Introduction
Gastric cancer (GC) is the fourth most prevalent
cancer (7.8% of all reported cancer cases) and the second major
cause of mortality (9.7% of all cancer-related deaths) from cancer
in 2008, accounting for 95% of primary malignant gastric neoplasms
(1). Although risk factors for
stomach carcinogenesis have been established, such as
Helicobacter pylori infection, natural carcinogens (e.g.,
nitrates), dietary pro-carcinogens, adenomatous polyps, and
heritable and family hazards, primary prevention remains a
challenge (2). Due to a lack of
evident or distinct symptoms in the early stages, and frequently
with only a few or minimum symptoms, this leads to even advanced
stage III or IV disease at the time of diagnosis. Despite the fact
that surgical resection is the first-line treatment option for GC
patients, and recent breakthroughs in systemic multi-line
chemotherapy or postoperative adjuvant radiation paired with
surgery have been found to improve patient survival (3), GC recurrence is common following
surgery. As a result, it is critical to identify useful biomarkers
that mediate the formation and progression of GC in order to
understand in an improved way the process of gastric carcinogenesis
and create effective treatments to enhance patient outcomes.
Galectin-1 (Gal-1) is a protein family member with a
preference for glycoconjugate-galactose residues (4). Gal-1 was shown to be overexpressed in
a variety of malignancies, including lung cancer (5), renal cancer (6), hepatocellular carcinoma (7) and ovarian cancer (8). According to evidence, altered Gal-1
expression in cancer tissues may be crucial for tumor development,
growth, proliferation, metastasis, angiogenesis, immune response
and resistance to systemic chemotherapy (9). The role of Gal-1 in GC remains mainly
unknown from a molecular perspective.
Endoplasmic reticulum stress (ERS) or the unfolded
protein response (UPR), a cellular adaptive mechanism that happens
in response to the disruption of ER homeostasis, can be brought on
by a variety of physiological or pathological conditions, including
nutrient deprivation, Ca2+ depletion, hypoxia,
inflammation activation and oxidative stress (10). The ER chaperone protein known as
glucose-regulated protein 78 (GRP78) is also a recognized indicator
of ERS. GRP78 separates from IRE1, PERK and ATF6 during ERS to
start three signaling pathways (11). UPR and GRP78 are overexpressed in
the early stages of ERS, which helps cells survive by lowering the
amount of protein that enters the ER, blocking protein synthesis
and lessening cellular stress. But when ERS is prolonged and
severe, the apoptotic marker protein C/EBP homologous protein
(CHOP) is increased (12). A
versatile protein called GRP78 is essential in both physiological
and pathological stress (13).
GRP78 was strongly activated in a number of malignancies, including
breast cancer, according to numerous studies (14,15),
lung cancer (16), thyroid
carcinoma (17), colorectal cancer
(18), hepatocellular cancer
(19) and prostate cancer
(20). GRP78 overexpression
contributed to tumor cell survival, growth, angiogenesis, and
resistance to treatment. In individuals with GC, overexpression of
GRP78 is associated with increased lymph node (LN) metastases and a
poor prognosis (21). Cancer cells
undergo growth suppression and apoptosis when exposed to GRP78
inhibitors, such as small molecules and specific binding peptides.
These results revealed that the progression of GC is mediated by
GRP78. However, the role of GRP78 in GC is not entirely
elucidated.
As a result, it was investigated whether GRP78
played a role in the development of the tumor that Gal-1 caused in
the current study. Positive connection was discovered between GRP78
and Gal-1. GRP78 was a target of Gal-1 in GC cells, according to
the Co-IP data. Given that GRP78 has been linked to the spread of
various malignancies, our database was utilized to confirm that
there is a strong link between the high levels of GRP78 expression
in GC tissues and the TNM stage. These findings suggested that the
Gal-1-GRP78 axis is a viable target for the development of GC
medications.
Materials and methods
Bioinformatic analysis
'Expression Plots' of the GEPIA database (http://gepia.cancer-pku.cn/) were used to compare the
expression levels of Gal-1 in tumor and normal tissues in GC
(22). Oncomine (https://www.oncomine.org/resource/login.html) was used
to simultaneously collect the mRNA expression levels of Gal-1 in GC
and normal tissues, and the results were as follows: Cho gastric
and DErrico gastric (23).
Mechanistically, the interacting proteins of Gal-1 were examined
using the BioGRID database (https://thebiogrid.org/) (24).
Patients and GC specimens
Human surgical clinical specimens, including
formalin-fixed paraffin-embedded human GC specimens (n=80, aging
from 43 to 77 years old) and fresh surgically removed GC tissues
(n=16; including 9 male and 7 female, aging from 46 to 67 years
old), were collected from Northern Jiangsu People's hospital,
clinical medical school, Yangzhou University (Yangzhou, China)
between July 2015 and May 2017. Prior to surgery, none of the
patients underwent preoperative chemotherapy. At least two
qualified pathologists determined that all tissues were GC.
According to the American Joint Committee on Cancer TNM staging
system (AJCC-8 TNM) for gastric carcinoma, the disease stage was
identified. Our hospital database and clinical records were used to
gather clinicopathological data on age, sex, tumor size, TNM stage,
degree of differentiation, histological grade, venous and nerve
invasion (Tables I and III). The present study was approved
(approval no. 2019KY-022) by The Northern Jiangsu People's
Hospital, the Clinical Medical School and the Ethics Committee of
Yangzhou University (Yangzhou, China). Written informed consent was
obtained from all participants before the study was carried
out.
 | Table IAssociations between Gal-1 expression
and clinicopathological features of 80 patients with gastric
cancer. |
Table I
Associations between Gal-1 expression
and clinicopathological features of 80 patients with gastric
cancer.
| Histopathological
parameters | Total number
(80) | Expression level of
Gal-1
| P-value |
|---|
| Low | High |
|---|
| Age, years | | | | 0.759 |
| <65 | 33 | 15 | 18 | |
| ≥65 | 47 | 23 | 24 | |
| Sex | | | | 0.408 |
| Male | 51 | 26 | 25 | |
| Female | 29 | 12 | 17 | |
| Tumor size | | | | 0.003 |
| <6 | 43 | 27 | 16 | |
| ≥6 | 37 | 11 | 26 | |
| Lauren type | | | | 0.797 |
| Intestinal
type | 73 | 35 | 38 | |
| Diffuse type | 7 | 3 | 4 | |
| Depth of
invasion | | | | 0.049 |
| T1, T2 | 29 | 18 | 11 | |
| T3, T4 | 51 | 20 | 31 | |
| Lymphonodus
metastasis | | | | 0.007 |
| N0, N1 | 40 | 25 | 15 | |
| N2, N3 | 40 | 13 | 27 | |
| Distant
metastasis | | | | 0.026 |
| Negative | 58 | 32 | 26 | |
| Positive | 22 | 6 | 16 | |
| TNM stage | | | | 0.015 |
| I-II | 35 | 22 | 13 | |
| III-IV | 45 | 16 | 29 | |
| Degree of
differentiation | | | | 0.849 |
| Highly | 43 | 20 | 23 | |
| Moderately and
poorly | 37 | 18 | 19 | |
| Histological
grade | | | | 0.617 |
| I | 8 | 4 | 4 | |
| II | 38 | 20 | 18 | |
| III | 34 | 14 | 20 | |
| Venous
invasion | | | | 0.866 |
| No | 45 | 21 | 24 | |
| Yes | 35 | 17 | 18 | |
| Nerve invasion | | | | 0.463 |
| No | 35 | 15 | 20 | |
| Yes | 45 | 23 | 22 | |
| Glucose-regulated
protein 78 expression | | | | 0.008 |
| Low | 38 | 24 | 14 | |
| High | 42 | 14 | 28 | |
 | Table IIIExpression of GRP78 in gastric cancer
with different clinicopathological variables using our own
cohort. |
Table III
Expression of GRP78 in gastric cancer
with different clinicopathological variables using our own
cohort.
| Histopathological
parameters | Total number
(80) | Expression level of
GRP78
| P-value |
|---|
| Low | High |
|---|
| Age, years | | | | |
| <65 | 33 | 17 | 16 | 0.547 |
| ≥65 | 47 | 21 | 26 | |
| Sex | | | | 0.917 |
| Male | 51 | 24 | 27 | |
| Female | 29 | 14 | 15 | |
| Tumor size | | | | 0.796 |
| <6 | 43 | 21 | 22 | |
| ≥6 | 37 | 17 | 20 | |
| Lauren type | | | | 0.184 |
| Intestinal
type | 73 | 33 | 40 | |
| Diffuse type | 7 | 5 | 2 | |
| Depth of
invasion | | | | 0.049 |
| T1, T2 | 29 | 18 | 11 | |
| T3, T4 | 51 | 20 | 31 | |
| Lymphonodus
metastasis | | | | 0.025 |
| N0, N1 | 40 | 24 | 16 | |
| N2, N3 | 40 | 14 | 26 | |
| Distant
metastases | | | | 0.084 |
| Negative | 58 | 31 | 27 | |
| Positive | 22 | 7 | 15 | |
| TNM stage | | | | 0.048 |
| I-II | 35 | 21 | 14 | |
| III-IV | 45 | 17 | 28 | |
| Degree of
differentiation | | | | 0.004 |
| Highly | 43 | 14 | 29 | |
| Moderately and
poorly | 37 | 24 | 13 | |
| Histological
grade | | | | 0.506 |
| I | 8 | 5 | 3 | |
| II | 38 | 19 | 19 | |
| III | 34 | 14 | 20 | |
| Venous
invasion | | | | |
| No | 45 | 23 | 22 | 0.463 |
| Yes | 35 | 15 | 20 | |
| Nerve invasion | | | | 0.284 |
| No | 35 | 19 | 16 | |
| Yes | 45 | 19 | 26 | |
Immunohistochemistry (IHC)
The GC samples and xenografted tumor tissues were
embedded in paraffin, sectioned into 4 m-thick pieces, and set on
glass slides after being fixed in a 10% formalin solution for 24 h
at room temperature. They were then rehydrated with an ethanol
gradient (100-50%) after being deparaffinized with xylene. After
that, the sections were heated in sodium citrate antigen retrieval
solution (cat. no. C1032; Beijing Solarbio Science & Technology
Co., Ltd.). The slides were then incubated with normal goat serum
for 30 min at room temperature after being submerged in hydrogen
peroxide (0.3%; OriGene Technologies, Inc.) to block endogenous
peroxidase activity. Following that, IHC staining was performed.
Anti-Gal-1 rabbit polyclonal antibody (1:200; cat. no. 11858-1-AP;
ProteinTech Group, Inc.) and anti-GRP78 antibody (1:200; cat. no.
ab21685; Abcam) were incubated on GC patients' sections, anti-Gal-1
rabbit polyclonal antibody (1:200; cat. no. 11858-1-AP; ProteinTech
Group, Inc.) and anti-Ki-67 rabbit monoclonal anti-body (1:400,
9027; Cell Signaling Technology) were incubated on xenografted
tumor tissues' sections for an overnight period at 4°C. The
following day, the slides were washed, incubated at 37°C for 30 min
with HRP-conjugated goat anti-rabbit IgG (1:100; cat. no. ab288151;
Abcam), and then stained with hematoxylin and
3,3'-diaminobenzidine, respectively. After that, the slides were
looked at and captured on camera with an Olympus BX53 fluorescent
microscope (Olympus Corporation). Based on the number of positive
cells and staining intensity, the Ki-67, GRP78 and Gal-1 staining
was graded. The staining intensity scores are as follows: The
staining intensity scores were as follows: 0 for colorless, 1 for
yellow, 2 for brown, and 3 for dark brown. According to the
percentage of positive cells, the scores were as follows: 0 for
0-5%, 1 for 5-25%, 2 for 25-50%, 3 for 50-75%, and 4 for >75%.
