
A total of 66 paired PCa tissues and adjacent
non-tumor tissues were obtained from patients who received surgery
at The Second Affiliated Hospital of Anhui Medical University
(Hefei, China) between May 2017 and May 2021. None of the patients
received preoperative therapy. All samples were confirmed by
experienced pathologists. The inclusion criteria were as follows:
i) Patients met the World Health Organization diagnostic criteria
for PCa (15); ii) patients had
the capacity to provide informed consent. The exclusion criteria
were as follows: i) Patient refusal; ii) patients with other
diseases; iii) patients received treatment before admission.
Tissues were snap-frozen in liquid nitrogen after resection and
stored at −80°C. Written informed consent was provided by the
participants. The present study was approved by the Ethics
Committee of The Second Affiliated Hospital of Anhui Medical
University (approval no. 2021-130). Patient information is shown in
Table SI.
Cell culture and transfection
Human PCa cell lines (PC-3, DU145, 22Rv1 and LNCaP)
and a human normal prostate epithelial cell line (RWPE-1) were
obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. Cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (FBS: Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at
37°C with 5% CO2. For cell transduce, LNCaP and 22Rv1
cells at 60-70% confluence were plated in six-well plates at a
density of 5×105 cells per well. The short hairpin RNA
(shRNA) against CASC11 (sh-CASC11) and the shRNA-negative control
(sh-NC) were designed and synthesized by Zorin, and were encoded
within a CMV-PURO-MCS vector (Zorin). The shRNA sequences were as
follows: sh-CASC11-1, 5′-GAT CCG CCC ACA TCA AGC CTT-3′;
sh-CASC11-2, 5′-GAT CCG CCT TCA TAT AAC AGC AGT-3′; sh-CASC11-3,
5′-GAT CCG AAC TCA CCA GCC AAG TT-3′ and sh-NC, 5′-CGT CTA CGT CCC
GTG ATA CAA TAA-3′. For gene overexpression, pcDNA3.1 vectors
(Zorin) were subcloned with CASC11 (OE-CASC11) and an empty vector
was used as a NC. LNCaP and 22Rv1 cells (5×105 cells per
well) were transfected with 2 µg OE-CASC11 or the empty
vector using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 48 h according to the
manufacturer's protocol. After 12 h, cells were subsequently used
for experiments. The sh-NC and sh-CASC11 particles were packaged in
293T cells (cat. no. CRL-3216; American Type Culture Collection)
using a 2nd generation system, with the ratio of lentiviral
construct, packaging plasmid and envelope plasmid as 1
µg:900 ng:100 ng. The 293T cells were then cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
in 5% CO2 at 37°C for 48 h. The cultured media were
centrifuged at 1,000 × g for 5 min at room temperature to remove
packaging cells and the supernatant containing viral particles was
collected. Lentiviral infection (multiplicity of infection, 10) was
performed to knock down CASC11 in LNCaP and 22Rv1 cells. For the
establishment of stable cell lines, cells were infected with
lentivirus along with polybrene (5 µg/ml) and screened with
2 µg/ml puromycin (MilliporeSigma) for 2 weeks.
Subsequently, surviving cells were maintained in complete medium
with puromycin (0.5 µg/ml) and the stable cell lines were
used for subsequent experiments. Transfection efficiency was
validated by reverse transcription-quantitative PCR (RT-qPCR).
LNCaP and 22Rv1 cells (1×106) were also
transfected with corresponding YBX1 small interfering RNA
(siRNA/si, 20 µM), si-NC (20 µM), empty vector (50
nM) and YBX1 overexpression plasmid (OE-YBX1, 50 nM) (Sangon
Biotech Co., Ltd.) within pcDNA3.1 vector (Sangon Biotech Co.,
Ltd.) using Lipofectamine 2000 transfection reagent at 37°C for 48
h. A total of 48 h following transfection, cells were subjected to
subsequent experiments. The target sequences were as follows:
YBX1-siRNA, 5′-CAG UUC AAG GCA GUA AAU AUG CA-3′; si-NC, 5′-CGU GAA
CUA AAG UCG AGU ACU AA-3′. Western blotting was used to validate
the efficiency of transfection.
RT-qPCR
Total RNA was extracted from cells and tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The A260/A280 ratio was used to assess RNA purity. A total
of 2 µg RNA was reverse transcribed into cDNA using the
PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. Expression levels were
determined by qPCR using SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd.). The thermocycling conditions were as follows: Initial
denaturation at 95°C for 30 sec; 40 cycles of 10 sec at 95°C, 30
sec at 60°C and 72°C for 15 sec; with a final extension cycle at
72°C for 5 min. GAPDH was used as an internal control. Fold-changes
were calculated using the 2−∆∆Cq method (16). The primer sequences were as
follows: CASC11, forward 5′-ACC CTA TGG AGA ACC GAG AC-3′ and
reverse 5′-GAG GAC CAA CTC AGT AGG AAA T-3′; GAPDH, forward 5′-AAT
GGG CAG CCG TTA GGA AA-3′ and reverse 5′-GCC CAA TAC GAC CAA ATC
AGA G-3′.
Cell viability assay
Cell viability was detected using the Cell Counting
Kit 8 (CCK-8; Sangon Biotech Co. Ltd.). Briefly, LNCaP and 22Rv1
cells were cultured in 96-well plates (2,000 cells/well). Cell
viability was measured every 24 h, whereby 10 µl CCK-8
solution was added to each well and incubated for 2 h at 37°C. For
assessing the biosafety of PBAE, different concentrations of PBAE
(0, 5, 10, 15, 20, 25 and 30 µg/ml) were added to LNCaP and
22Rv1 cells for 24 h at 37°C. The absorbance was measured
spectrophotometrically at an optical density of 450 nm using a
SpectraMax i3X (Molecular Devices, LLC). The experiments were
repeated three times.
Cell colony formation assay
A total of 500 cells were grown in each well of a
6-well plate for ~2 weeks at 37°C and 5% CO2 until
colony formation was evident. Cells were fixed with methanol for 10
min at room temperature and stained with 0.5% crystal violet for 10
min at room temperature, and images were captured for cell
counting. The colonies were counted using ImageJ software (version
1.5; National Institutes of Health). The experiments were repeated
three times.
Cell migration assay
The cell migration assay was conducted using a
24-well plate with chamber inserts (pore size, 8 µm;
Corning, Inc.). Cells (1×105) in 200 µl
serum-free medium were added to the upper chamber, whereas the
lower chamber contained 800 µl medium supplemented with 10%
FBS. After 24 h of incubation at 37°C and 5% CO2, cells
on the lower surface of the membrane were fixed with 4%
paraformaldehyde for 20 min at room temperature and stained with
Giemsa for 15 min at room temperature. The images were acquired
with an inverted light microscope (Olympus Corporation) and counted
using ImageJ software. The experiments were repeated three
times.
Wound healing assay
The wound healing assay was conducted using a 6-well
plate. When the cell monolayers reached 80% confluence, they were
scratched using a 200-µl pipette tip. Subsequently, cells
were cultivated in fresh serum-free medium at 37°C and 5%
CO2. Images of cell migration were captured at the same
locations at 0 and 24 h using an inverted light microscope (Olympus
Corporation), and the wound area was estimated using ImageJ
software. Wound healing rate was determined as follows: Wound
healing rate (%)=[(wound distance at 0 h-wound distance at 24
h)/(wound distance at 0 h)] ×100. The experiments were repeated
three times.
5-ethynyl-2′-deoxyuridine (EdU)
proliferation assay
To measure cell proliferation, cells were cultured
in 24-well plates and treated with EdU for 4 h according to the
protocol of the EdU Kit (Guangzhou RiboBio Co., Ltd.). Cells were
then fixed with 4% paraformaldehyde for 30 min at room temperature
and permeabilized with 0.5% Triton X-100 for 10 min at room
temperature (MilliporeSigma). Images were captured using a
fluorescence microscope. The experiments were repeated three
times.
RNA immunoprecipitation (RIP)
RIP experiments were performed using a Magna RIP RNA
Binding Protein Immunoprecipitation Kit (cat: no. 17-700;
MilliporeSigma) according to the manufacturer's instructions.
Briefly, 1×107 cells were collected, centrifuged at 4°C
for 5 min at 1,000 × g, washed with pre-cooled phosphate-buffered
saline (PBS) and lysed in 800 µl RIP buffer
(MilliporeSigma). Subsequently, 50 µl protein A/G magnetic
beads (cat. no. 26162; Pierce; Thermo Fisher Scientific, Inc.) were
resuspended in 1 ml RIP wash buffer before being incubated for 1 h
at room temperature with YBX1 (cat. no. ab76149; 1:50; Abcam) or
IgG (cat. no. ab200699; 1:1,000; Abcam) antibodies. The
beads-antibody complex was then mixed with RIP buffer and 100
µl cell lysate at 4°C overnight. The beads were then washed
with RIP wash buffer three times. Subsequently, proteinase K was
added to each immunoprecipitated product and incubated for 30 min
at 55°C to digest the protein. Following centrifugation at 4°C for
5 min at 1,000 × g, total RNA was extracted for agarose gel
electrophoresis on 2% agarose gels and RT-qPCR analysis. Bands were
visualized by staining with ethidium bromide (MilliporeSigma). The
experiments were repeated three times.
RNA pull-down and mass spectrometry
(MS)
CASC11 sense (5′-GGG AAG GGA CAA CAC TAA GCA AAA-3′)
and antisense (5′-AAC GGA CGG AAA CAT AAA AAG GGC-3′) were
amplified in vitro using the MAXIscript™ T7 Transcription
Kit (Thermo Fisher Scientific, Inc.) and labeled via
desulfurization biotinylation using a Pierce™ RNA 3′ End
Desthiobiotinylation kit (Thermo Fisher Scientific, Inc.) before
being incubated with 40 µl streptavidin magnetic beads (cat.
no. 88816; Thermo Fisher Scientific, Inc.) for 2 h at room
temperature. Cell extracts were prepared from 1×107
cells in 900 µl Pierce IP Lysis Buffer (Thermo Scientific,
Inc.), then mixed with biotin-labeled CASC11 at 4°C for 1 h. Biotin
Elution Buffer (Thermo Fisher Scientific, Inc.) was added to
collect the RNA complex pulled down. All experiments were carried
out as recommended by the manufacturer of the Pierce Magnetic
RNA-Protein Pull-Down kit (Thermo Fisher Scientific, Inc.), and the
proteins that engaged with the sense or antisense CASC11 were
identified. After elution, lncRNA-associated proteins were
separated by SDS-PAGE on 10% gels and stained with silver for 2 min
at room temperature. Subsequently, the retrieved proteins were
analyzed using MS. MS analysis was performed on a Q Exactive mass
spectrometer (Thermo Scientific, Inc.) that was coupled to Easy nLC
(Proxeon Biosystems, now Thermo Fisher Scientific) for 60 min. The
mass spectrometer was operated in positive ion mode.
