Introduction
Thyroid carcinoma (TC) is the most frequent
malignant neoplasm of the endocrine system and its incidence rate
has been steadily increasing with an annual growth rate of 4.5-6.6%
across the world (1). In 2018
alone, according to the Surveillance, Epidemiology and End Results
Program from the National Cancer Institute, nearly 54,000 new cases
of TC were registered, accounting for 3.1% of all new cases of
cancer during this year so far (2).
Papillary TC (PTC) is the most common type of
thyroid malignancy, accounting for >80% of all thyroid cancers
(3), and an even higher proportion
of 95.1% was estimated in the Chinese population (4). Over the past decade, the diagnosis
level of PTC has markedly improved due to the wide use of
fine-needle aspiration cytology (FNAC) together with the detection
of B-Raf proto-oncogene, serine/threonine kinase
(BRAF)V600E mutation in clinical practice (5). FNAC is routinely used as the main tool
in the preoperative evaluation of thyroid nodules. However, ~15–30%
of FNA specimens were reported to give inconclusive results, which
are read as ‘indeterminate’ or ‘suspicious malignancy’, offering a
challenge in terms of interpretation and clinical management
(6). As for the
BRAFV600E mutation, despite a high specificity (1.00,
95% CI: 0.98-1.00) for PTC, BRAFV600E mutation has a low
overall sensitivity [(0.40, 95% CI: 0.32-0.48) or (0.60, 95% CI:
0.556-0.634)], limiting its diagnostic value as a single screening
test (7,8). Although most patients with PTC have an
excellent prognosis (9), the
prevalence of PTC still raises concern due to a recurrence rate of
almost 30% and cause-specific mortality of 8.6% for a three-decade
period (10). Therefore, it becomes
particularly urgent and necessary to screen novel tumor markers and
new therapeutic targets for PTC.
TRAF2- and NCK-interacting protein kinase (TNIK) is
one of the germinal center kinase family members, localized in
chromosome region 3q26, where gene amplification often occurs in
various cancers (11,12). Accumulating evidence suggested that
TNIK, as a protein interacting with transcription factor 4 (TCF4)
(13,14), is involved in extensive biochemical
pathways, such as the JNK, Wnt/β-catenin or PI3K/Akt pathways,
which are shared between embryogenesis and tumorigenesis (11,13,15).
TNIK is essential for the transactivation of Wnt signal target
genes (13,16) and its expression is associated with
poor prognosis of patients with hepatocellular (17), colorectal (18) and pancreatic (19) cancers. Amplification of the TNIK
gene is detectable in 7% of gastric cancers (11) and 50% of lung carcinomas (12), and TNIK is reportedly one of several
putative driver oncogenes (20).
Furthermore, TNIK-targeted treatments were revealed to be potential
therapies for colorectal cancer (21), synovial sarcoma (22), lung cancer (23), osteosarcoma (24), prostate cancer (25) and breast cancer (26). Therefore, in the present study, the
expression of TNIK and its activated form, phosphorylated (p)-TNIK,
were preliminary investigated and compared among PTC samples,
benign thyroid tumors and normal thyroid tissues, and the results
revealed that both TNIK and p-TNIK were upregulated in PTC compared
to benign tumors and normal tissues. The expression of p-TNIK was
positively associated with extrathyroidal extension in patients
with PTC.
Patients and methods
Public datasets
The expression pattern of TNIK in multiple cancer
types was downloaded from the Gene Expression Profiling Interactive
Analysis (GEPIA) dataset (http://gepia.cancer-pku.cn/). The expression of TNIK
in PTC tissues compared with that in normal tissues was downloaded
from the public The Cancer Genome Atlas (TCGA) dataset (https://ualcan.path.uab.edu/analysis.html).
Patients
Patients who were diagnosed with primary PTC and
underwent subtotal thyroidectomy or radical resection at the
Department of Otolaryngology Head and Neck Surgery at the Fourth
Hospital of Hebei Medical University (Shijiazhuang, China) between
June 2021 and June 2022 were enrolled in the present study. Samples
from patients who suffered from benign thyroid tumors and underwent
subtotal thyroidectomy during the same period were collected as
controls. Furthermore, isthmuses of the thyroid gland of patients
who underwent tracheotomy, without benign or malignant thyroid
tumors, between October 2015 and June 2022 were considered normal
thyroid tissues and included in the study.