Staining intensity and the percentage of positive cell values were
combined to create the final score. Cases with a score of ≥5 were
considered positive expression, while those with a score of ≤4 were
considered negative expression.
Cell culture
The normal gastric epithelial cells GES-1 and GC
cell lines (HGC-27 and AGS) were obtained from the Cell Bank of
Type Culture Collection of Chinese Academy of Sciences, Shanghai
Institute of Cell Biology, Chinese Academy of Science. Cell lines
were cultured in RPMI-1640 medium supplemented with 10% fetal
bovine serum (both from Gibco; Thermo Fisher Scientific, Inc.)
containing 100 units/ml penicillin and 100 mg/ml streptomycin (cat.
no. SV30010; Hyclone; Cytiva) at 37°C in a humidified incubator
containing 5% CO2.
Cell transfection
Lentiviral vectors (called Lv-GAL-1) that were based
on the GV358 (Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin) lentiviral
vector from Shanghai Genechem Co., Ltd., were created. The negative
control was an empty vector. Using two lentiviral vector-based
short hairpin (sh)RNAs, RNA interference was used to knock down
Gal-1 and GRP78 (Shanghai Genechem Co., Ltd.). The following target
sequences were created for Gal-1: shGal-1 forward, 5′-CAC CAT CGT
GTG CAA CAG CAA-3′ and reverse, 5′-TTG CTG TTG CAC ACG ATG GTG-3′.
The non-targeting shRNA (forward, 5′-TTC TCC GAA CGT GTC ACG T-3′
and reverse, 5′-ACG TGA CAC GTT CGG AGA A-3) was used as a
non-target shRNA control. The following target sequences were
created for GRP78: shGRP78 forward, 5′-GAG CGC ATT GAT ACT AGA
AAT-3′ and reverse, 5′-ATT TCT AGT ATC AAT GCG CTC-3′. The empty
vector GV654 was used as the lentiviral negative control. For the
purpose of producing lentiviral particles, 20 µg the
aforementioned vectors were co-transfected with 15 µg
pHelper 1.0 (envelope plasmid) and 10 µg pHelper 2.0
(pack-aging plasmid) in the 2nd generation transfection system into
293T cells (cat. no. GNHu17; Shanghai Institute of Cell Biology,
Chinese Academy of Sciences) cultured in DMEM (Hyclone; Cytiva)
with 10% FBS (Hyclone; Cytiva) at 37°C with 5% CO2 using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.),
following the manufacturer′s instructions. The packaging plasmids
were co-transfected 48 h later, and the virus-containing
supernatant was collected. The lentivirus was then purified by
ultracentrifugation at 30,000 × g for 2 h at 4°C. Then, the
lentivirus (Lv-GAL-1, the empty vector GV358, Gal-1-shRNA, or
NC-shRNA) was added and co-cultured with the target cells for 24 h
at 37°C in the presence of polybrene (5 g/ml; Shanghai GeneChem
Co., Ltd.) at a multiplicity of infection of 10. This was conducted
after the GC cell lines had been plated into 24-well plates with
1×104 cells per well. After that, the medium was changed
in preparation for another 48 h of incubation at 37°C and 5%
CO2. Then, the stably transfected cell lines were chosen
using a 25 g/ml puromycin treatment for two weeks at 37°C. In the
rescue tests, Gal-1 overexpressing AGS cells were transfected with
GRP78 shRNA or NC shRNA. To establish stable transfected cell
lines, 25 g/ml neomycin was then added to the media for 2 weeks.
The pcDNA3.1-GRP78 and the negative control (pcDNA3.1-NC) were
provided by Shanghai GenePharma Co., Ltd. pcDNA3.1-GRP78 or
pcDNA3.1-NC plasmids were transfected using
Lipofectamine® 2000, and the control and Gal-1 knockdown
HGC-27 cells were then cultured for an additional two days before
being selected using G418 reagent (Sigma-Aldrich; Merck KGaA). For
the ensuing experiments, the cells were gathered.
Reverse transcription-quantitative PCR
(RT-qPCR)
The TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from the
cell lysate. Using the RevertAid First Strand cDNA Synthesis kit
according to the manufacturer's instructions and oligos(dT) from
Thermo Fisher Scientific, Inc., the concentration of total RNA was
determined spectrophotometrically and cDNA was synthesized. qPCR
was carried out in accordance with the manufacturer's instructions
using the SYBR Green I kit (cat. no. rk21203; ABclonal Biotech Co.,
Ltd.) and the StepOnePlus Real-time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The primer sequences were as
follows: Gal-1 forward, 5′-GCT GAA CCT GGG CAA AGA CAG-3′ and
reverse, 5′-GTT GAG GCG GTT GGG GAA CTT-3′ and GAPDH forward,
5′-CCA CTC CTC CAC CTT TGA C-3′ and reverse, 5′-ACC CTG TTG CTG TAG
CCA-3′. The thermocycling conditions were as follows: Initial
denaturation at 95°C for 2 min; followed by 40 cycles at 95°C for 5
sec and 60°C for 30 sec. The relative mRNA levels of the target
gene were determined using the 2−ΔΔCq method, with GAPDH
serving as the internal control (25).
Western blot analysis
RIPA lysis buffer (cat. no. R0020; Beijing Solarbio
Science & Technology Co., Ltd.) was used to lyse cells to
obtain protein samples for 30 min at 4°C. The samples were
centrifuged at 13,000 × g for 20 min at 4°C before the supernatant
was collected and the protein concentration was measured using a
BCA kit (cat. no. P0012; Beyotime Institute of Biotechnology).
Samples were boiled for 5 min after being mixed with a 5X loading
buffer solution. Equivalent extracted amounts of protein (20
µg protein per lane) from GC tissues or whole cells were
then separated with 10% SDS-PAGE gel and electro-transferred onto
polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific,
Inc.). They were blocked using 5% skimmed milk for 1 h at room
temperature and then incubated overnight at 4°C with various
primary antibodies, including rabbit anti-Gal-1 (1:3,000; cat. no.
ab108389), rabbit anti-GRP78 (1:3,000; cat. no. ab21685; both from
Abcam), rabbit anti-GAPDH (1:5,000; cat. no. AC001) and mouse-anti
GAPDH (1:5,000; cat. no. AC002; both from ABclonal Biotech Co.,
Ltd.); the two latter antibodies were used as internal controls.
Lastly, signals were detected with an enhanced chemiluminescence
substrate (cat. no. P10300; New Cell & Molecular Biotech Co.,
Ltd.) after incubating with secondary antibody HRP-conjugated goat
anti-rabbit (1:5,000; cat. no. AP132P; MilliporeSigma) or goat
anti-mouse antibodies (1:5,000; cat. no. AS003; ABclonal Biotech
Co., Ltd.). The intensity of western blot bands was quantified
using ImageJ (version 1.51; National Institutes of Health).
Cell Counting Kit-8 (CCK-8) and colony
formation assays
A total of 1,000 cells/well were plated in a 96-well
plate and cultured overnight at 37°C. After 24 h, 10 µl
CCK-8 (Beyotime Institute of Biotechnology) reagent was added to
each well at 24, 48 and 72 h and incubated for 2 h. Absorbance was
read at 450 nm using an automatic plate reader. Triplicates of each
experiment were independently performed. Additionally, a colony
formation experiment was run to assess the capacity for cell
proliferation. Six-well plates were seeded with 5×102 GC
cells per well and incubated at 37°C for 10 days. The colonies were
stained with 0.1% crystal violet at room temperature for 10 min
after being fixed with 4% paraformaldehyde at room temperature for
10 min. The number of visible colonies (consisting of >50 cells)
of three replicates was counted manually after the plates dried
up.
Wound healing assay
In order to create a confluent monolayer, HGC-27 and
AGS cells were seeded into six-well plates and cultured in
serum-free RPMI-1640 medium. At day 2, when the cells grew to
80-90% confluency as a monolayer, the monolayer was gently
scratched with a 200-µl pipette tip, with the tip being
perpendicular to the bottom of the plate during the operation. The
recovered tissues were then rinsed three times with PBS to remove
debris after scratching. To culture the remaining cells, the medium
containing 3% FBS was also changed. After the scratching, images
were captured at 0, 24 and 48 h under a light microscope
(magnification, ×200). ImageJ program examined the wound closure
(version 1.51; National Institutes of Health).
Transwell migration and invasion
assays
The migration and invasion assays were performed
using Transwell chamber (Corning, Inc.). For the migration assay,
1×104 transfected cells per well (serum-starved for 24
h) were suspended in 200 µl of serum-free RPMI-1640 medium
and plated in the upper chamber of a 24-well Boyden chamber. The
lower chamber contained RPMI-1640 medium with 10% FBS. For the
invasion assay, Transwell (Corning, Inc.) chambers were first
coated with Matrigel (cat. no. 356234; BD Biosciences) that was
diluted at 1:30 with serum-free RPMI-1640 medium at 37°C for 30
min. Cells (1×104) suspended in 200 µl of
serum-free medium were seeded in the upper chamber of a microporous
(8-µm pores) Transwell insert. In the bottom of the chamber,
500 µl RPMI-1640 medium containing 10% FBS was added. After
incubation for 48 h at 37°C, the chamber was removed, the cells
which had not passed through the chamber membrane were removed
using a wet cotton swab, and cells that migrated or invaded through
the membrane were fixed with 4% formaldehyde at room temperature
for 15 min and stained with 1% crystal violet staining solutions
for 5 min at room temperature and images were captured under a
light microscope (magnification, ×200). Cell numbers in five
randomly selected fields were counted.
Xenotransplantation experiment
A total of 10 4-week-old BALB/c male nude mice
(weight, 18-22 g) were purchased from GemPharmatech Co. Ltd. and
raised in a pathogen-free laminar flow cabinet throughout the
experiments under the following conditions: Controlled humidity
(30-40%), a constant temperature of 25°C, a 12-h light/dark cycle
and free access to food and water. The ethical approval (approval
no. 202111020) to perform the animal experiments was obtained from
the Ethics Committee for Animal Experiments of the Yangzhou
University (Yangzhou, China). The experimental protocol was
performed in accordance with the Laboratory Animal Guideline for
Ethical Review of Animal Welfare (26). 4-week-old male BALB/c nude mice
were randomly divided into Ctrl and OE-Gal-1 groups (n=5 in both
groups). Under isoflurane inhalation anesthesia (1-2%),
~1×106 AGS cells of stably transfected strains
Ctrl/OE-Gal-1 resuspended in 100 µl PBS were subcutaneously
into the left armpit of the mice. The health and behaviour of the
mice were monitored every 2 days to determine if there were
difficulties eating or drinking, unrelieved pain or distress
without recovery. If the tumor reached 2,000 mm3, the
animal would be euthanized as a humane endpoint. The following
formula was used to calculate the tumor volume (V) every week:
V=(Width2 × Length)/2. Four weeks post-inoculation, all
the mice were sacrificed by cervical dislocation under anesthesia.