Fluorescence in situ hybridization
(FISH)
LNCaP and 22Rv1 cells were cultured in confocal
dishes (Corning, Inc.). Oligonucleotide modified Cy3-labeled probes
for human lncRNA CASC11 (5′-TTA TGC GGT TGA ATA GTC ACC TCTG-3′)
were designed and obtained from Shanghai GenePharma Co., Ltd. A
cy3-labeled 18S rRNA was used as a positive control (Shanghai
GenePharma Co., Ltd.). Experimental procedures were carried out
according to the instructions included in the Rib™ FISH kit
(Guangzhou Ribobio Co., Ltd.). Briefly, the cell suspension
(1×104) was pipetted onto autoclaved glass slides. The
slides were washed in PBS and fixed in 4% paraformaldehyde for 10
min at room temperature and permeabilized with 0.3% Triton X-100
for 5 min at 4°C. Hybridization was performed at 37°C overnight in
a dark moist chamber. After being washed three times in saline
sodium citrate buffer, the coverslips were sealed with parafilm
containing DAPI. The images were acquired using a confocal
microscope (LSM900; Carl Zeiss AG). The experiments were repeated
three times.
Western blot analysis
Western blotting was carried out based on a standard
protocol. Briefly, cells were washed with chilled PBS and lysed
with RIPA lysis buffer (Beyotime Institute of Biotechnology) The
concentration of each protein was determined using a BCA protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.). Total protein
extracts (20 µg) were separated by SDS-PAGE on 10% gels and
transferred onto PVDF membranes. The membranes were blocked with 5%
nonfat dry milk at room temperature for 2 h and blotted with
primary antibodies against Cyclin A2 (cat. no. 67955; 1:1,000; Cell
Signaling Technology, Inc.), CDK2 (cat. no. 18048; 1:1,000; Cell
Signaling Technology, Inc.), CDK4 (cat. no. 12790; 1:1,000; Cell
Signaling Technology, Inc.), p53 (cat. no. 2527; 1:1,000; Cell
Signaling Technology, Inc.), p21 (cat. no. 2947; 1:1,000; Cell
Signaling Technology, Inc.), YBX1 (cat. no. ab76149; 1:1,000;
Abcam) and GAPDH (cat. no. ab8245; 1:2,000; Abcam) at 4°C
overnight. The membranes were then washed three times with PBS and
incubated with goat anti-rabbit IgG secondary antibody (cat. no.
ab7090; 1:10,000; Abcam) and goat anti-mouse IgG secondary antibody
(cat. no. ab6789; 1:10,000; Abcam) for 1 h at room temperature.
Signals from membranes were measured using ECL Substrate (Bio-Rad
Laboratories, Inc.). GAPDH was used as an internal reference.
Semi-quantitative analysis was performed using ImageJ software. The
experiments were repeated three times.
Flow cytometry
For cell cycle distribution analysis,
1×105 cells were collected and fixed in 70% pre-cooled
ethanol overnight at 4°C. Subsequently, cells were treated with
RNase A (Beyotime Institute of Biotechnology) at 37°C for 30 min
and stained with 500 µl propidium iodide (Beyotime Institute
of Biotechnology) at room temperature for 30 min in the dark. The
percentage of cells in each cycle phase was calculated by flow
cytometry (BD FACSCanto II; BD Biosciences). Data were analyzed by
FlowJo version 10 (FlowJo LLC). The experiments were repeated three
times.
RNA-sequencing (RNA-seq) processing
Total RNA was extracted from sh-NC and sh-CASC11
cells using TRIzol. A NanoDrop (NanoDrop; Thermo Fisher Scientific,
Inc.) was used to measure the quantity and quality of RNA. The DNA
library was constructed using an Illumina Nextera XT DNA Library
Prep Kits (Illumina, Inc.), based on the KAPA stranded RNA-Seq
Library, and enrichment of RNA was performed with oligo(dT)
magnetic beads. Agilent 4200 (Agilent Technologies, Inc.) was used
to detect the quality of the constructed library. To select cDNA
fragments of the preferred 200 bp in length, the library fragments
were purified using the AMPure XP system (Beckman Coulter, Inc.).
Libraries were validated by electrophoresis, pooled, and sequenced
on an Illumina NovaSeq 6000 (150 base pairs, paired ends; Illumina,
Inc.). The RNA sequencing was performed by HaploX Biotechnology.
Differentially expressed mRNAs were identified according to
|log2(Fold Change)|>1 and P<0.05 by R package EdgeR (version
3.6.3; http://bioconductor.org/packages/edgeR/).
Preparation of poly (β-amino ester)
(PBAE)/si-CASC11 nanocomplexes
PBAE was purchased from Xi'an Ruixi Biotechnology
Co., Ltd. CASC11 siRNA (siCASC11; sequence: 5′-GCC CAC AUC AAG CCU
UCA U-3′) was synthesized by Shanghai GenePharma Co., Ltd. Seven
complexes with different ratios (1, 5, 10, 20, 40, 60 and 80 of
PBAE to si-CASC11 by weight) were prepared. Subsequently, PBAE
solutions of different concentrations were vortexed along with a
si-CASC11 solution to obtain PBAE/si-CASC11 nanocomplexes (17,18).
PBAE/si-CASC11 nanocomplexes were mixed with loading buffer and
underwent electrophoresis on 2% agarose gels; si-CASC11 alone was
used as a control. Electrophoresis was performed at 130 V for 15
min and the gel was imaged on the Tanon Gel image system (Tanon
Science & Technology Co., Ltd). The particle size of the
nanocomplexes was determined using a particle size potentiometer
(Nano ZS90; Malvern Panalytical; Spectris Plc).
Transmission electron microscopy
(TEM)
PBAE/siRNA nanoparticles were formed and then
droplets of the sample (5 µl) were applied to hydrophilized
carbon-covered copper grids (300 mesh) for 30 min. The sample was
subsequently rinsed with contrasting material (1% uranyl acetate,
pH 4.5). The remaining stain solution was removed with a filter
paper and air-dried. TEM microstructure was determined using a high
contrast TEM (JEM2010; JEOL, Ltd.) at 120 kV.
In vivo experiments
A total of 36 (n=6 mice/group; weight, 20-25 g; age,
4-6 weeks) male BALB/C nude mice were purchased from Weitong Lihua
Laboratory Animal Technology Co., Ltd. and were maintained in a
specific pathogen-free-grade research center (temperature, 28°C;
humidity, 50%; light/dark cycle, 10/14-h cycle), with free access
to food and water. The mice were divided into the following groups:
sh-NC group; sh-CASC11-1 group; sh-CASC11-2 group; saline group;
si-CASC11 group; PBAE/si-CASC11 group. The animal experiment was
approved by the Animal Research Ethics Committee of The Second
Affiliated Hospital of Anhui Medical University (approval no.
2021080). A total of 5×106 sh-NC and sh-CASC11 22Rv1
cell lines were injected subcutaneously into the nude mice. The
weight and tumor size were measured every 4 days. Tumor volume was
calculated as follows: Volume (mm3)=0.5 ×
width2 × length. After 4 weeks, the mice were euthanized
by cervical dislocation. For comparing the antitumor effect of
si-CASC11 and PBAE/si-CASC11, 5×106 22Rv1 cell lines
were injected subcutaneously into the nude mice and the xenograft
tumors developed 4 weeks after injection. Saline, si-CASC11 or
PBAE/si-CASC11 nanocomplexes were injected intratumorally (50
µl) into the three groups, twice a week for 3 weeks. The
mice were euthanized by cervical dislocation and sacrifice was
confirmed when the mice had stopped breathing and did not respond
to stimulation. Finally, tumor tissues were carefully resected,
images were captured and they were assessed using hematoxylin and
eosin (H&E) staining and immunochemistry (IHC).
H&E staining
Mouse tissues were fixed in 10% formalin solution
for 12 h at room temperature and paraffin-embedded, then cut and
mounted on slides (4 µm). In a descending alcohol series,
the tissue sections were dewaxed and rehydrated. The slides were
then stained with hematoxylin for 2 min at room temperature.
Subsequently, the slides were washed in water for 10 min and then
stained with eosin for 1 min at room temperature after
differentiation in acid alcohol. Images were obtained using a light
microscope (Olympus Corporation).
IHC
Tumor tissues were embedded in paraffin and
sectioned (4 µm) after being fixed with 4% paraformaldehyde
at 4°C for 24 h. Dewaxed sections were rehydrated with ethanol at
room temperature for 15 min and then incubated with 3%
H2O2 for 10 min to inhibit endogenous
peroxidases. For antigen retrieval, citrate buffer (pH 6.0) was
applied for 20 min at 95°C. Slides were incubated with primary
antibodies overnight at 4°C, against Ki-67 (cat. no. ab15580;
1:200; Abcam). Subsequently, the sections were incubated with
HRP-conjugated secondary antibody (cat. no. ab6721; 1:1,000; Abcam)
for 40 min at 37°C. The sections were rinsed three times using TBS
with 0.1% Tween-20 for 5 min. DAB (MilliporeSigma) was also applied
at room temperature for 15 min and the tissues were observed under
a light microscope (BX53; Olympus Corporation). The experiments
were repeated three times.
Bioinformatics analysis
The expression levels of CASC11 in PCa were obtained
from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/) and Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE46602 and
GSE55945 datasets were assessed using the GEO database (19,20).
The expression of CASC family members in prostate tumors and normal
tissue samples, as well as their association with Gleason scores,
were analyzed by Gene Expression Profiling Interactive Analysis
(http://gepia.cancer-pku.cn/). Kyoto
Encyclopedia of Genes and Genomes (KEGG) analysis (https://www.genome.jp/kegg/) was used to identify the
biological functions of CASC11. P<0.05 and |log2(fold
change)|>1 were set as the cut-off criteria. The heatmap plot
was generated using R heatmap package (http://CRAN.R-project.org/package=pheatmap).
Statistical analysis
GraphPad Prism 8 (GraphPad Software, Inc.) was used
for statistical analysis. Unpaired Student's t-test was used for
the comparisons between two groups. Paired Student's t-test was
used to analyze differences between paired tumor and paracancerous
tissues. For comparisons between three or more groups, one-way
ANOVA was performed followed by Dunnett's or Tukey's multiple
comparison test. Data are presented as the mean ± SD, or as scatter
dot plots, box plots or violin plots, where the lines represent the
median. Two-tailed P<0.05 was considered to indicate a
statistically significant difference.