The use of tissues for this study was approved by
the Research Ethics Committee of the Fourth Hospital of Hebei
Medical University (Shijiazhuang, China), and conformed to all
relevant ethical regulations for human research subjects in
accordance with the Declaration of Helsinki. Written informed
consent was obtained from all participants before any clinical
samples were collected. The excluded patients were breastfeeding
patients, those with other benign or malignant tumors, or severe
cardiovascular or renal diseases.
A total of 202 patients with PTC were assigned to
the mRNA analysis by reverse transcription-quantitative (RT-q)PCR
to compare the relative mRNA expression of TNIK in PTC tissue and
their matched adjacent tissue. Furthermore, 202 PTC tissues, 150
PTC-adjacent tissues, 100 benign thyroid tumor tissues accompanied
by PTC (termed as benign tumor A), 100 benign thyroid tumors not
accompanied by PTC (termed as benign tumor B) and 100 normal
thyroid tissues were subjected to immunohistochemistry (IHC) to
detect the TNIK and p-TNIK protein levels. For RT-qPCR analysis, a
strip of tumor tissue (without any adjacent tissue as much as
possible) and corresponding adjacent tissues (at least 1 cm apart
from the PTC tissue) were collected. Immediately after excision,
the tissue samples were stored in M5 HiPer RNA stay (Mei5bio) and
placed in a −80°C refrigerator. The paraffin-embedded specimens for
IHC analysis were produced by the Pathology Department of the
Fourth Hospital of Hebei Medical University (Shijiazhuang,
China).
RNA extraction and RT-qPCR
RNA was extracted from PTC tissues and corresponding
adjacent tissues using the Eastep® SuperTotal RNA
Extraction Kit (Promega Corp.) according to the manufacturer's
protocol. RNA concentration and quality were assessed using a
NanoDrop® One spectrophotometer (Thermo Fisher
Scientific, Inc.). The GoScript™ Reverse Transcription Mix (Promega
Corp.) was used to generate cDNA from RNA according to the
manufacturer's instructions. The amplification reaction was
performed using GoTaq® qPCR Master Mix (Promega Corp.)
according to the manufacturer's protocol. The qPCR procedure was as
follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 sec
and 60°C for 1 min. qPCR was performed using a QuantStudio DX
Real-Time PCR system (Thermo Fisher Scientific, Inc.) with each
reaction run in triplicate. The relative target gene mRNA
expression was determined using the comparative Ct method (27), with GAPDH as the endogenous control.
The primer sequences were as follows: TNIK forward,
5′-GGTAGAAGAACGGTCAAGGCTCAAC-3′ and reverse,
5′-GGCTGAACTCCACTAATGCTGAAGG-3′; GAPDH forward,
5′-AATCCCATCACCATCTTCCA-3′ and reverse,
5′-TGGACTCCACGACGTACTCA-3′.
Immunohistochemistry (IHC)
According to the manufacturer's instructions, IHC
staining was performed using the UltraSensitive TMSP (Rabbit) IHC
Kit (cat. no. PV-9001; OriGene Technologies, Inc.). First, the
antigens were retrieved by autoclaving them at 121°C for 2 min in
pH 6.0 citrate buffer (Fuzhou Maixin Biotech. Co., Ltd.). The
slides were then incubated in a solution of endogenous peroxidase
blocker (OriGene Technologies, Inc.) for 15 min at room temperature
in order to quench endogenous peroxidase activity. Thereafter, the
slides were incubated with a rabbit polyclonal antibody against
TNIK at 1:150 dilution (cat. no. ab224252; Abcam) or p-TNIK
(Ser764) at 1:300 dilution (cat. no. bs-5598R; BIOSS) overnight at
4°C. Next, the slides were incubated with reaction strengthening
fluid (OriGene Technologies, Inc.) for 20 min at 37°C, followed by
incubation with enhanced enzyme-labeled goat anti-rabbit IgG
polymer (cat. no. PV-9001; OriGene Technologies, Inc.) at 37°C for
20 min. Color development was performed with DAB (OriGene
Technologies, Inc.) for 5 min at room temperature. The slides were
counterstained with Harris hematoxylin, after which they were
dehydrated using a series of increasing alcohol concentrations, and
finally mounted with cover slips.