The method of anesthesia used for the mice was CO2
asphyxiation (CO2 was introduced into the chamber at a
rate of 40-70% of the chamber volume per min to minimize distress).
Dilated pupils were then used to verify death. Then, the tumors
were removed, weighed and fixed for further histopathological
analyses. Immunohistochemistry was performed on xenografted tumor
tissues with anti-Gal-1 rabbit polyclonal antibody (1:200; cat. no.
11858-1-AP; ProteinTech Group, Inc.) and anti-Ki-67 rabbit
monoclonal antibody (1:400, 9027; Cell Signaling Technology).
Co-immunoprecipitation (Co-IP)
assays
The cell lysate was harvested from HGC-27 cells
using ice-cold non-denaturing lysis buffer with the addition of
proteinase inhibitor cocktail Complete Mini and phosphatase
inhibitor cocktail PhosSTOP (both from Roche Diagnostics). The
total protein of the lysates was measured by the BCA protein assay
kit (Beyotime Institute of Biotechnology) analyzed by an Eppendorf
Master Photometer.
A total of 500 µl of cell lysates with a
concentration of 1 µg/µl were used for Co-IP
experiments performed with the Pierce Co-IP kit according to the
manufacturer's instructions (Thermo Fisher Scientific, Inc.). A
total of 10 µg of the monoclonal Gal-1 antibody (1:50) or
GRP78 antibody (1:50; cat. no. sc166490; Santa Cruz Biotechnology,
Inc.) were briefly incubated with 50 µl AminoLink Plus
Coupling Resin and covalently coupled. Moreover, 10 µg
non-specific IgG was set as a control. The purified protein lysate
(500 µl) was pre-cleared by incubation with 80 µl of
the Control Agarose Resin slurry (Thermo Fisher Scientific, Inc.)
for 30 min at 4°C on a rotator. The antibody-conjugated agarose
resin was incubated with 500 µl of the pre-cleared cell
lysates and rotated overnight at 4°C. The resin was washed, and the
protein complexes bound to the antibody were eluted. Subsequent
proteins were separated in a 10% SDS-PAGE gel, transferred to a
PVDF membrane, and detected using Gal-1 and GRP78 MAbs.
Immunofluorescence staining
HGC-27 cells were cultured for 24 h on coverslips in
24-well plates. After that, cells were washed three times and fixed
in 4% paraformaldehyde at room temperature for 10 min. The cells
were permeabilized with 0.1% Triton X-100 for 15 min. After
blocking with 5% bovine serum albumin (Beyotime Institute of
Biotechnology) at room temperature for 60 min, the cells were
incubated with the following antibodies for 60 min at 37°C: Rabbit
anti-human GRP78 (1:300; cat. no. 11587-1-AP) and mouse anti-human
Gal-1 (1:100; cat. no. 60223-1-Ig; both from ProteinTech Group,
Inc.). The cells were washed with PBS three times and incubated for
1 h in the dark at room temperature with the following secondary
antibodies: Donkey Anti-Rabbit IgG H&L (1:200; cat. no.
ab150062), Goat Anti-Mouse IgG H&L (1:200; cat. no. ab150113;
both from Abcam). For the experiments in which ER stress of HGC-27
cells were activated with tunicamycin (2 µg/ml; cat. no.
T8480; Beijing Solarbio Science & Technology Co., Ltd.). HGC-27
cells and tunicamycin-activated HGC-27 cells were incubated with
Rabbit anti-human GRP78 (1:300; cat. no. 11587-1-AP) overnight at
4°C and then were incubated with Donkey Anti-Rabbit IgG H&L
(1:200; cat. no. ab150062) at RT for 1 h. After being washed three
additional times, the cells were counterstained with DAPI (Beyotime
Institute of Biotechnology) at the concentration of 5 µg/ml
for 5 min. The results were examined by laser scanning confocal
microscopy (Leica Microsystems GmbH).
Statistical analysis
At least three times each of the data were
replicated. The data was examined using the statistical analysis
program SPSS 24.0 (IBM Corp.). To compare two sets of independent
samples, two-tailed unpaired Student's tests were utilized
(Figs. 2-5). In Gal-1 and GRP78 expression in GC
tissues, the data with normal distribution, and the paired 2-tailed
Student's t-test was used to evaluate the significance of
differences between two groups (Figs.
1E and G and 7B). One-way
ANOVA and Tukey's post hoc test were used to examine measurement
data between three or more groups (Figs. 1I and 6B-E). The expression of Gal-1 of GRP78
was associated with clinicopathological characteristics using a
chi-squared test. The relationship between the expression of Gal-1
or GRP78 and patient prognosis was examined using the Kaplan-Meier
method and log-rank test. Non-parametric Gal-1 and GRP78 expression
were correlated using Spearman's rank correlation analysis.
Statistics were judged significant at P<0.05.
Results
Gal-1 is upregulated in GC and its
upregulation is associated with a poor prognosis for GC
patients
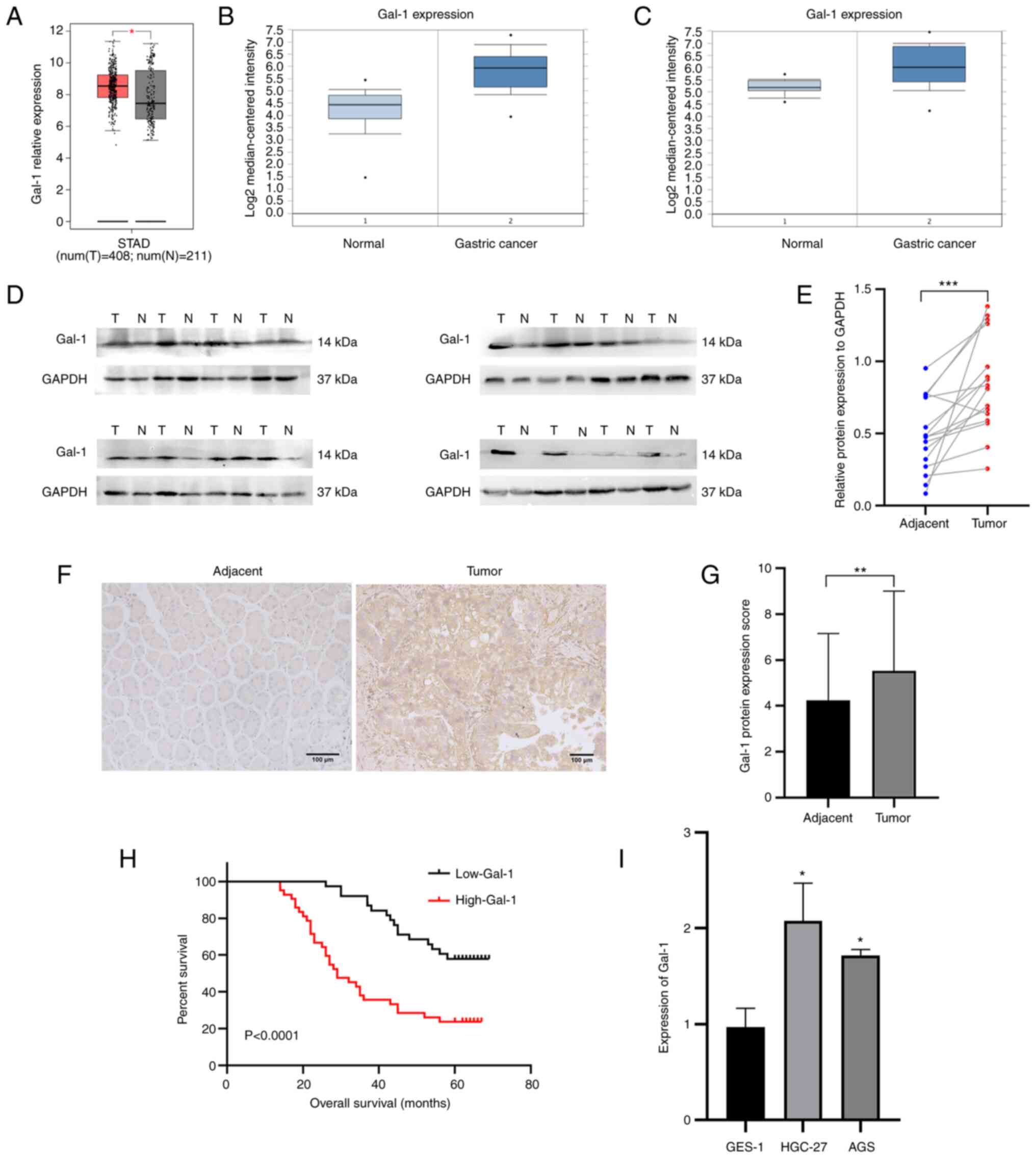
The mRNA expression levels of Gal-1 were first
examined in GC tissues from the GEPIA, whose RNA sequencing
expression data of tumors and normal samples from TCGA public
databases (http://gepia.cancer-pku.cn/) and Oncomine database in
order to evaluate the significant role of Gal-1 in GC tissue. Gal-1
mRNA expression was significantly upregulated in 408 GC samples
when compared with 211 instances of the normal tissues (Fig. 1A). In the two distinct validation
datasets, the geometric mean of Gal-1 expression was considerably
higher in GC tissues than in normal stomach samples (Fig. 1B and C). In addition, it was found
that the 16 GC patients had Gal-1 protein levels. When GC tissues
were compared with the nearby normal gastric tissues, it was
identified that the protein level of Gal-1 was upregulated
(Fig. 1D and E). IHC analysis was
used to find visible Gal-1 expression levels in a developed TMA
that comprised 80 matched GC and nearby normal tissues in order to
understand in an improved way the role of Gal-1 in the formation of
GC. When GC tissue was compared with the nearby non-cancerous
tissues, Gal-1 was more highly expressed in the GC tissue (Fig. 1F and G). According to the results
of the RT-qPCR investigation, Gal-1 was expressed significantly
higher in GC cell lines compared with GES-1 cells (Fig. 1I). Moreover, the association
between Gal-1 expression and clinicopathological characteristics
was investigated, showing that higher Gal-1 expression was
associated with the tumor size (P=0.003), depth of invasion
(P=0.049), LN metastasis (P=0.007), distant metastasis (P=0.026),
TNM stage (P=0.015) and GRP78 expression (P=0.008). However, Gal-1
expression was not associated with age (P=0.759), sex (P=0.408),
Lauren type (P=0.797), degree of differentiation (P=0.849),
histological grade (P=0.617), venous invasion (P=0.866) and nerve
invasion (P=0.463) (Table I). The
associations between Gal-1 expression and the overall survival (OS)
of 80 patients with GC were evaluated in order to look into the
connection between the expression of Gal-1 and the prognosis of GC.