Results
LncRNA CASC11 expression is upregulated
in PCa
Bioinformatics analysis was performed to determine
the expression of CASC11 in PCa in TCGA dataset. Among the CASC
family, the expression levels of CASC11 in TCGA dataset were
increased in prostate tumor tissues compared with those in normal
prostate tissues (Fig. 1A). In
addition, CASC11 expression was elevated in tumor samples from
patients with PCa and high Gleason scores (Fig. 1B). Moreover, the R package was used
to analyze two GEO datasets (GSE46602 and GSE55945) to identify the
expression of CASC11; the results revealed that CASC11 had a
tendency to increase in tumor specimens compared with in normal
tissues. (Fig. 1C and D). To
further verify the oncogenic potential of CASC11, the mRNA
expression levels of CASC11 were analyzed in prostate tumor samples
obtained from The Second Affiliated Hospital of Anhui Medical
University, and the results revealed that the expression levels
were significantly upregulated in tumor samples compared with those
in normal prostate specimens (Fig.
1E). Consistent with these data, analysis of PCa cell lines
(22Rv1, DU145, PC-3 and LNCaP) and RWPE-1 non-tumorigenic prostate
epithelial cells revealed that CASC11 was upregulated in cancer
cells compared with in RWPE-1 cells (Fig. 1F). These data indicated that CASC11
may have a carcinogenic role in PCa. Since CASC11 is expressed
highest in LNCaP and 22Rv1 cells, these two cell lines were used
for subsequent analyses. Furthermore, FISH assay revealed that
CASC11 was mainly distributed in the nucleus of LNCaP and 22Rv1
cells (Fig. 1G).
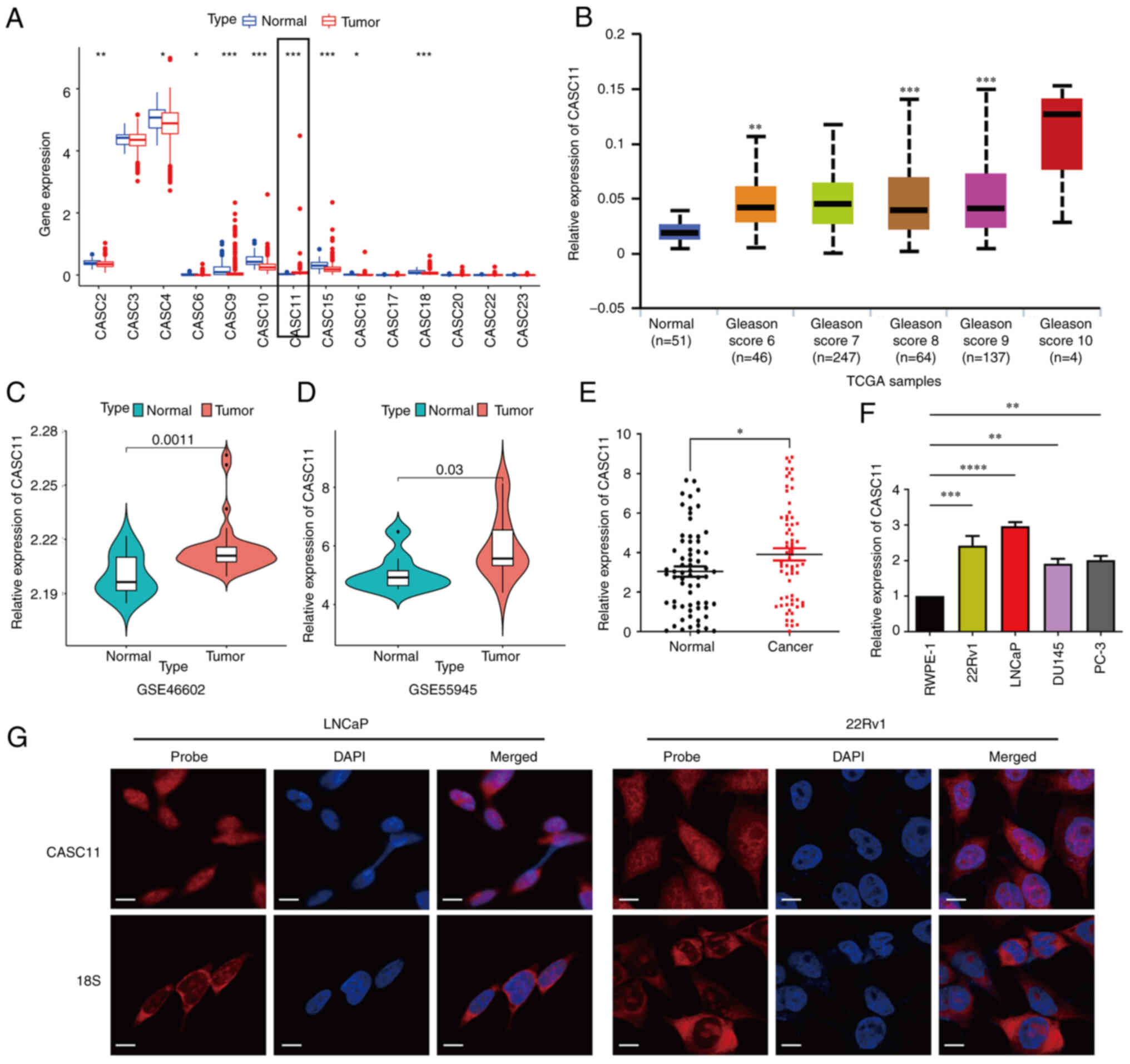 | Figure 1Long non-coding RNA CASC11 expression
is upregulated in PCa. (A) Differential expression profile of
non-coding CASC gene family members in PCa samples and normal
tissues. (B) Expression of CASC11 in prostate tumor and normal
tissue samples with different Gleason scores in TCGA dataset. (C
and D) Expression of CASC11 in prostate tumor and normal tissues in
GSE46602 and GSE55945 datasets. (E) Expression of CASC11 in 66
paired prostate tumor and normal tissue samples. (F) Expression
levels of CASC11 in PCa cell lines (LNCaP, PC-3, DU145 and 22Rv1)
and a prostate epithelial cell line (RWPE-1). (G) Fluorescence
in situ hybridization analysis of CASC11 in LNCaP and 22Rv1
cells. DAPI was used to stain the nuclei, and 18S was used as a
positive control for cytoplasmic staining. Scale bar, 50 µm.
*P<0.05, **P<0.01,
***P<0.001, ****P<0.0001 vs. normal or
as indicated. CASC11, cancer susceptibility candidate 11; PCa,
prostate cancer; TCGA, The Cancer Genome Atlas. |
CASC11 promotes the proliferation and
migration of PCa cells in vitro
Loss-of-function and gain-of-function assays were
conducted in PCa cells to explore the biological function of
CASC11. CASC11 was knocked down in LNCaP and 22Rv1 cells using a
highly efficient shRNA-lentivirus and an overexpression plasmid
system was used to upregulate the expression of CASC11 in these
cell lines (Fig. 2A and B).
Notably, of the three shRNAs, sh-CASC11-1 and sh-CASC11-2 displayed
the highest efficiency; therefore, these were used for further
experiments. CCK-8, colony formation and EdU assays revealed that
knockdown of CASC11 significantly inhibited the proliferative
capacity of PCa cells. By contrast, overexpression of CASC11
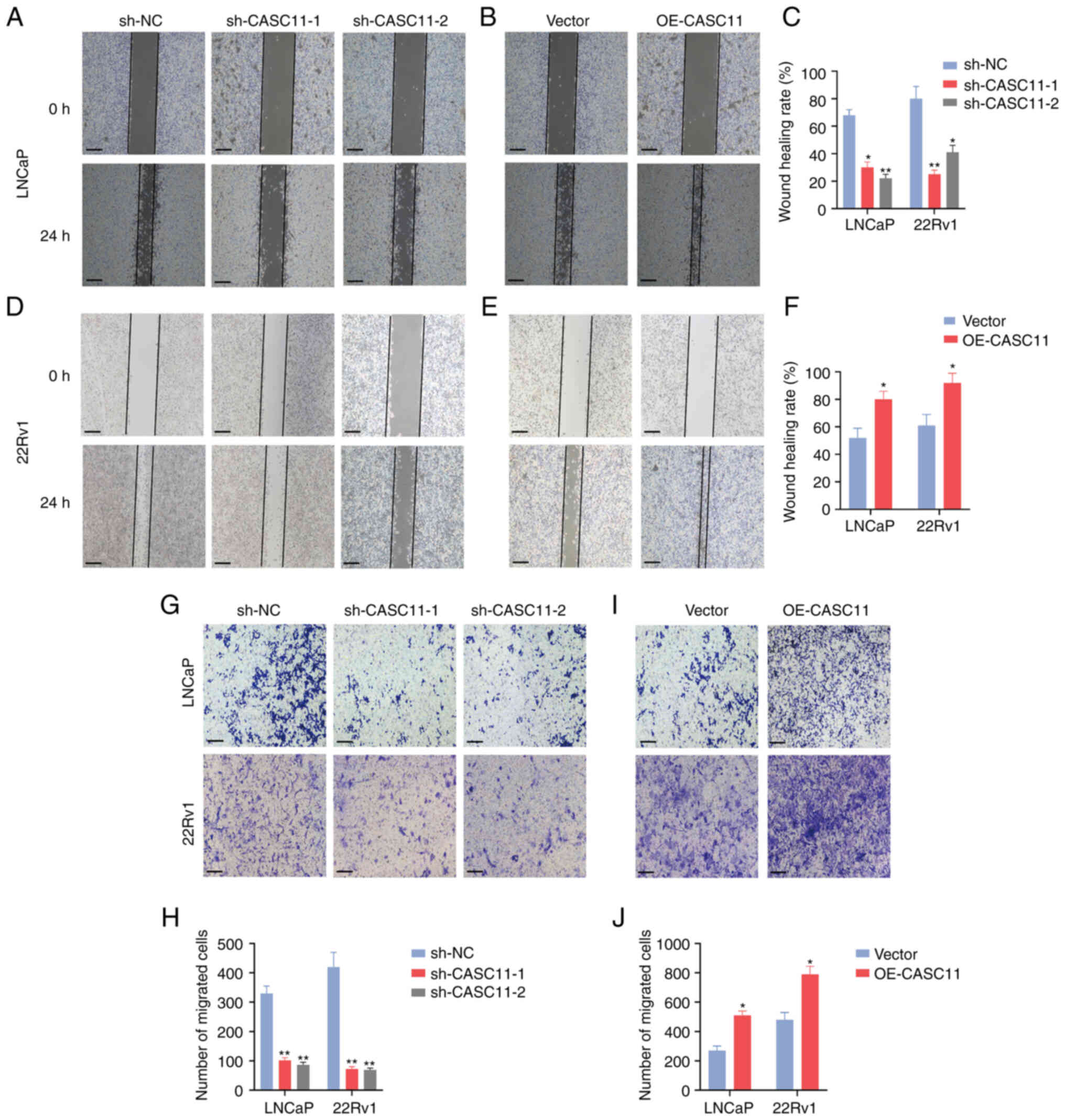
significantly enhanced the proliferation of PCa cells (Fig. 2C-N). Furthermore, wound healing and
Transwell assays were performed to assess cell motility; the
results revealed that CASC11 knockdown inhibited the migratory
ability of PCa cells, whereas CASC11 overexpression promoted cell
migration (Fig. 3A-J). Taken
together, these data indicated that CASC11 could promote the
proliferation and migration of PCa cells.