IHC staining was evaluated according to a previously
reported scoring method (28). All
slides were scored by three experienced pathologists blinded to the
clinical data. The staining intensity was scored as 0 (negative), 1
(weak), 2 (moderate) or 3 (strongly positive). The staining
extensity was scored as 0 (0–25% of the tumor cells stained), 1
(26–50%), 2 (51–75%) or 3 (76–100%). The sum of the intensity and
extensity scores, potentially ranging from 0 to 6, was calculated,
and the average of five fields (magnification, ×400) was used to
determine the TNIK and p-TNIK staining score for each patient.
Expression was classified as low when staining scores were ≤2 and
cases were defined as high when staining scores were ≥3. Cases were
defined as positive when the staining intensity was more than
weakly positive (weak + moderate + strong) and >10% of tumor
cells were positive.
Statistical analysis
All statistical analyses were performed using SPSS
version 21.0 software for Windows (IBM Corp.). Measurement data
were expressed as the mean ± standard deviation and analyzed by
Student's t-test. One-way ANOVA was performed for multiple
comparisons and post-hoc testing was used to compare two groups
among multiple comparisons. Matched tissue comparisons were
performed with a paired t-test, while comparisons between
non-matched samples were performed by two-samples t-tests. The
χ2 test was used to evaluate the association of
expression with the clinicopathological parameters. Fisher's exact
test was performed to replace the χ2 test in cases with
low numbers. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of TNIK and p-TNIK is
upregulated in PTC tissues
To explore the potential role of TNIK involved in
PTC tumorigenesis, the expression pattern of TNIK in multiple
cancer types from the GEPIA dataset was first analyzed. The results
indicated that TNIK was significantly upregulated in TC (THCA)
compared to normal tissues (Fig.
1A). Subsequently, the public TCGA dataset was analyzed and it
was also found that the mRNA expression of TNIK was markedly
increased in PTC tissues compared with that in normal tissues
(Fig. 1B). To further investigate
the relative expression of TNIK in PTC tissues and matched adjacent
tissues, the expression of TNIK in 202 paired PTC and adjacent
tissues was comparatively analyzed using RT-qPCR. The results
indicated that the relative mRNA expression of TNIK in PTC tissues
was 4.47±6.16, which was markedly higher compared with the relative
expression of mRNA in adjacent tissues 2.57±5.83 (P=0.0015)
(Fig. 1C). Next, IHC analysis was
performed to detect the protein levels of TNIK and p-TNIK in PTC
tissues, PTC adjacent tissues, benign thyroid tumors and normal
tissues. A total of 202 paired samples of fresh PTC and adjacent
tissues were obtained. However, not every adjacent tissue block was
sufficient to be made into a wax block, and in the subsequent IHC
analysis, 202 PTC and 150 adjacent tissue wax blocks were available
for the analysis of TNIK and p-TNIK. The IHC results indicated that
the level of TNIK and p-TNIK in PTC tissues was markedly elevated
compared with that in benign thyroid tumors A, benign thyroid
tumors B, adjacent tissue and normal tissues (Fig. 1D and E). There was no significant
difference in TNIK and p-TNIK between benign thyroid tumors A and
benign thyroid tumors B, suggesting that the elevated TNIK and
p-TNIK levels in PTC tissues did not implicate the adjacent benign
thyroid tumors. Similarly, there was no significant difference
between PTC adjacent tissue and normal tissues, indicating that the
upregulation of TNIK and p-TNIK in PTC tissue did not include the
adjacent tissue.