According to Kaplan-Meier analysis, poor OS in GC patients was
positively connected with increased Gal-1 protein expression
(P<0.0001; Fig. 1H). The
factors affecting OS (P<0.05) were shown by univariate
regression analysis to be the tumor size, distant metastases, TNM
stage (stages I and II vs. stages III and IV), Gal-1 expression
(low vs. high), and GRP78 expression (low vs. high). The same
findings from multivariate regression analysis were obtained
regarding the independent risk factors for GC progression
(P<0.05, Table II): tumor
size, distant metastases, TNM stage (stages I and II vs. stages III
and IV), Gal-1 expression (low vs. high) and GRP78 expression (low
vs. high). These findings suggested that Gal-1 is increased in GC
and that GC patients with high levels of Gal-1 have a bad
prognosis.
 | Table IIPrognostic factors on overall
survival were analyzed by univariate and multivariate Cox's
proportional hazards models in 80 patients with gastric cancer. |
Table II
Prognostic factors on overall
survival were analyzed by univariate and multivariate Cox's
proportional hazards models in 80 patients with gastric cancer.
| Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 0.739
(0.344-1.584) | 0.437 | | |
| Sex | 0.847
(0.416-1.724) | 0.647 | | |
| Tumor size | 2.565
(1.257-5.236) | 0.010 | 2.350
(1.237-4.463) | 0.009 |
| Lauren type | 1.854
(0.474-7.253) | 0.375 | | |
| Lymphonodus
metastasis | 0.849
(0.308-2.339) | 0.751 | | |
| Distant
metastases | 3.440
(1.372-8.625) | 0.008 | 3.345
(1.682-6.648) | 0.001 |
| Depth of
invasion | 1.131
(0.461-2.771) | 0.788 | | |
| TNM stage | 4.376
(1.159-16.521) | 0.029 | 4.014
(1.757-7.170) | 0.001 |
| Degree of
differentiation | 0.617
(0.289-1.314) | 0.210 | | |
| Histological
grade | 0.961
(0.535-1.726) | 0.893 | | |
| Venous
invasion | 0.870
(0.390-1.938) | 0.733 | | |
| Nerve invasion | 0.970
(0.471-1.996) | 0.934 | | |
| Galectin-1 | 2.414
(1.033-5.641) | 0.042 | 2.224
(1.161-4.262) | 0.016 |
| Glucose-regulated
protein 78 | 2.614
(1.304-5.238) | 0.007 | 2.228
(1.191-4.170) | 0.012 |
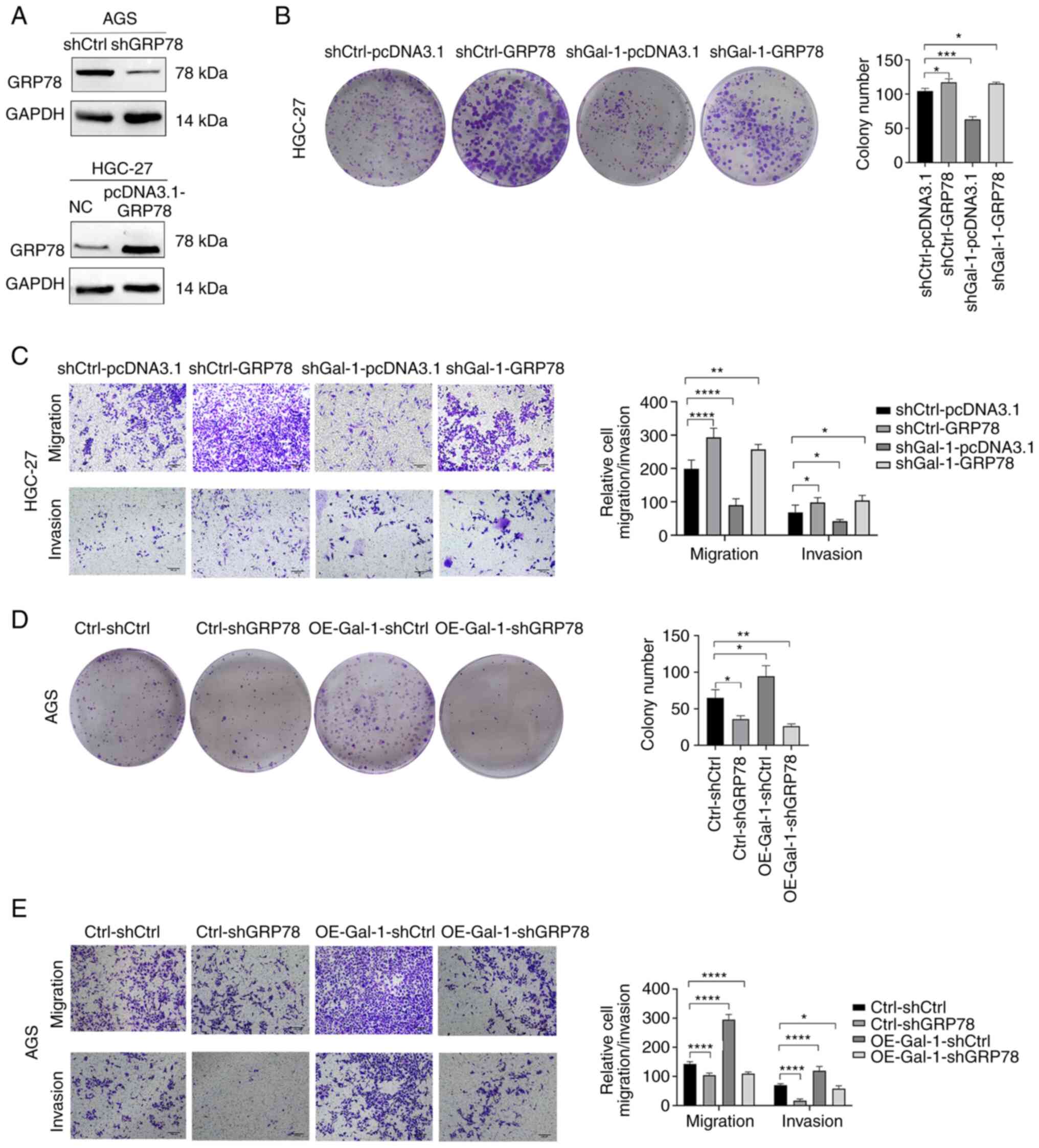
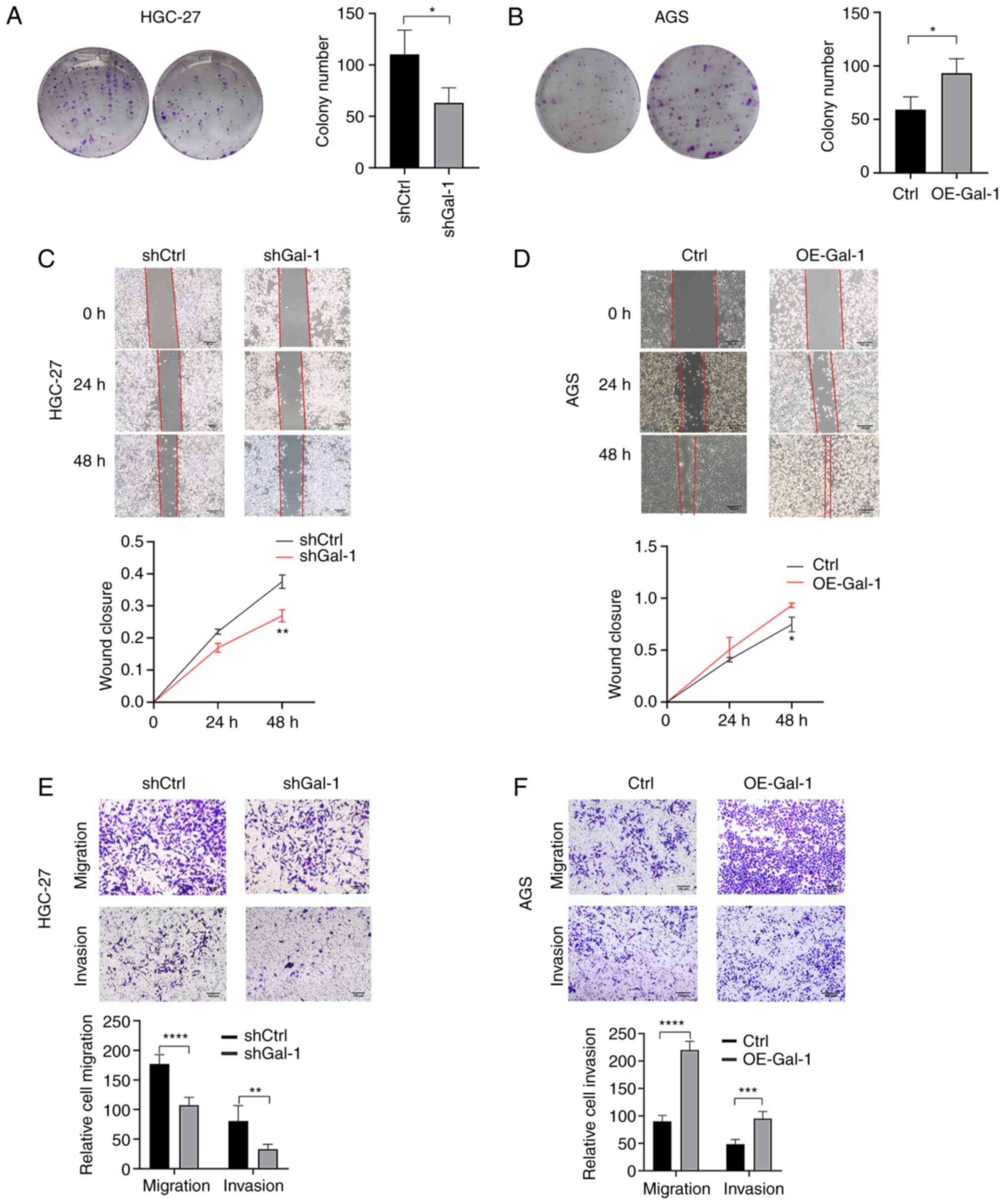
Gal-1 regulates proliferation, migration
and invasion of gastric carcinoma
Based on the expression levels of Gal-1 in GC cell
lines, Gal-1 stably overexpressing cells were generated in AGS
cells and the GC cell line HGC-27 was established with