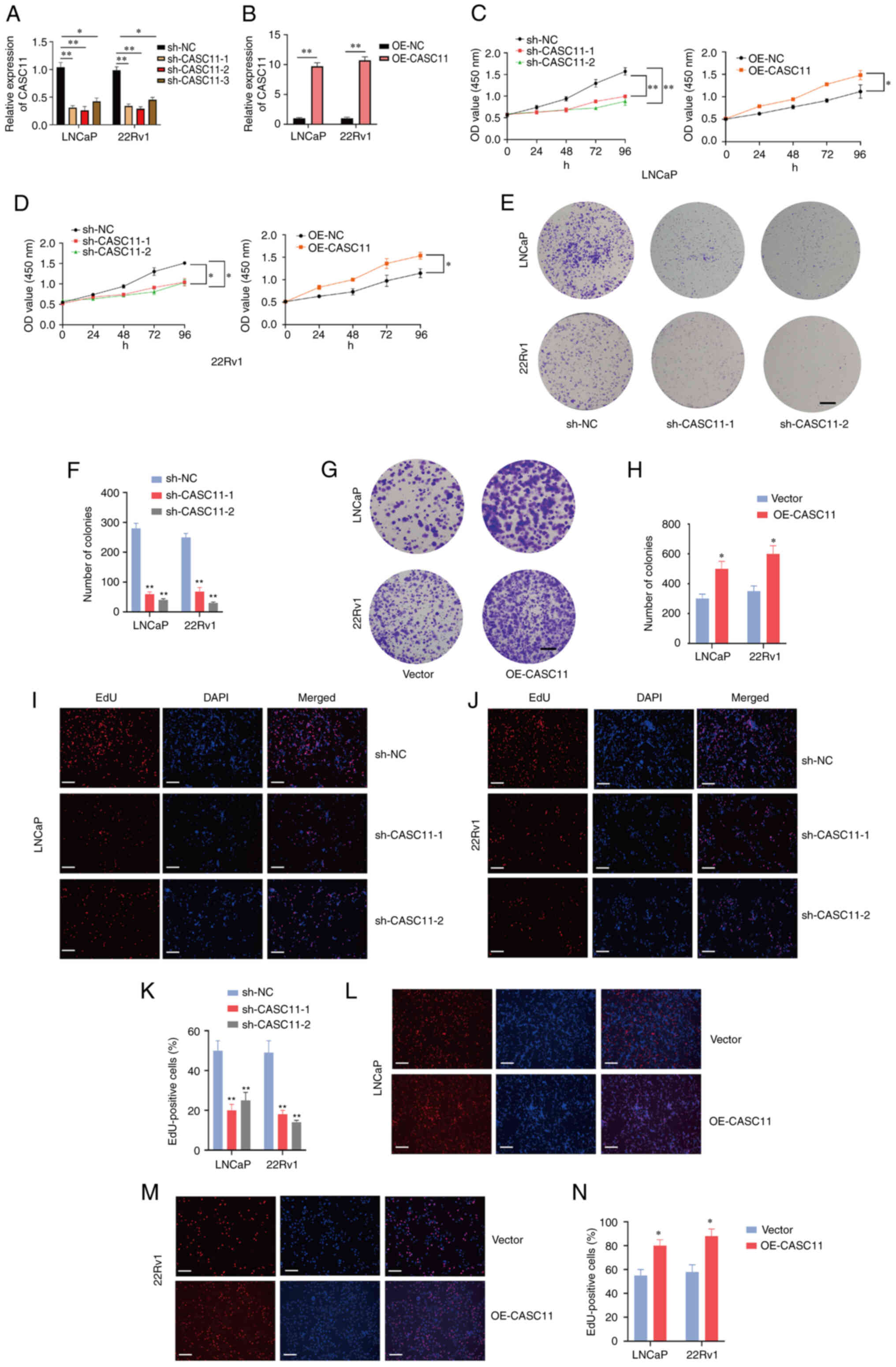 | Figure 2CASC11 promotes proliferation of PCa
cells. Expression levels of CASC11 were (A) significantly
downregulated in sh-CASC11-infected PCa cells and (B) significantly
upregulated in OE-CASC11-transfected cells. (C and D) Cell Counting
Kit-8 assay was used to investigate the proliferative effects of
CASC11 knockdown or overexpression on PCa cells. (E-H) Colony
formation assay was applied to investigate the proliferative
ability of PCa cells. Scale bar, 500 µm. (I-N) EdU assay was
applied to investigate the proliferative effects of CASC11
knockdown or overexpression in PCa cells. Scale bar, 100 µm.
*P<0.05, **P<0.01 vs. sh-NC or vector,
or as indicated. CASC11, cancer susceptibility candidate 11; EdU,
5-ethynyl-2′-deoxyuridine; NC, negative control; OE-CASC11, CASC11
overexpression plasmid; PCa, prostate cancer; sh, short
hairpin. |
CASC11 promotes PCa cell progression by
affecting G1/S transition of the cell cycle
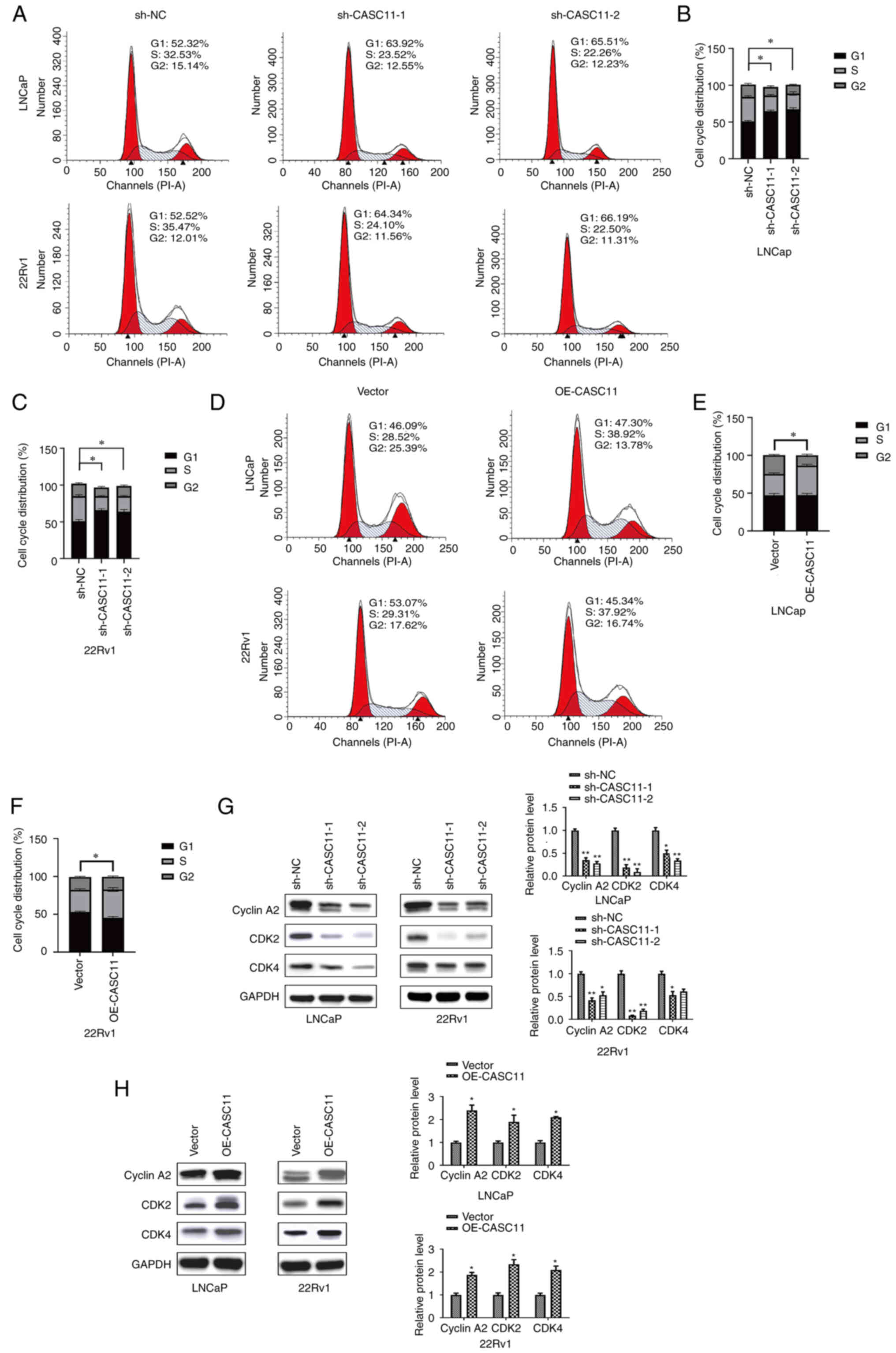
In order to further explore the underlying mechanism
of action of CASC11, flow cytometry was used to analyze the cell
cycle progression of PCa cells. Compared with in the control group,
the proportion of cells in G1 phase was significantly
increased, whereas the proportion of cells in S phase was decreased
after CASC11 knockdown (Fig.
4A-C). By contrast, the overexpression of CASC11 led to an
increase in the number of cells in S phase (Fig. 4D-F). The results of flow cytometry
indicated that CASC11 could affect cell cycle progression in PCa;
CASC11 knockdown caused cell cycle arrest at G1 phase,
whereas CASC11 overexpression induced cell cycle arrest at S phase.
Subsequently, the expression levels of three proteins associated
with the G1/S phase were assessed. The results revealed
that the expression levels of CyclinA2, CDK2 and CDK4 were
downregulated after CASC11 knockdown and upregulated in
CASC11-overexpressed PCa cells (Fig.
4G and H). These data indicated that CASC11 promoted PCa cell
progression by mediating the G1/S transition in the cell
cycle.