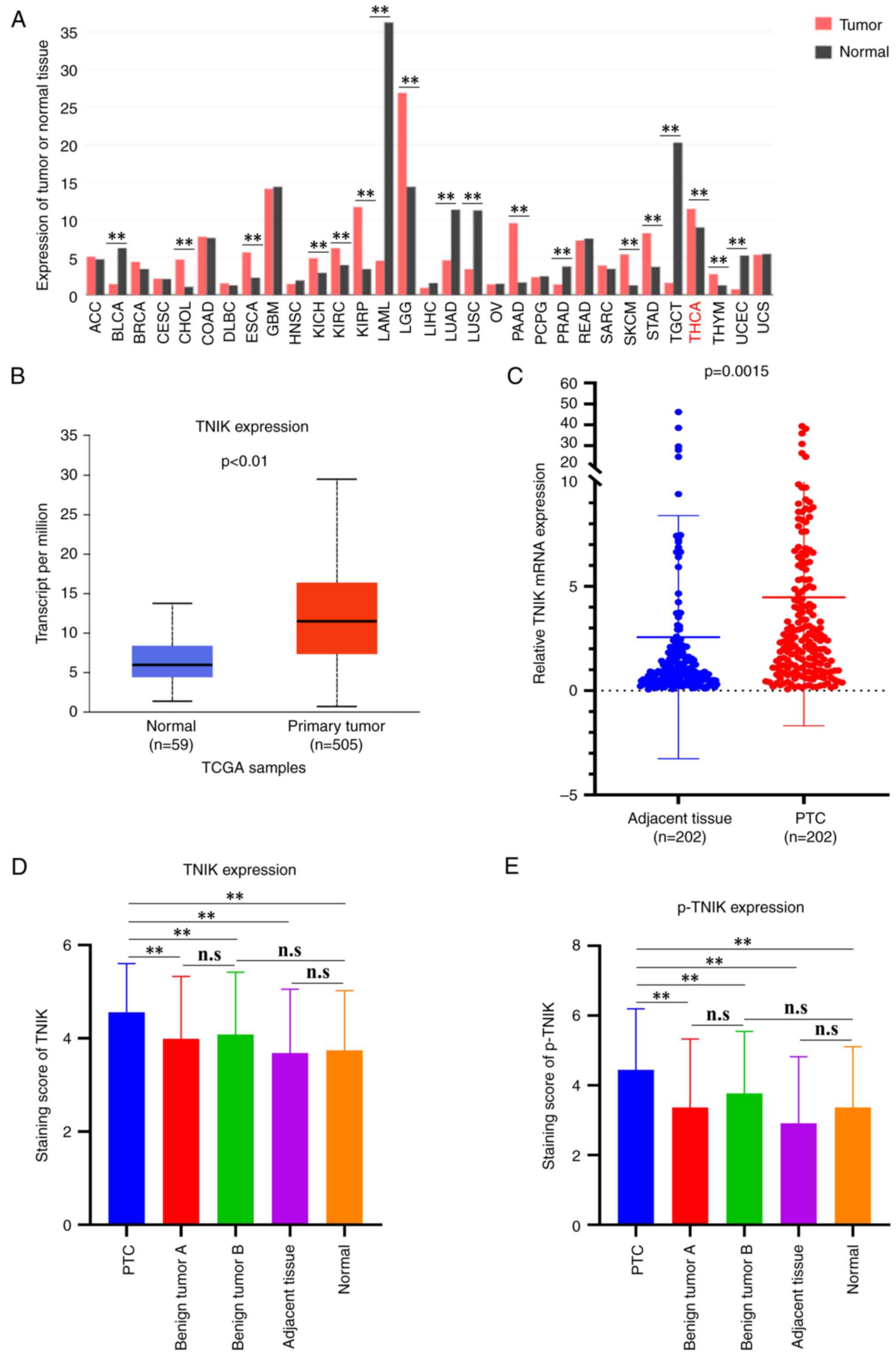 | Figure 1.TNIK and p-TNIK are upregulated in
PTC tissue. (A) The expression of TNIK mRNA in diverse cancers was
determined from GEPIA dataset. (B) The expression of TNIK mRNA in
PTC was determined from the TCGA dataset. (C) The mRNA expression
of TNIK was detected by reverse transcription-quantitative PCR in
202 pairs of PTC tissues and matched adjacent tissues. (D) The
protein expression of TNIK was analyzed by IHC in PTC tissues,
benign thyroid tumors, adjacent tissues and normal tissues. (E) The
protein levels of p-TNIK were analyzed by IHC in PTC tissues,
benign thyroid tumors, adjacent tissues and normal tissues.