lentivirus-mediated Gal-1 knockdown. By using immunofluorescence
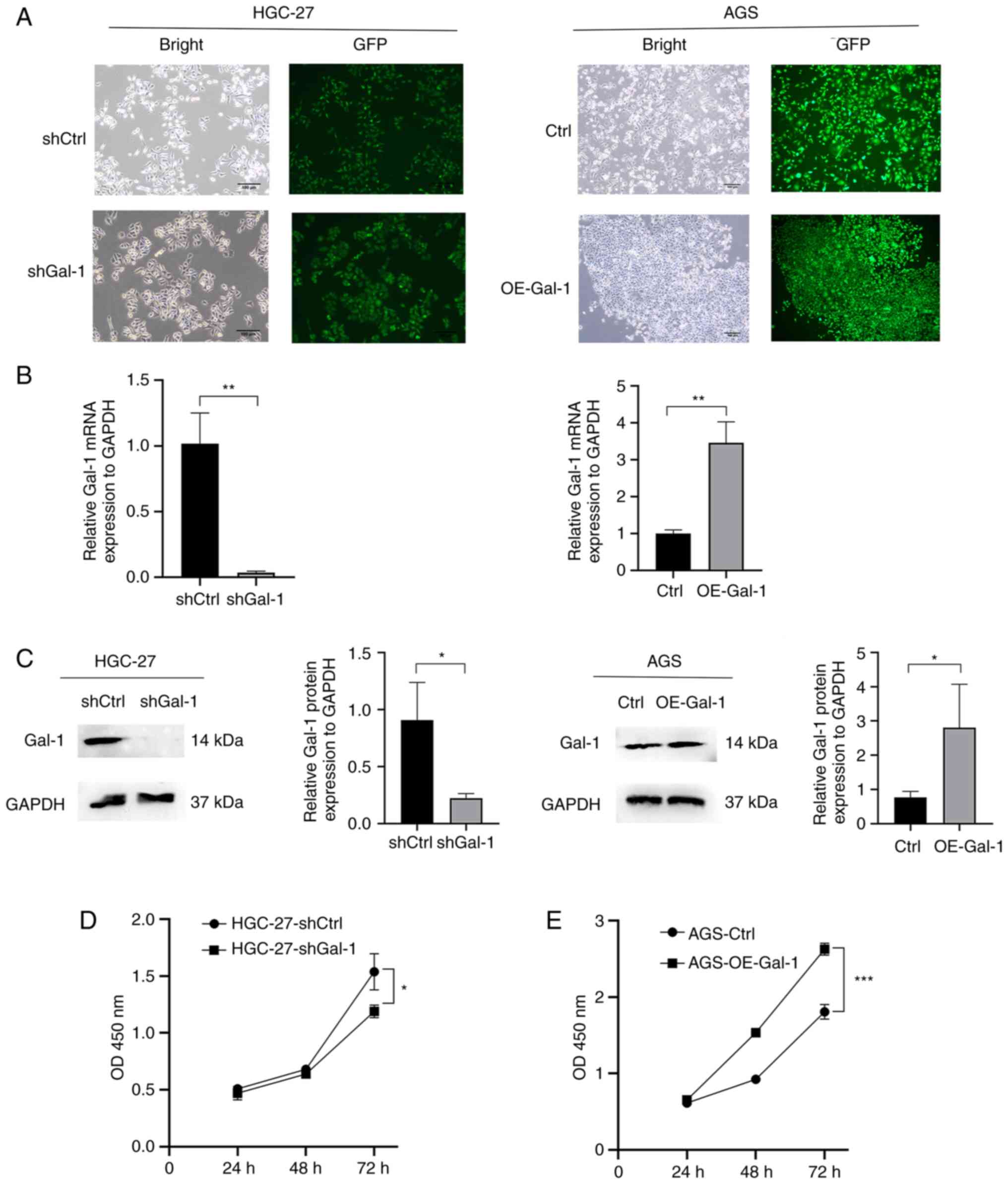
analysis, RT-qPCR and western blotting, the effectiveness of Gal-1
knockdown and overexpression in HGC27 and AGS cells was verified
(Fig. 2A-C). The potential of
Gal-1 to influence cell proliferation was examined using the CCK-8
and colony formation assays. As revealed in Figs. 2D and 3A, cell proliferation in the shGal-1
group in HGC-27 cells was considerably lower than that in the
control group (P<0.05). In addition, Transwell experiments and
wound healing assays both showed that decreasing Gal-1
significantly reduced cell migration in the HGC-27 cell line
(Fig. 3C and E). In addition, the
Matrigel invasion experiments showed that blocking Gal-1
significantly reduced the ability to invade (Fig. 3E). The overexpression of Gal-1 in
the AGS cell line, on the other hand, had the opposite impact of
Gal-1 knockdown, as observed in Fig.
2E, leading to an increase in tumor cell proliferation,
migration and invasion when compared with the control group
(Fig. 3B, D and F; P<0.05).
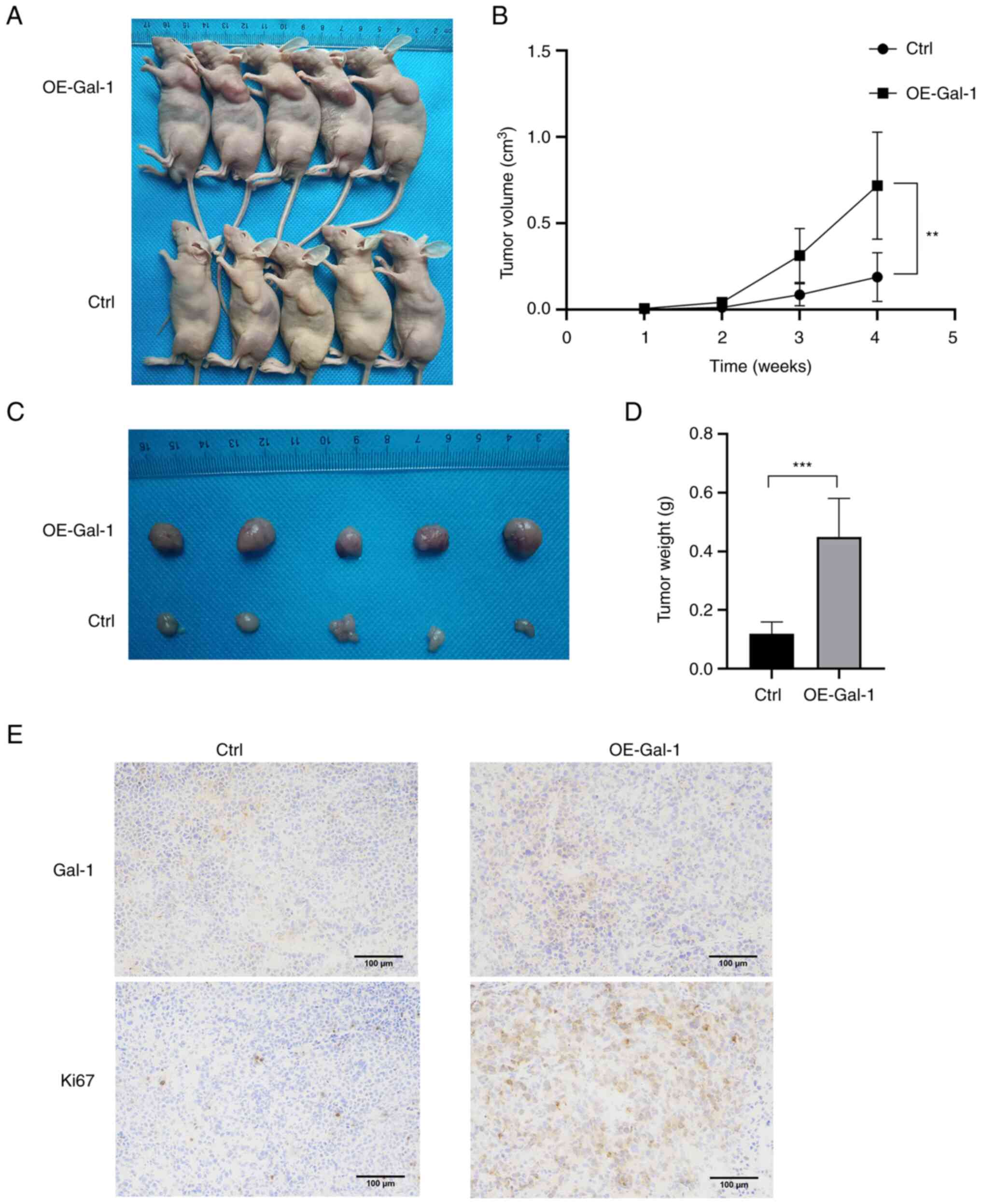
Overexpression of Gal-1 promotes the
growth of tumors in nude mice
The xenograft tumor model was used to further
explore the role of Gal-1 in vivo. Nude mice were injected
with AGS cells that had been stably transfected with either the
Ctrl or the OE-Gal-1 lentivirus, and the xenograft tumor growth was
observed every week after the tumor injection. When compared with
the Ctrl group, overexpression of Gal-1 almost caused the tumor to
expand (Fig. 4A and B). The most
significant tumor volume was 1.183 cm3 in the OE-Gal-1
group and 0.384 cm3 in the Ctrl group when the mice were
euthanized four weeks later (Fig.
4C). The two groups of tumors were then weighed. Compared with
the Ctrl group, the average weight of the OE-Gal-1 group was
significantly larger (0.450±0.130 g vs. 0.120±0.039 g; P<0.05)
(Fig. 4D). According to IHC
examination, transfection with the lentiviral overexpression vector
may successfully increase the level of Gal-1 expression in tumor
tissues (Fig. 4E). After that,
Ki-67 staining was employed to determine the capacity of tumors for
proliferation. The findings demonstrated that tumor proliferative
capacity was boosted by Gal-1 overexpression (Fig. 4E). The results demonstrated that
overexpression of Gal-1 promoted GC xenograft tumor growth in
vivo.