CASC11 knockdown inhibits PCa cell
tumorigenesis in vivo
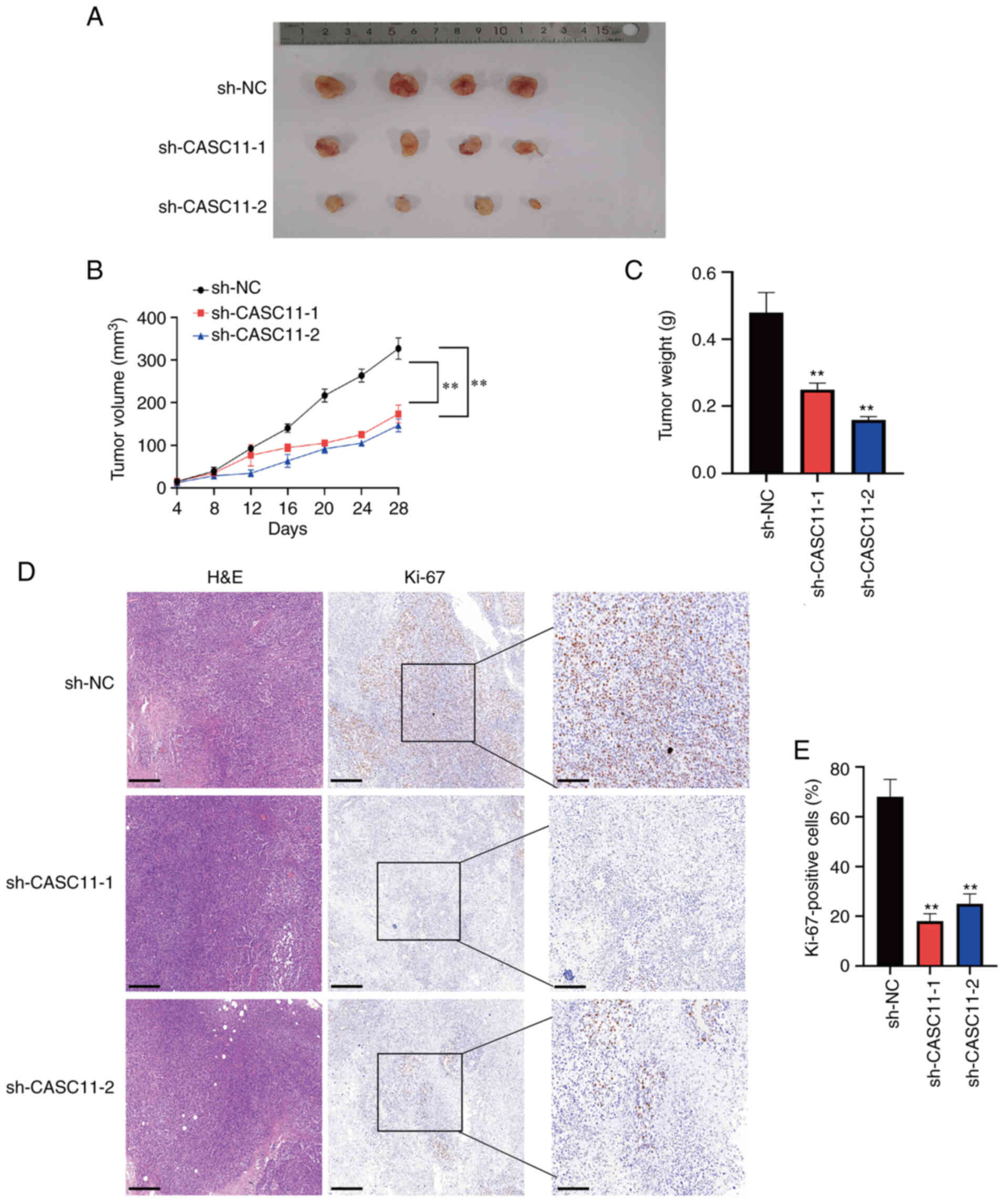
To verify the effects of CASC11 knockdown on PCa
tumorigenesis in vivo, 22Rv1 cells transduced with
sh-CASC11-1, sh-CASC11-2 or sh-NC were injected subcutaneously into
nude mice. As presented in Fig.
5A, tumors implanted in the sh-CASC11 group were smaller than
those in the sh-NC group. Additionally, the tumor volume and tumor
weight were significantly lower in the sh-CASC11 group (Fig. 5B and C). Immunohistochemical
staining of Ki-67 indicated that tumor proliferation was markedly
reduced in the sh-CASC11 group (Fig.
5D and E).
CASC11 promotes PCa progression by
attenuating p53 signaling
To further understand the underlying mechanism of
the effects of CASC11 on PCa tumorigenesis, RNA-seq analysis of
22Rv1 cells following CASC11 gene silencing was performed and a
heat map displayed the differentially expressed genes between sh-NC
and sh-CASC11 groups (Fig. 6A). As
shown in Fig. S1A, KEGG pathway
analysis demonstrated that CASC11 was associated with the 'cell
cycle' and 'p53 signaling' pathways (ranking 1 and 2, P<0.05),
which are closely related to cell proliferation and apoptosis. To
validate the role of p53 signaling, the expression levels of p53
and p21, which is a downstream gene of p53 signaling, were examined
by western blotting. CASC11 silencing induced an increase in p53
and p21 expression, as expected (Fig.
6B). According to these results, it was suggested that impaired
p53 signaling may contribute to the biological function of CASC11
in regulating PCa progression.
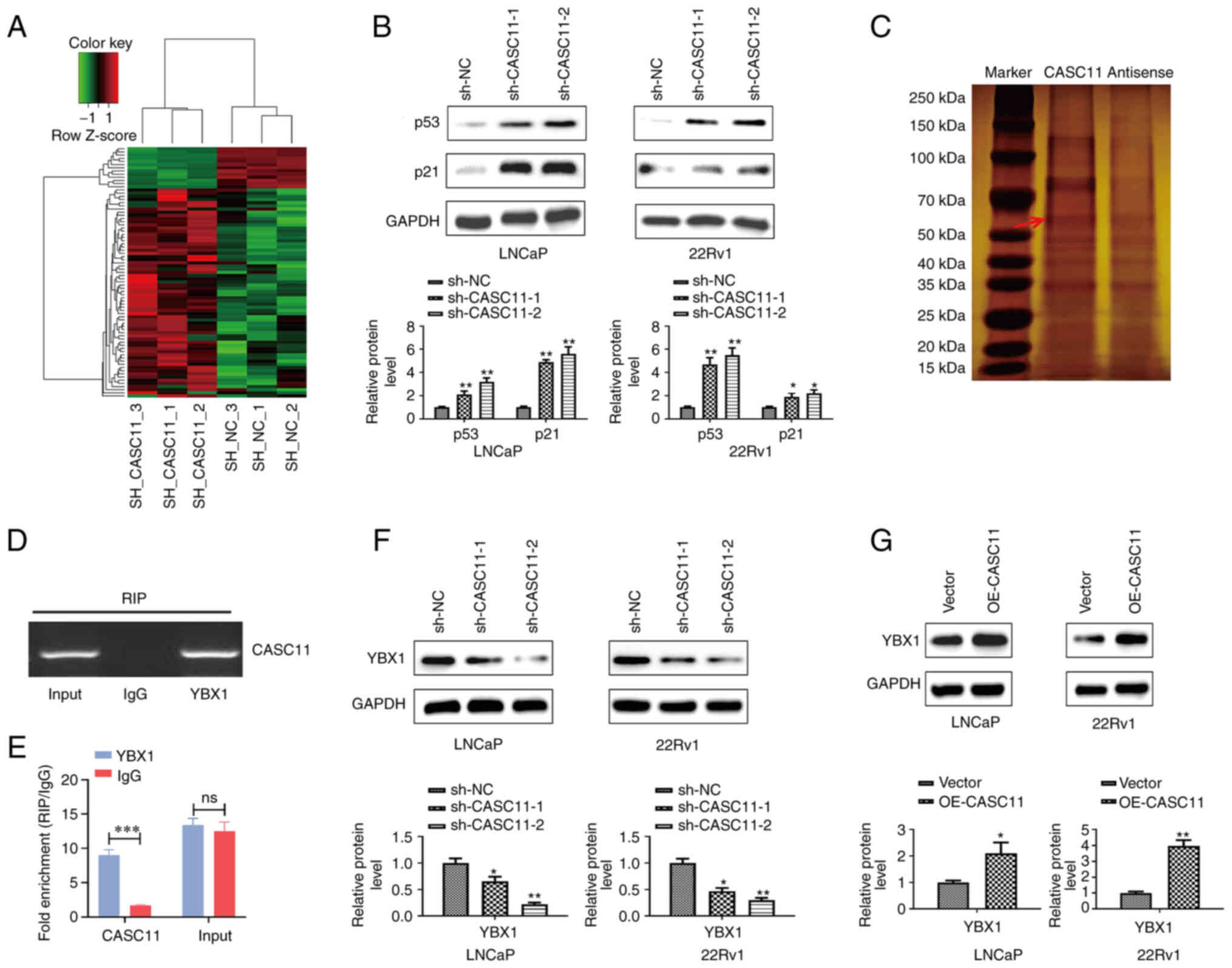 | Figure 6CASC11 suppresses p53 signaling in
prostate cancer cells by binding with YBX1. (A) Heat map displaying
differentially expressed genes in sh-CASC11 groups and sh-NC groups
by RNA sequencing. (B) Protein expression levels of p53 and p21
were measured by western blotting when CASC11 was knocked down in
LNCaP and 22Rv1 cells. (C) SDS-PAGE gel stained with silver to show
separated proteins. (D and E) Using the YBX1 or IgG antibody,
reverse transcription-quantitative PCR was used to examine RNA
enrichment in RIP assay. (F and G) Protein expression levels of
YBX1 were measured by western blotting when CASC11 was knocked down
or overexpressed in LNCaP and 22Rv1 cells. *P<0.05,
**P<0.01, ***P<0.001 vs. sh-NC or
vector, or as indicated. CASC11, cancer susceptibility candidate
11; NC, negative control; ns, not significant; OE-CASC11, CASC11
overexpression plasmid; RIP, RNA immunoprecipitation; sh, short
hairpin. |
CASC11 suppresses p53 signaling in PCa
cells through interaction with YBX1
Analysis of protein interactions with CASC11 using
RNA-protein pull-down was used to better elucidate the pathway
governing the CASC11 regulatory network. By silver staining of
SDS-PAGE gels, it was revealed that CASC11 was specifically bound
to protein bands ranging in size from 30 to 150 kDa (Fig. 6C), and the primary protein involved
in CASC11 interaction was identified as YBX1 by MS (Fig. S1B). Furthermore, the interaction
between YBX1 and CASC11 was further assessed by RIP. RIP assay
confirmed that CASC11 could bind YBX1 (Fig. 6D and E). Moreover, a downregulation
in the protein expression levels of YBX1 was detected upon CASC11
knockdown in PCa cells, whereas the opposite results were detected
in cells where CASC11 was overexpressed (Fig. 6F and G). In general, these results
indicated that CASC11 inhibited p53 signaling through binding with
YBX1.
YBX1 knockdown reverses the carcinogenic
effects of CASC11 promotion on PCa cells
For further investigating whether CASC11 exerted its
biological function by binding to YBX1, a rescue assay between
CASC11 and YBX1 was performed. YBX1 expression levels were
validated via western blot when YBX1 was knocked down or
overexpressed (Fig. S2). Colony
formation and Transwell assays demonstrated that YBX1 knockdown
significantly reversed the OE-CASC11-induced cell proliferation and
migration of PCa cells (Fig.
7A-D). Furthermore, western blotting revealed that knockdown of
YBX1 reversed the OE-CASC11-induced promotion of Cyclin A2, CDK2
and CDK4 protein expression (Fig.
7E), whereas the expression levels of p53 and p21 were returned
to almost basal levels in response to YBX1 overexpression (Fig. 7F). These findings indicated that
the CASC11/YBX1/p53 axis may exert role in PCa.
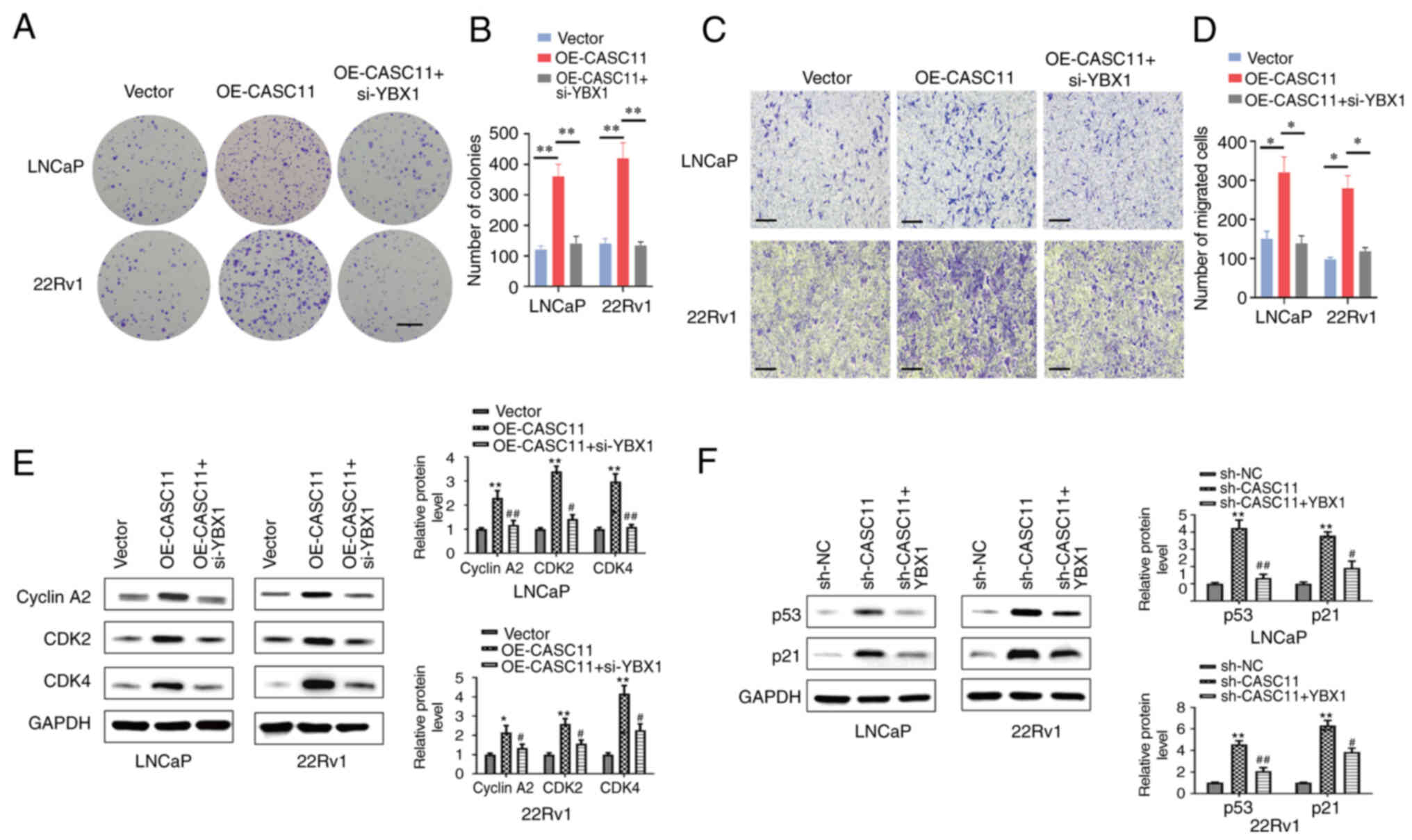 | Figure 7Knockdown of YBX1 in prostate cancer
cells reverses the carcinogenic effects of CASC11. YBX1 knockdown
reversed the OE-CASC11-induced (A and B) proliferation (scale bar,
500 µm) and (C and D) migration (scale bar, 100 µm)
of LNCaP and 22Rv1 cells. *P<0.05,
**P<0.01. (E) Western blot analysis demonstrated that
YBX1 knockdown abolished the OE-CASC11-induced promotion of Cyclin
A2, CDK2 and CDK4 proteins. (F) Western blot analysis demonstrated
that YBX1 overexpression reversed the sh-CASC11-induced increase in
the expression levels of p21 and p53 proteins.
*P<0.05, **P<0.01 vs. the control
group; #P<0.05, ##P<0.01 vs. the
OE-CASC11 or sh-CASC11 group. CASC11, cancer susceptibility
candidate 11; NC, negative control; OE-CASC11, CASC11
overexpression plasmid; sh, short hairpin; YBX1, Y-box binding
protein 1. |
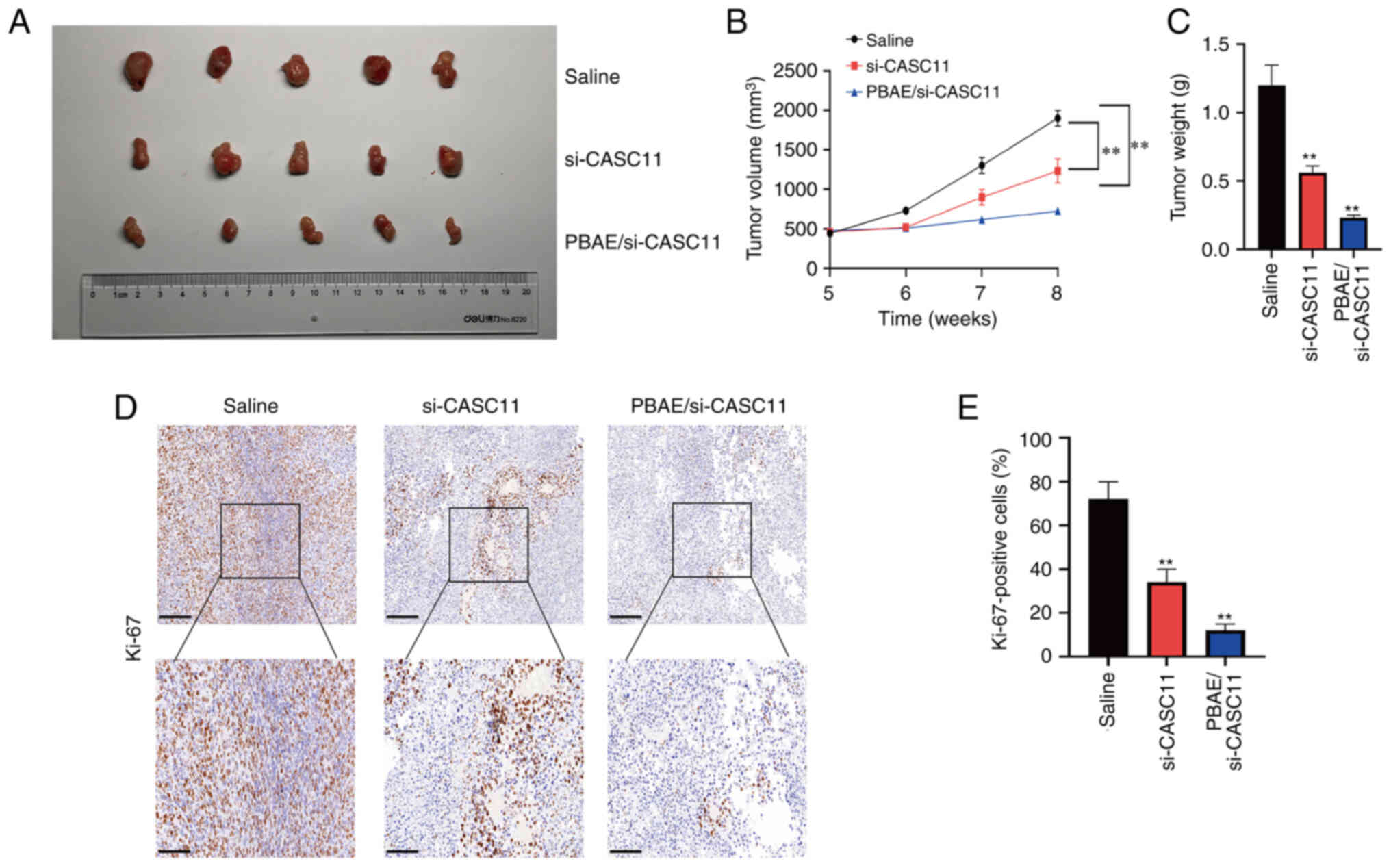
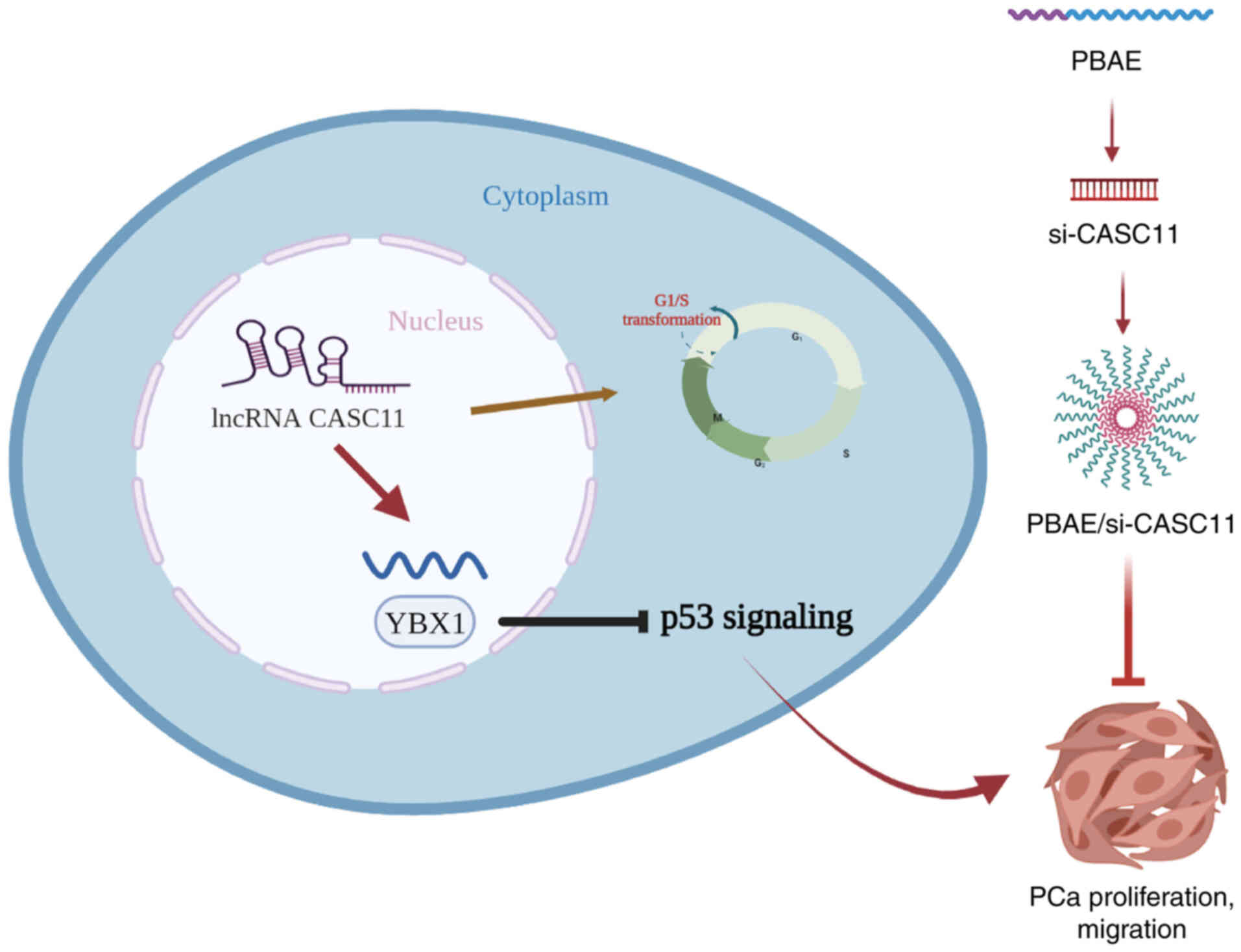
PBAE/si-CASC11 nanocomplexes inhibit the
growth of PCa
The aforementioned results revealed that CASC11 may
regulate the progression of PCa by interacting with the YBX1/p53
axis, thus highlighting the importance of CASC11 in the development
of PCa. Nanoparticles encapsulating siRNA have emerged as promising
small molecule inhibitor substitutes. Because of its high
transfection efficiency and low toxicity, PBAE is an ideal
candidate for targeted delivery (21). As shown in Fig. S3A, CCK-8 results confirmed that
PBAE exhibited no obvious toxicity when used at different
concentrations in PCa cells. Therefore, PBAE was used as a carrier
to deliver si-CASC11 and to suppress CASC11 expression.