GRP78 oncoprotein is identified as a
Gal-1-binding partner
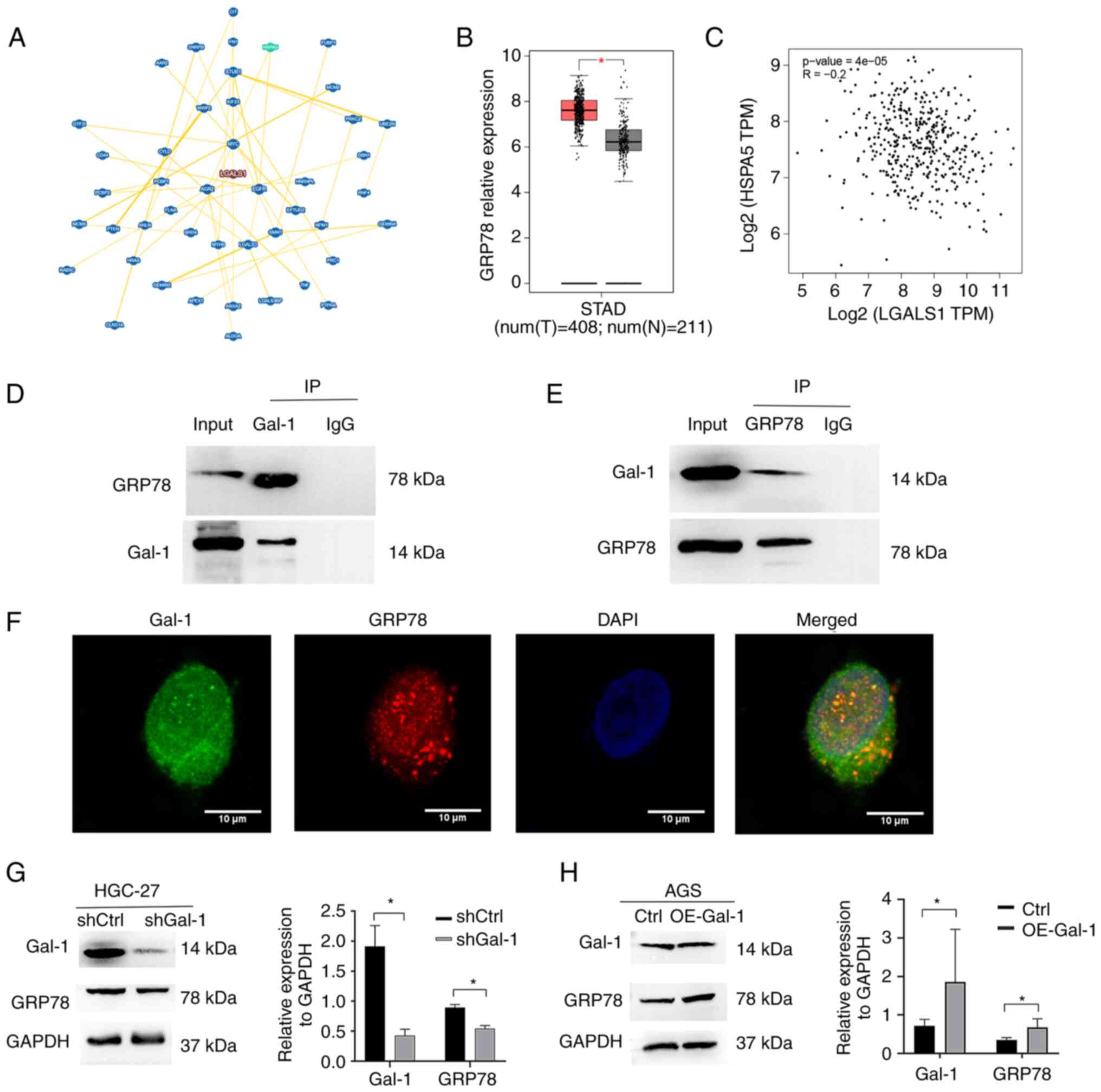
The interacting proteins of Gal-1 were examined
using the BioGRID database (https://thebiogrid.org/) in order to understand in an
improved way the fundamental molecular mechanisms behind the
tumor-promoting activity of Gal-1 in GC. According to the BioGRID
database, Gal-1 is linked to 296 different proteins. A total of 45
proteins were ultimately identified as interacting with Gal-1 by
high throughput and physical interactions after the search area was
restricted and the minimum 3 evidence was chosen (Fig. 5A). Following validation, the heat
shock protein GRP78, which is mostly localized at the ER and is
encoded by HSPA5, was chosen for more examination. It is well
recognized that GRP78 is involved in the migratory, invasion,
angiogenesis and immune evasion processes that contribute to the
advancement of GC (27-29). The mRNA expression of GRP78,
similar to that of Gal-1, was significantly overexpressed in the
TCGA GC cohort (Fig. 5B). In the
TCGA GC cohort, there was also no discernible relationship between
the genes for Gal-1 and GRP78 (Fig.
5C). Therefore, the present study concentrated on the
interaction between the GRP78 and Gal-1 proteins. First, the
function of the ER in HGC-27 cells cultivated in our lab was
examined using western blotting and immunofluorescence staining
assays. As expected, the level of GRP78, the marker for ERS, was
increased markedly after treated with Tunicamycin (ERS inducer, 2
µg/ml) for 24 h (29),
(Fig. S1). Subsequently, by using
a reciprocal Co-IP experiment, it was examined whether endogenous
GRP78 and Gal-1 had a functional connection that increased GC
development by controlling GRP78. AminoLink Plus Coupling Resin was
used to immunoprecipitate the Gal-1 protein complex, and anti-Gal-1
or anti-GRP78 antibodies were used for the immunoblot. An
anti-Gal-1-specific antibody easily identified a positive GRP78
signal in the Co-IP complex that was brought down (Fig. 5D). Gal-1 was also observed in the
immunoprecipitated material that the anti-GRP78 antibody was able
to draw down (Fig. 5E). In order
to ascertain whether GRP78 had the same subcellular location as
Gal-1, an immunofluorescence staining test was carried out. HGC-27
cells underwent dual immunostaining for GRP78 and Gal-1 to look for
subcellular localization (Fig.
5F). In HGC-27 cells, GRP78 was largely expressed in the
cytoplasm and nucleus, where Gal-1 is also found. Recombinant
lentiviruses were employed to over- and under-express Gal-1 in AGS
and HGC-27 cells in order to examine if Gal-1 affects GRP78
expression and activity. In the Lv-shGal-1 group (HGC-27), it was
revealed that the expression of GRP78 was significantly
downregulated compared with the control group (Fig. 5G). Additionally, it was identified
that AGS cells with high levels of Gal-1 overexpressed GRP78
protein (Fig. 5H). This
information taken together suggested a connection between GRP78 and
Gal-1.
Gal-1 influences GC proliferation,
invasion and migration through GRP78
To determine if GRP78 is essential for
Gal-1-mediated GC development, a functional rescue test was carried
out. First, western blotting verified the effectiveness of GRP78
overexpression and knockdown in AGS and HGC-27 cells (Fig. 6A). The next step was to
co-transfection of HGC-27 cells with shGal-1 lentivirus and pcDNA
3.1-GRP78. The colony formation experiment revealed that ectopic
GRP78 expression restored the ability of Gal-1 knockdown cells
(shGal-1-pcDNA 3.1 group) to form colonies to baseline levels in
HGC27 cells (Fig. 6B). Data from
cell migration and invasion studies further showed that cells
co-transfected with pcDNA 3.1-GRP78 had a limited ability to block
cell migration and invasion by shGal-1 (Fig. 6C). When OE-Gal-1 AGS cell
proliferation, migration, and invasion were compared with an empty
vector control, GRP78 knockdown significantly reduced all three
(Fig. 6D and E). These results
supported the hypothesis that the Gal-1/GRP78 axis controls the
course of GC by mediating the action of Gal-1 tumor-promoting in
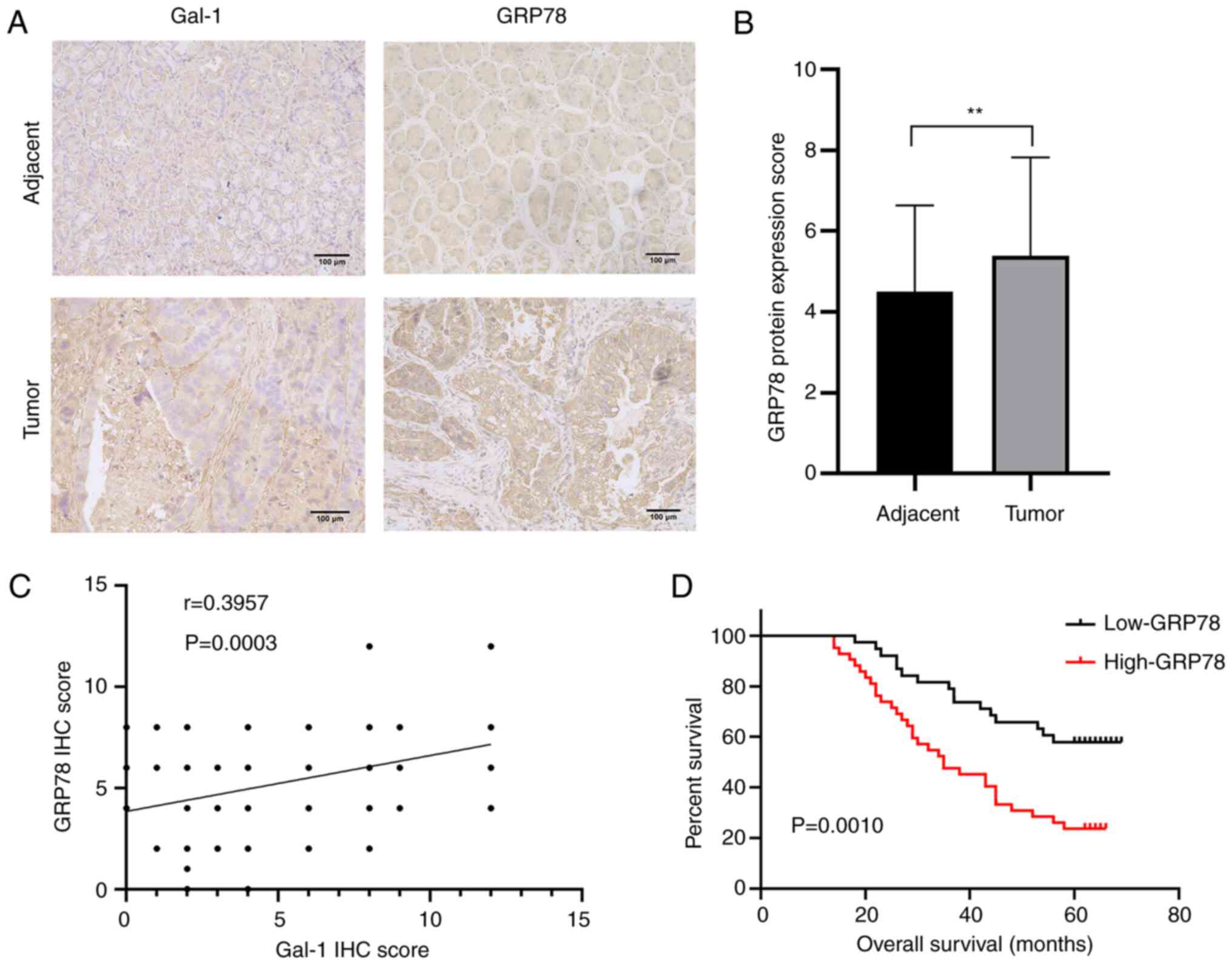
GC. In addition, IHC was performed to clarify the connection
between the levels of Gal-1 and GRP78 protein expression in 80 GC
samples and matched non-tumor tissues. GRP78 protein levels were
significantly greater in GC tissues than in matching para-cancerous
tissues, according to results of GRP78 protein expression in 80
pairs of GC tumors and matched non-tumor tissues (Fig. 7A and B). Additionally, Gal-1
expression in GC tissues was significantly and favorably linked
with GRP78 protein level expression (R=0.3957, P<0.01; Fig. 7C). Additionally, the GRP78
expression in our sample group was measured and was found to be in
contrast to the pathological characteristics in the dichotomized
GRP78 status. According to the summary in Table III, GRP78 expression was
positively connected with the degree of differentiation (P=0.004),
lymphonodus metastasis (P=0.025), TNM stage (P=0.048) and depth of
invasion (P=0.049). High GRP78 protein expression was found to be
positively connected with poor OS in GC patients according to a
Kaplan-Meier analysis (P=0.0010; Fig.
7D). GRP78 expression was strongly associated with the overall
five-year survival rate in both univariate and multivariate
analyses (Table II,
P<0.05).
Discussion
Our earlier research (30,31)
has extensively discussed the relationship between Gal-1 expression
and GC in the present study. The analysis of Gal-1 expression in GC
using multiple experimental techniques and databases was carried
out for the first time. The present findings revealed that Gal-1
plays a tumor-promoting role in GC and that it is upregulated and
crucial for enhancing GC proliferation and invasion via its
interaction with GRP78.