Nanocomplexes with varying proportions of PBAE and si-CASC11 were
prepared based on their weight ratios. When the weight ratio of
PBAE and si-CASC11 exceeded 40, relatively homogeneous
nanoparticles formed. PBAE/si-CASC11 of 80:1 showed excellent
performance with 2% si-CASC11 remaining, indicating a high loading
efficiency of 98% (Fig. S3B).
Additionally, both transmission electron microscopy and particle
size potentiometer measurements indicated that the particle size of
PBAE/si-CASC11 nanocomplexes was ~130 nm (Fig. S3C and D). Subsequently,
subcutaneous xenograft tumor models were generated to further
evaluate the efficacy of PBAE/si-CASC11 nanocomplexes on PCa
progression in vivo. The knockdown efficiency of si-CASC11
in subcutaneous tumors was confirmed by RT-qPCR (Fig. S3E). The results revealed that the
tumor size and weight of mice treated with PBAE/si-CASC11
nanocomplexes were significantly decreased compared with those
treated with saline (Fig. 8A-C).
Similarly, IHC revealed a substantial decrease in Ki-67 expression
(Fig. 8D and E). H&E staining
of the main organs, including the liver and kidney, further
revealed that no notable morphological changes occurred in the
groups, indicating a good biosecurity (Fig. S4). Therefore, PBAE/si-CASC11
nanocomplexes may be effective in suppressing PCa development and
exhibited excellent therapeutic outcome on limiting tumor growth. A
schematic diagram summarizing the findings of the present study is
presented in Fig. 9.
Discussion
PCa is one of the major types of cancer affecting
human health. Despite significant advances in therapeutic options,
the mortality rate of PCa has not been markedly improved (22,23).
Numerous studies have confirmed that lncRNAs serve vital roles in
carcinogenesis. Dysregulation of lncRNAs have been reported to be
strongly associated with tumor development; therefore, it is
essential to understand the function and governing mechanisms of
lncRNAs (24,25). Tumor-specific treatments may target
certain lncRNAs and a number of lncRNA-associated molecular
functions have been described in PCa, such as targeting other RNAs,
activation of the androgen receptor, and the modulation of
epigenetic status through interactions with transcriptional
regulators (26). The development
of potentially effective alternative therapies for advanced PCa may
be enhanced by targeting lncRNAs with cancer-specific expression
patterns.
In the present study, the role of CASC11 in PCa was
determined through TCGA and GEO dataset analyses, and was validated
in PCa samples. The present results demonstrated that CASC11 was
specifically upregulated in PCa tissues/cells and was implicated in
cell cycle progression. The present study confirmed that CASC11 was
closely associated with the progression of PCa and may be a
potential therapeutic target for PCa.
Functionally, the present study assessed whether
CASC11 was a key modulator of PCa tumorigenesis. Loss- and
gain-of-function assays were conducted in two PCa cell lines, and
the results of cell proliferation, colony formation and cell
migration assays indicated the oncogenic properties of CASC11 in
PCa cells. In vivo experiments further confirmed that
knockdown of CASC11 could inhibit PCa progression. Hence, the
present study demonstrated that CASC11 may exert oncogenic effects
on PCa progression.
The cell cycle is a major regulatory mechanism for
maintaining cellular physiological activity. Changes in different
phases of the cell cycle are critical for cell proliferation,
differentiation and senescence (27,28).
Cell cycle progression analysis and western blotting revealed that
CASC11 induced G1/S transition and influenced the
expression of cell cycle-related proteins. These findings indicated
that CASC11 facilitated PCa cell proliferation by impacting cell
cycle progression. As determined by RNA-seq and KEGG analyses
between the sh-NC and sh-CASC groups, it was revealed that cell
cycle signaling was the top enriched signaling pathway and it was
suggested that the p53 signaling pathway may be involved in the
molecular mechanism of CASC11. p53 functions as a tumor suppressor,
triggering cell cycle arrest and apoptosis by stimulating
downstream targets (29,30). It has been reported that several
lncRNAs interact with the p53 signaling network, and serve as
upstream regulators or downstream effectors of the apoptotic
process (31). Mitra et al
(32) previously reported that
p53-responsive lncRNAs modulated the chemotherapy response through
regulation of the nuclear p53 pathway. Chen et al (33) demonstrated that lncRNA RMRP
promoted colorectal cancer cell growth by inactivating p53. In
addition, a genome-wide transcriptome study previously reported
that a pathway web containing 16 p53-targeted lncRNAs could form a
strongly diagnostic tumor suppressor signature (34). Further analyses of expression
levels in the present study confirmed that CASC11 caused
inactivation of downstream p53 genes thus suggesting that the p53
gene may be regulated by CASC11.
Results from RNA pull-down and RIP assays identified
an interaction between YBX1 and CASC11, which could mediate p53
transcriptional repression. YBX1 is a pivotal transcription factor
that also serves as a multi-functional RNA-binding protein, which
has been reported to be markedly overexpressed in a wide range of
tumors (35). In addition, a
number of cellular processes, along with tumor progression, have
been shown to be mediated by YBX1 (36,37).
In order to determine whether YBX1 affects cell progression and
migration in PCa, a siRNA was used to silence the protein
expression levels of YBX1. The experimental results indicated that
suppressing YBX1 partially reversed CASC11-induced PCa progression
and cell cycle-related protein expression. As a result, the present
findings suggested that CASC11 may mediate PCa progression by
interacting with YBX1. In addition, the upregulation of p53 caused
by silencing CASC11 was reversed by YBX1 overexpression. These
findings provide further evidence to support that CASC11/YBX1
binding may be critical to regulate p53 suppression.
The present study reported that CASC11 mediated the
progression and migration of PCa by interacting with the YBX1/p53
axis, highlighting the critical role of CASC11 in the progression
of PCa. Although evidence indicated that inhibition of CASC11 could
reduce tumor proliferation, there are currently no small molecule
inhibitors that target CASC11. High-throughput screening has been
used to explore novel therapeutic candidates; however, some
compounds have displayed less than encouraging results and the
efficacy and specificity need to be improved (38,39).
Therefore, extensive research into potent and specific CASC11
inhibitors is urgently required. Nanoparticles carrying siRNA have
attracted attention as promising alternatives for cancer therapy
(17,40,41).
In the present study, a PBAE carrier was generated to deliver
si-CASC11 and inhibit CASC11 expression. PBAE/si-CASC11
nanoparticles were revealed to be effective in blocking CASC11
expression and in enhancing the anti-tumor effects of CASC11
knockdown. The effectiveness and safety of PBAE/si-CASC11
nanoparticles need further clinical investigation.
In conclusion, the present study demonstrated that
CASC11 was a valuable regulatory factor associated with the
proliferation and migration of PCa cells. CASC11 contributed to PCa
development and progression by interacting with YBX1 and
suppressing p53 signaling. The present findings indicated that
CASC11 may be a promising therapeutic target in PCa. Furthermore,
PBAE/si-CASC11 nanocomplexes were generated, which may provide
novel insights into the treatment of PCa.
Supplementary Data
Availability of data and materials
The sequencing data during the current study are not
publicly available due to other unpublished research but are
available from the corresponding author on reasonable request.
Other data generated or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
XS, WM and LY designed experiments. XS, SX and LY
undertook statistical analyses. XS, YZ, LJ, XL, JZ, WM and BZ
performed the experiments. XS, SX, and LY confirm the authenticity
of all the raw data. XS, WM and YZ wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was provided by the
participants. The present study was approved by the Ethics
Committee of The Second Affiliated Hospital of Anhui Medical
University (approval no. 2021-130). The animal experiment was
approved by the Animal Research Ethics Committee of The Second
Affiliated Hospital of Anhui Medical University (approval no.
2021080).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by the National Natural Science
Foundation of China (grant nos. 81972409 and 81672549).