The current study used data from the Oncomine and
GEPIA databases to observe Gal-1 expression profiling in GC. The
findings demonstrated that samples from GC patients had
significantly higher levels of Gal-1 mRNA. Gal-1 protein expression
was also found in both malignant and non-cancerous tissues. In line
with a previous study by the authors (30), it was showed that the expression of
Gal-1 was higher in GC tissues than in nearby non-cancerous tissues
in our database. The tumor size, depth of invasion, LN metastasis,
distant metastasis, TNM stage and GRP78 expression were all
associated with high levels of Gal-1 expression. Additionally, an
association between increased Gal-1 expression and lower patient
survival rates was identified. It was determined that high
expression of Gal-1 may be used as a poor prognostic factor for GC
by analyzing the link between higher expression of Gal-1 and poor
survival rate in our cohort.
A number of studies have revealed that targeting
Gal-1 is a promising therapeutic approach to slow the progression
of cancer in addition to the diagnostic and prognostic utility of
this protein (31-33). Gal-1 knockdown in GC cells was
found to considerably impede the ability of HGC-27 cells to
proliferate, migrate, and invade, according to the following in
vitro investigations of the study. However, exogenous
overexpression of Gal-1 facilitated AGS tumor cell motility,
invasion and proliferation. Additional in vivo animal
investigations revealed that, as compared with the Ctrl group, GC
cells overexpressing Gal-1 grew larger in a nude mouse model of the
disease. These findings indicated that Gal-1 promotes tumor growth
in GC.
Gal-1 is widely expressed by various tissues,
including natural killer cells and endothelial cells, and is
encoded by the LGALS1 gene (34).
Gal-1 has been implicated in a number of biological processes,
including cell proliferation, differentiation, migration, invasion,
apoptosis, homeostasis and vascular embryogenesis in a number of
malignancies, according to previous studies (35-37).
In other words, depending on the kind of cell and the context of
the cell, Gal-1 can bind and cross-link glycoconjugates on the cell
surfaces or ECM and regulate a variety of biological processes,
including immunosuppression, angiogenesis and metastasis.
Additionally, it has been established through IHC that initial
tumor sections have significant levels of Gal-1 expression
throughout (38-40). Gao et al (41) revealed that Gal-1 was overexpressed
in breast cancer and hypothesized that the interaction between the
N-terminus of Gal-1 and the FKH domain of FOXP3 prevented FOXP3
from having tumor-suppressive effects.
On the other hand, Gal-1 can bind to substances that
boost their ability to promote carcinogenesis and metastasis in
carcinoma. Gal-1 was assessed in hepatocellular carcinoma (HCC)
tissues, according to Leung et al (42); the amount of serum Gal-1 is
associated with HBV and liver cirrhosis. Gal-1 additionally
increases HCC cell motility, proliferation and growth through the
overexpression of RER1. It was also showed that the combination of
sorafenib and the particular Gal-1 inhibitor OTX008 inhibits tumor
growth in an animal model in a synergistic manner. A recent study
revealed that the HGSC-secreted factor cathepsin L triggered Gal-1
secretion autocrine, which enhances angiogenesis, proliferation and
migration in the omental microvascular endothelial cell metastasis
of HGSC via activating the ERK1/2 pathway (43). Kucińska et al (44) revealed that extracellular Gal-1 and
Gal-3 interact directly with the sugar residues on fibroblast
growth factor receptor 1 (FGFR1), inducing phosphorylation of FGFR1
and initiating downstream kinase ERK1/2 and PLCγ signaling pathway,
which results in cell proliferation and avoidance of apoptosis.
In the present study, GRP78 was identified as a
potential interaction partner in Gal-1-mediated tumor progression
utilizing the BioGRID database and Co-IP analysis to clarify the
mechanism by which Gal-1 drives GC formation. GRP78 is linked to a
number of processes that contribute to the growth of cancer,
including cell growth, angiogenesis, apoptosis, chemoresistance,
cell survival and metabolism (13). Additionally, GRP78 was
overexpressed on the plasma membrane of several cancers, including
endometrial, hepatocellular, prostate and breast cancer. In a
different biological setting, GRP78 proteins could have a dual role
as oncogenes or tumour suppressors. By reducing the activity of the
VEGF/VEGFR2 pathway, for instance, the inhibition of GRP78 reduced
the growth of colon cancer (45).
High surface expression of GRP78 decreased colon cancer tumor
growth and proliferation while increasing the potential to invade
due to higher levels of NRF-2 and heme oxygenase-1 expression (HO1)
(46).
On the contrary, Yuan et al (47) showed that the overexpression of
GRP78 caused an enhancement of proliferation, invasion and
migration and increased the percentage of cells in the S phase of
the cell cycle due to a rise in matrix metal-loproteinase-2 and 9
(MMP-2 and MMP-9) secretion level. It was revealed that GRP78 could
be physically interacting with IGF1-R and redistributed from the ER
to the surface of hepatoma cells by IGF-I, promoting cellular
proliferation, migration and invasion activity (48). Several studies reported that
various ligands could interact with GRP78, eliciting diverse
signaling responses. Qian et al (49) showed that SCNN1B exhibited
post-tumor-suppressive function through ubiquitination-dependent
degradation of GRP78 by regulating the UPR signaling pathway. In a
previous study, it was identified that the interaction between
GRP78 and a2-macroglobulin (a2M) increased prostate cancer cell
survival and migration/invasion via activating the PAK-2/MAPK and
PI3K pathways, in which GRP78 functioned as an a2M receptor
(50). The aberrant expression of
GRP78 in GC is explained in the current study by a unique
mechanism. It was revealed that GRP78 interacts with Gal-1 using
bioinformatic techniques and Co-IP tests. The co-localization of
Gal-1 and GRP78 in the cytoplasm and nucleus was revealed by
immunofluorescence labeling.
Furthermore, it was demonstrated that ectopic
production of Gal-1 in AGS cells had the opposite effect on the
expression of GRP78 from knocking down Gal-1 in HGC-27 cells, which
reduced the expression of GRP78. As previously indicated, increased
expression of GRP78 encourages the growth, invasion, and migration
of tumors. As a result, it was suggested that GRP78 may play a
crucial part in the progression of GC caused by Gal-1. Furthermore,
ectopic GRP78 expression significantly increased the proliferation,
migration and invasion of Gal-1 knockdown GC cell line, according
to function rescue assays. The function of Gal-1 overexpression was
consistently hampered by silencing GRP78. These findings
demonstrated the critical part GRP78 plays in tumor growth caused
by Gal-1. Notably, this GRP78 rescue function was insufficient,
suggesting that Gal-1 may potentially advance GC through additional
targets. According to previous studies (42-44),
it was hypothesized that Gal-1 that is upregulated promotes the
production of the GRP78 protein, transports it from the ER to the
nucleus, and anchors. Nevertheless, more information about the
precise mechanism has to be provided.
It was identified that the expression of GRP78 was
upregulated in GC in our clinical sample sequence. Additionally,
the degree of differentiation, lymphonodus metastasis, TNM stage
and depth of invasion were statistically linked with the
expression, which was consistent with previous results (21). In GC samples, GRP78 and Gal-1
expression levels are strongly associated, and patients with high
GRP78 expression had a bad prognosis.
There were a number of limitations to the present
study. First, track of OS of patients was just kept; we did not
track their survival free from disease. Second, more samples should
be included in the present investigation. The inability to identify
the precise chemical mechanism by which Gal-1 interacts with GRP78
to advance GC along the cellular signaling pathway is another
drawback.
In conclusion, it was determined that the protein
Gal-1 plays a crucial role in mediating migration, invasion and
proliferation. The connection between Gal-1 and GRP78, which
results in the proliferation, invasion and migration of GC cells,
is now understood in an improved way thanks to the present study.
The present research may lead to the identification of new
therapeutic targets for the Gal-1 and GRP78 pathways for the
treatment of GC and may also provide guidance for existing
targets.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ, D-RW and MA conceived the study. YW, DT and WW
provided methodology. Q-NS, C-YZ, H-HZ and X-DZ conducted
experimental investigation. C-YZ provided resources. QZ, DT and WW
prepared and wrote the original draft. H-HZ and D-RW acquired
funding. QZ and D-RW confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The studies involving human participants were
reviewed and approved (approval no. 2019KY-022) by the Ethics
Committee of the Northern Jiangsu People's hospital, the Clinical
Medical School, Yangzhou University Ethics Committee (Yangzhou,
China). Written informed consent was obtained from all
participants. The animal study was reviewed and approved (approval
no. 202111020) by the Ethics Committee of Yangzhou University
(Yangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professor Liu-Hua
Wang (Yangzhou Key Laboratory of Basic and Clinical Transformation
of Digestive and Metabolic Diseases, Yangzhou, Jiangsu 225001, P.R.
China) for providing technical support.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81972269).
References
|
1
|
Machlowska J, Baj J, Sitarz M, Maciejewski
R and Sitarz R: Gastric cancer: Epidemiology, risk factors,
classification, genomic characteristics and treatment strategies.
Int J Mol Sci. 21:40122020. View Article : Google Scholar :
|
|
2
|
Wong MCS, Huang J, Chan PSF, Choi P, Lao
XQ, Chan SM, Teoh A and Liang P: Global incidence and mortality of
gastric cancer, 1980-2018. JAMA Netw Open. 4:e21184572021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Joshi SS and Badgwell BD: Current
treatment and recent progress in gastric cancer. CA Cancer J Clin.
71:264–279. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Camby I, Le Mercier M, Lefranc F and Kiss
R: Galectin-1: A small protein with major functions. Glycobiology.
16:137R–157R. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carlini MJ, Roitman P, Nuñez M, Pallotta
MG, Boggio G, Smith D, Salatino M, Joffé ED, Rabinovich GA and
Puricelli LI: Clinical relevance of galectin-1 expression in
non-small cell lung cancer patients. Lung Cancer. 84:73–78. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang CS, Tang SJ, Chung LY, Yu CP, Ho JY,
Cha TL, Hsieh CC, Wang HH, Sun GH and Sun KH: Galectin-1
upregulates CXCR4 to promote tumor progression and poor outcome in
kidney cancer. J Am Soc Nephrol. 25:1486–1495. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang PF, Li KS, Shen YH, Gao PT, Dong ZR,
Cai JB, Zhang C, Huang XY, Tian MX, Hu ZQ, et al: Galectin-1
induces hepatocellular carcinoma EMT and sorafenib resistance by
activating FAK/PI3K/AKT signaling. Cell Death Dis. 7:e22012016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schulz H, Schmoeckel E, Kuhn C, Hofmann S,
Mayr D, Mahner S and Jeschke U: Galectins-1, -3, and -7 are
prognostic markers for survival of ovarian cancer patients. Int J
Mol Sci. 18:12302017. View Article : Google Scholar :
|
|
9
|
Elola MT, Wolfenstein-Todel C, Troncoso
MF, Vasta GR and Rabinovich GA: Galectins: Matricellular
glycan-binding proteins linking cell adhesion, migration, and
survival. Cell Mol Life Sci. 64:1679–1700. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boyce M and Yuan J: Cellular response to
endoplasmic reticulum stress: A matter of life or death. Cell Death
Differ. 13:363–373. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hetz C: The unfolded protein response:
Controlling cell fate decisions under ER stress and beyond. Nat Rev
Mol Cell Biol. 13:89–102. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo B and Lee AS: The critical roles of
endoplasmic reticulum chaperones and unfolded protein response in
tumorigenesis and anticancer therapies. Oncogene. 32:805–818. 2013.
View Article : Google Scholar
|
|
13
|
Ibrahim IM, Abdelmalek DH and Elfiky AA:
GRP78: A cell's response to stress. Life Sci. 226:156–163. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cook KL, Soto-Pantoja DR, Clarke PA, Cruz
MI, Zwart A, Wärri A, Hilakivi-Clarke L, Roberts DD and Clarke R:
Endoplasmic reticulum stress protein GRP78 modulates lipid
metabolism to control drug sensitivity and antitumor immunity in
breast cancer. Cancer Res. 76:5657–5670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Serrano-Negrón JE, Zhang Z, Rivera-Ruiz
AP, Banerjee A, Romero-Nutz EC, Sánchez-Torres N, Baksi K and
Banerjee DK: Tunicamycin-induced ER stress in breast cancer cells
neither expresses GRP78 on the surface nor secretes it into the
media. Glycobiology. 29:5992019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ahmadi A, Khansarinejad B, Hosseinkhani S,
Ghanei M and Mowla SJ: miR-199a-5p and miR-495 target GRP78 within
UPR pathway of lung cancer. Gene. 620:15–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao G, Kang J, Xu G, Wei J, Wang X, Jing
X, Zhang L, Yang A, Wang K, Wang J, et al: Tunicamycin promotes
metastasis through upregulating endoplasmic reticulum stress
induced GRP78 expression in thyroid carcinoma. Cell Biosci.
10:1152020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chern YJ, Wong JCT, Cheng GSW, Yu A, Yin
Y, Schaeffer DF, Kennecke HF, Morin G and Tai IT: The interaction
between SPARC and GRP78 interferes with ER stress signaling and
potentiates apoptosis via PERK/eIF2α and IRE1α/XBP-1 in colorectal
cancer. Cell Death Dis. 10:5042019. View Article : Google Scholar
|
|
19
|
Luo C, Xiong H, Chen L, Liu X, Zou S, Guan
J and Wang K: GRP78 promotes hepatocellular carcinoma proliferation
by increasing FAT10 expression through the NF-κB pathway. Exp Cell
Res. 365:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Zhang Y, Lin F, Shi X, Xiang L
and Li L: Shh overexpression is correlated with GRP78 and AR
expression in primary prostate cancer: Clinicopathological features
and outcomes in a Chinese cohort. Cancer Manag Res. 12:1569–1578.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ogawa H, Kaira K, Takahashi K, Shimizu A,
Altan B, Yoshinari D, Asao T and Oyama T: Prognostic role of
BiP/GRP78 expression as ER stress in patients with gastric
adenocarcinoma. Cancer Biomark. 20:273–281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45(W1):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oughtred R, Rust J, Chang C, Breitkreutz
BJ, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, et
al: The BioGRID database: A comprehensive biomedical resource of
curated protein, genetic, and chemical interactions. Protein Sci.
30:187–200. 2021. View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
MacArthur Clark JA and Sun D: Guidelines
for the ethical review of laboratory animal welfare People's
Republic of China national standard GB/T 35892-2018[Issued 6
February 2018 effective from 1 September 2018. Animal Model Exp
Med. 3:103–113. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang S, Hong H, Li L, He D, Xu Z, Zuo S,
Han J, Wu Q, Dai Z, Cai W, et al: Plasminogen kringle 5 suppresses
gastric cancer via regulating HIF-1α and GRP78. Cell Death Dis.
8:e31442017. View Article : Google Scholar
|
|
28
|
Fu Z, Wang X, Zhou H, Li Y, Chen Y, Wang Z
and Liu L: GRP78 positively regulates estrogen-stimulated cell
growth mediated by ER-α36 in gastric cancer cells. Mol Med Rep.
16:8329–8334. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong Q, Zhang Z and Liang Z: Upregulating
miR-637 aggravates endoplasmic reticulum stress-induced apoptosis
in gastric cancer cells by suppressing calreticulin. Anim Cells
Syst (Seoul). 24:267–274. 2020. View Article : Google Scholar
|
|
30
|
You X, Wang Y, Wu J, Liu Q, Chen D, Tang D
and Wang D: Prognostic significance of galectin-1 and vasculogenic
mimicry in patients with gastric cancer. Onco Targets Ther.
11:3237–3244. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
You X, Wu J, Wang Y, Liu Q, Cheng Z, Zhao
X, Liu G, Huang C, Dai J, Zhou Y, et al: Galectin-1 promotes
vasculogenic mimicry in gastric adenocarcinoma via the Hedgehog/GLI
signaling pathway. Aging (Albany NY). 12:21837–21853. 2020.
View Article : Google Scholar
|
|
32
|
Nambiar DK, Aguilera T, Cao H, Kwok S,
Kong C, Bloomstein J, Wang Z, Rangan VS, Jiang D, von Eyben R, et
al: Galectin-1-driven T cell exclusion in the tumor endothelium
promotes immunotherapy resistance. J Clin Invest. 129:5553–5567.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He YS, Hu YQ, Xiang K, Chen Y, Feng YT,
Yin KJ, Huang JX, Wang J, Wu ZD, Wang GH and Pan HF: Therapeutic
potential of galectin-1 and galectin-3 in autoimmune diseases. Curr
Pharm Des. 28:36–45. 2022. View Article : Google Scholar
|
|
34
|
Arciniegas E, Carrillo LM, Rojas H, Pineda
J, Ramírez R, Reyes O, Chopite M and Rocheta A: Plump endothelial
cells integrated into pre-existing venules contribute to the
formation of 'mother' and 'daughter' vessels in pyogenic granuloma:
Possible role of galectin-1, -3 and -8. Scars Burn Heal.
7:20595131209866872021.PubMed/NCBI
|
|
35
|
Camby I, Belot N, Lefranc F, Sadeghi N, de
Launoit Y, Kaltner H, Musette S, Darro F, Danguy A, Salmon I, et
al: Galectin-1 modulates human glioblastoma cell migration into the
brain through modifications to the actin cytoskeleton and levels of
expression of small GTPases. J Neuropathol Exp Neurol. 61:585–596.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kovács-Sólyom F, Blaskó A, Fajka-Boja R,
Katona RL, Végh L, Novák J, Szebeni GJ, Krenács L, Uher F, Tubak V,
et al: Mechanism of tumor cell-induced T-cell apoptosis mediated by
galectin-1. Immunol Lett. 127:108–118. 2010. View Article : Google Scholar
|
|
37
|
Horiguchi N, Arimoto K, Mizutani A,
Endo-Ichikawa Y, Nakada H and Taketani S: Galectin-1 induces cell
adhesion to the extracellular matrix and apoptosis of non-adherent
human colon cancer Colo201 cells. J Biochem. 134:869–874. 2003.
View Article : Google Scholar
|
|
38
|
Kim HJ, Jeon HK, Cho YJ, Park YA, Choi JJ,
Do IG, Song SY, Lee YY, Choi CH, Kim TJ, et al: High galectin-1
expression correlates with poor prognosis and is involved in
epithelial ovarian cancer proliferation and invasion. Eur J Cancer.
48:1914–1921. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Saussez S, Cucu DR, Decaestecker C,
Chevalier D, Kaltner H, André S, Wacreniez A, Toubeau G, Camby I,
Gabius HJ and Kiss R: Galectin 7 (p53-induced gene 1): A new
prognostic predictor of recurrence and survival in stage IV
hypopharyngeal cancer. Ann Surg Oncol. 13:999–1009. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chiang WF, Liu SY, Fang LY, Lin CN, Wu MH,
Chen YC, Chen YL and Jin YT: Overexpression of galectin-1 at the
tumor invasion front is associated with poor prognosis in
early-stage oral squamous cell carcinoma. Oral Oncol. 44:325–334.
2008. View Article : Google Scholar
|
|
41
|
Gao Y, Li X, Shu Z, Zhang K, Xue X, Li W,
Hao Q, Wang Z, Zhang W, Wang S, et al: Nuclear galectin-1-FOXP3
interaction dampens the tumor-suppressive properties of FOXP3 in
breast cancer. Cell Death Dis. 9:4162018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Leung Z, Ko FCF, Tey SK, Kwong EML, Mao X,
Liu BHM, Ma APY, Fung YME, Che CM, Wong DKH, et al: Galectin-1
promotes hepatocellular carcinoma and the combined therapeutic
effect of OTX008 galectin-1 inhibitor and sorafenib in tumor cells.
J Exp Clin Cancer Res. 38:4232019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pranjol MZI, Zinovkin DA, Maskell ART,
Stephens LJ, Achinovich SL, Los' DM, Nadyrov EA, Hannemann M,
Gutowski NJ and Whatmore JL: Cathepsin L-induced galectin-1 may act
as a proangiogenic factor in the metastasis of high-grade serous
carcinoma. J Transl Med. 17:2162019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kucińska M, Porębska N, Lampart A, Latko
M, Knapik A, Zakrzewska M, Otlewski J and Opaliński Ł: Differential
regulation of fibroblast growth factor receptor 1 trafficking and
function by extracellular galectins. Cell Commun Signal. 17:652019.
View Article : Google Scholar
|
|
45
|
Kuo LJ, Hung CS, Chen WY, Chang YJ and Wei
PL: Glucose-regulated protein 78 silencing down-regulates vascular
endothelial growth factor/vascular endothelial growth factor
receptor 2 pathway to suppress human colon cancer tumor growth. J
Surg Res. 185:264–272. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chang YJ, Chen WY, Huang CY, Liu HH and
Wei PL: Glucose-regulated protein 78 (GRP78) regulates colon cancer
metastasis through EMT biomarkers and the NRF-2/HO-1 pathway.
Tumour Biol. 36:1859–1869. 2015. View Article : Google Scholar
|
|
47
|
Yuan XP, Dong M, Li X and Zhou JP: GRP78
promotes the invasion of pancreatic cancer cells by FAK and JNK.
Mol Cell Biochem. 398:55–62. 2015. View Article : Google Scholar
|
|
48
|
Yin Y, Chen C, Chen J, Zhan R, Zhang Q, Xu
X, Li D and Li M: Cell surface GRP78 facilitates hepatoma cells
proliferation and migration by activating IGF-IR. Cell Signal.
35:154–162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qian Y, Wong CC, Xu J, Chen H, Zhang Y,
Kang W, Wang H, Zhang L, Li W, Chu ESH, et al: Sodium channel
subunit SCNN1B suppresses gastric cancer growth and metastasis via
GRP78 degradation. Cancer Res. 77:1968–1982. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bastida-Ruiz D, Wuillemin C, Pederencino
A, Yaron M, Martinez de Tejada B, Pizzo SV and Cohen M: Activated
α2-macroglobulin binding to cell surface GRP78 induces
trophoblastic cell fusion. Sci Rep. 10:96662020. View Article : Google Scholar
|