Abbreviations:
|
PCa
|
prostate cancer
|
|
lncRNA
|
long non-coding RNA
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GEO
|
Gene Expression Omnibus
|
|
CASC11
|
cancer susceptibility candidate 11
|
|
RIP
|
RNA immunoprecipitation
|
|
FISH
|
fluorescence in situ
hybridization
|
|
shRNA
|
short hairpin RNA
|
|
NC
|
negative control
|
|
PBAE
|
poly (β-amino ester)
|
|
siRNA
|
small interfering RNA
|
|
EdU
|
5-ethynyl-2′-deoxyuridine
|
|
PBS
|
phosphate-buffered saline
|
|
IHC
|
immunohistochemistry
|
|
H&E
|
hematoxylin and eosin
|
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marhold M, Kramer G, Krainer M and Le
Magnen C: The prostate cancer landscape in Europe: Current
challenges, future opportunities. Cancer Lett. 526:304–310. 2022.
View Article : Google Scholar
|
|
3
|
Rebello RJ, Oing C, Knudsen KE, Loeb S,
Johnson DC, Reiter RE, Gillessen S, Van der Kwast T and Bristow RG:
Prostate cancer. Nat Rev Dis Primers. 7:92021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu Y, Freedland SJ and Ye D: Prostate
cancer and prostatic diseases best of asia, 2019: Challenges and
opportunities. Prostate Cancer Prostatic Dis. 23:197–198. 2020.
View Article : Google Scholar
|
|
5
|
Bridges MC, Daulagala AC and Kourtidis A:
LNCcation: lncRNA localization and function. J Cell Biol.
220:e2020090452021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z,
Du H, Ren D, Dai Y and Peng X: m(6) A modification of lncRNA PCAT6
promotes bone metastasis in prostate cancer through
IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med. 11. pp.
e4262021, View Article : Google Scholar
|
|
7
|
Wu G, Hao C, Qi X, Nie J, Zhou W, Huang J
and He Q: LncRNA SNHG17 aggravated prostate cancer progression
through regulating its homolog SNORA71B via a positive feedback
loop. Cell Death Dis. 11:3932020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ghildiyal R, Sawant M, Renganathan A,
Mahajan K, Kim EH, Luo J, Dang HX, Maher CA, Feng FY and Mahajan
NP: Loss of long noncoding RNA NXTAR in prostate cancer augments
androgen receptor expression and enzalutamide resistance. Cancer
Res. 82:155–168. 2022. View Article : Google Scholar
|
|
9
|
Wang B, Xu W, Hu C, Liu K, Chen J, Guo C
and Yuan C: Critical roles of the lncRNA CASC11 in tumor
progression and cancer metastasis: The biomarker and therapeutic
target potential. Genes Dis. 9:325–333. 2022. View Article : Google Scholar :
|
|
10
|
Zhang Z, Zhou C, Chang Y, Zhang Z, Hu Y,
Zhang F, Lu Y, Zheng L, Zhang W and Li X and Li X: Long non-coding
RNA CASC11 interacts with hnRNP-K and activates the WNT/β-catenin
pathway to promote growth and metastasis in colorectal cancer.
Cancer Lett. 376:62–73. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song H, Liu Y, Li X, Chen S, Xie R, Chen
D, Gao H, Wang G, Cai B and Yang X: Long noncoding RNA CASC11
promotes hepatocarcinogenesis and HCC progression through
EIF4A3-mediated E2F1 activation. Clin Transl Med. 10:e2202020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng N, Wu J, Yin M, Xu J, Wang Y, Chen
X, Nie Z and Yin J: LncRNA CASC11 promotes cancer cell
proliferation in hepatocellular carcinoma by inhibiting
miRNA-188-5p. Biosci Rep. Apr 30–2019.Epub ahead of print.
View Article : Google Scholar
|
|
13
|
Cui Y, Shen G, Zhou D and Wu F: CASC11
overexpression predicts poor prognosis and regulates cell
proliferation and apoptosis in ovarian carcinoma. Cancer Manag Res.
12:523–529. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Capik O, Sanli F, Kurt A, Ceylan O, Suer
I, Kaya M, Ittmann M and Karatas OF: CASC11 promotes aggressiveness
of prostate cancer cells through miR-145/IGF1R axis. Prostate
Cancer Prostatic Dis. 24:891–902. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Humphrey PA, Moch H, Cubilla AL, Ulbright
TM and Reuter VE: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part b: Prostate and bladder
tumours. Eur Urol. 70:106–119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Mao W, Wang K, Xu B, Zhang H, Sun S, Hu Q,
Zhang L, Liu C, Chen S, Wu J, et al: ciRS-7 is a prognostic
biomarker and potential gene therapy target for renal cell
carcinoma. Mol Cancer. 20:1422021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yan X, Pan Q, Xin H, Chen Y and Ping Y:
Genome-editing prodrug: Targeted delivery and conditional
stabilization of CRISPR-Cas9 for precision therapy of inflammatory
disease. Sci Adv. 7:eabj06242021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mortensen MM, HØyer S, Lynnerup AS,
Ørntoft TF, SØrensen KD, Borre M and Dyrskjøt L: Expression
profiling of prostate cancer tissue delineates genes associated
with recurrence after prostatectomy. Sci Rep. 5:160182015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arredouani MS, Lu B, Bhasin M, Eljanne M,
Yue W, Mosquera JM, Bubley GJ, Li V, Rubin MA, Libermann TA and
Sanda MG: Identification of the transcription factor single-minded
homologue 2 as a potential biomarker and immunotherapy target in
prostate cancer. Clin Cancer Res. 15:5794–5802. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang C, Xue Z, Liu Y, Xiao J, Chen J,
Zhang L, Guo J and Lin W: Delivery of anticancer drug using
pH-sensitive micelles from triblock copolymer MPEG-b-PBAE-b-PLA.
Mater Sci Eng C Mater Biol Appl. 84:254–262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamada Y and Beltran H: The treatment
landscape of metastatic prostate cancer. Cancer Lett. 519:20–29.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adamaki M and Zoumpourlis V: Prostate
cancer biomarkers: From diagnosis to prognosis and precision-guided
therapeutics. Pharmacol Ther. 228:1079322021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hua JT, Chen S and He HH: Landscape of
noncoding RNA in prostate cancer. Trends Genet. 35:840–851. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tan YT, Lin JF, Li T, Li JJ, Xu RH and Ju
HQ: LncRNA-mediated posttranslational modifications and
reprogramming of energy metabolism in cancer. Cancer Commun (Lond).
41:109–120. 2021. View Article : Google Scholar
|
|
26
|
Xu YH, Deng JL, Wang G and Zhu YS: Long
non-coding RNAs in prostate cancer: Functional roles and clinical
implications. Cancer Lett. 464:37–55. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ahmad I, Fakhri S, Khan H, Jeandet P,
Aschner M and Yu ZL: Targeting cell cycle by β-carboline alkaloids
in vitro: Novel therapeutic prospects for the treatment of cancer.
Chem Biol Interact. 330:1092292020. View Article : Google Scholar
|
|
28
|
Dyshlovoy SA, Tarbeeva D, Fedoreyev S,
Busenbender T, Kaune M, Veselova M, Kalinovskiy A, Hauschild J,
Grigorchuk V, Kim N, et al: Polyphenolic compounds from lespedeza
bicolor root bark inhibit progression of human prostate cancer
cells via induction of apoptosis and cell cycle arrest.
Biomolecules. 10:4512020. View Article : Google Scholar :
|
|
29
|
Duffy MJ, Synnott NC, O'Grady S and Crown
J: Targeting p53 for the treatment of cancer. Semin Cancer Biol.
79:58–67. 2022. View Article : Google Scholar
|
|
30
|
Kim MP, Li X, Deng J, Zhang Y, Dai B,
Allton KL, Hughes TG, Siangco C, Augustine JJ, Kang Y, et al:
Oncogenic KRAS recruits an expansive transcriptional network
through mutant p53 to drive pancreatic cancer metastasis. Cancer
Discov. 11:2094–2111. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang A, Xu M and Mo YY: Role of the
lncRNA-p53 regulatory network in cancer. J Mol Cell Biol.
6:181–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mitra S, Muralidharan SV, Di Marco M,
Juvvuna PK, Kosalai ST, Reischl S, Jachimowicz D, Subhash S,
Raimondi I, Kurian L, et al: Subcellular distribution of p53 by the
p53-responsive lncRNA NBAT1 determines chemotherapeutic response in
neuroblastoma. Cancer Res. 81:1457–1471. 2021. View Article : Google Scholar
|
|
33
|
Chen Y, Hao Q, Wang S, Cao M, Huang Y,
Weng X, Wang J, Zhang Z, He X, Lu H and Zhou X: Inactivation of the
tumor suppressor p53 by long noncoding RNA RMRP. Proc Natl Acad Sci
USA. 118:e20268131182021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sanchez Y, Segura V, Marin-Bejar O, Athie
A, Marchese FP, Gonzalez J, Bujanda L, Guo S, Matheu A and Huarte
M: Genome-wide analysis of the human p53 transcriptional network
unveils a lncRNA tumour suppressor signature. Nat Commun.
5:58122014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lyabin DN, Eliseeva IA and Ovchinnikov LP:
YB-1 protein: Functions and regulation. Wiley Interdiscip Rev RNA.
5:95–110. 2014. View Article : Google Scholar
|
|
36
|
Gandhi M, Groß M, Holler JM, Coggins SA,
Patil N, Leupold JH, Munschauer M, Schenone M, Hartigan CR,
Allgayer H, et al: The lncRNA lincNMR regulates nucleotide
metabolism via a YBX1-RRM2 axis in cancer. Nat Commun. 11:32142020.
View Article : Google Scholar
|
|
37
|
Zheng X, Zhang J, Fang T, Wang X, Wang S,
Ma Z, Xu Y, Han C, Sun M, Xu L, et al: The long non-coding RNA
PIK3CD-AS2 promotes lung adenocarcinoma progression via
YBX1-mediated suppression of p53 pathway. Oncogenesis. 9:342020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Itsumi M, Shiota M, Sekino Y, Ushijima M,
Kashiwagi E, Takeuchi A, Inokuchi J, Kajioka S, Uchiumi T and Eto
M: High-throughput screen identifies 5-HT receptor as a modulator
of AR and a therapeutic target for prostate cancer. Prostate.
80:885–894. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Adams J, Casali A and Campbell K:
Sensitive high-throughput assays for tumour burden reveal the
response of a drosophila melanogaster model of colorectal cancer to
standard chemotherapies. Int J Mol Sci. 22:51012021. View Article : Google Scholar :
|
|
40
|
Yari H, Gali H and Awasthi V:
Nanoparticles for targeting of prostate cancer. Curr Pharm Des.
26:5393–5413. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mezghrani B, Ali LMA, Richeter S, Durand
JO, Hesemann P and Bettache N: Periodic mesoporous ionosilica
nanoparticles for green light photodynamic therapy and
photochemical internalization of siRNA. ACS Appl Mater Interfaces.
13:29325–29339. 2021. View Article : Google Scholar : PubMed/NCBI
|