**P<0.01. n.s., no significance; p-TNIK, phosphorylated TRAF2-
and NCK-interacting kinase; IHC, immunohistochemistry; TCGA, The
Cancer Genome Atlas; ACC, adrenocortical carcinoma; BLCA, bladder
urothelial carcinoma; BRCA, breast invasive carcinoma; CESC,
cervical squamous cell carcinoma and endocervical adenocarcinoma;
CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC,
lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal
carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck
squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney
renal clear cell carcinoma; KIRP, kidney renal papillary cell
carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade
glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian
serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma;
PTC, papillary thyroid carcinoma; READ, rectum adenocarcinoma;
SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach
adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid
carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial
carcinoma; UCS, uterine carcinosarcoma. |
Association of TNIK and p-TNIK levels
with clinicopathological characteristics of patients with PTC
To determine the clinical significance of TNIK and
p-TNIK in PTC, the relationship between TNIK or p-TNIK levels and
clinicopathological parameters was analyzed. The expression of TNIK
in patients with PTC was not associated with gender, age, tumor
size, multifocality, extrathyroidal extension, lymph node (LN)
metastasis or TNM stage (P>0.05). p-TNIK expression was more
frequently observed in the extrathyroidal extension group
(χ2=4.199, P=0.040), while there was no association
between p-TNIK and gender, age, tumor size, multifocality, LN
metastasis or TNM stage (P>0.05; Table I).
 | Table I.Association of TNIK and p-TNIK levels
with clinicopathological characteristics of patients with papillary
thyroid carcinoma. |
Table I.
Association of TNIK and p-TNIK levels
with clinicopathological characteristics of patients with papillary
thyroid carcinoma.
|
|
| TNIK |
|
| p-TNIK |
|
|---|
|
|
|
|
|
|
|
|
|---|
| Characteristic | n | High (n=168) | Low (n=34) | P-value | n | High (n=172) | Low (n=30) | P-value |
|---|
| Gender |
|
|
| 0.388 |
|
|
| 0.056 |
|
Male | 60 | 52 (86.7) | 8 (13.3) |
| 56 | 52 (92.9) | 4 (7.1) |
|
|
Female | 142 | 116 (81.7) | 26 (18.3) |
| 146 | 120 (82.2) | 26 (17.8) |
|
| Age, years |
|
|
| 0.083 |
|
|
| 0.579 |
|
<55 | 154 | 132 (85.7) | 22 (14.3) |
| 168 | 142 (84.5) | 26 (15.5) |
|
|
≥55 | 48 | 36 (75.0) | 12 (25.0) |
| 34 | 30 (88.2) | 4 (11.8) |
|
| Tumor size, cm |
|
|
| 0.846 |
|
|
| 0.380 |
|
<1 | 110 | 92 (83.6) | 18 (16.4) |
| 120 | 100 (83.3) | 20 (16.7) |
|
| ≥1 | 92 | 76 (82.6) | 16 (17.4) |
| 82 | 72 (87.8) | 10 (12.2) |
|
| Multifocality |
|
|
| 0.092 |
|
|
| 0.788 |
|
Single | 142 | 114 (80.3) | 28 (19.7) |
| 144 | 122 (84.7) | 22 (15.3) |
|
|
Multifocal | 60 | 54 (90.0) | 6 (10.0) |
| 58 | 50 (86.2) | 8 (13.8) |
|
| Extrathyroidal
extension |
|
|
| 0.381 |
|
|
| 0.040 |
|
Positive | 70 | 56 (80) | 14 (20) |
| 74 | 68 (91.9) | 6 (8.1) |
|
|
Negative | 132 | 112 (84.8) | 20 (15.2) |
| 128 | 104 (81.3) | 24 (18.7) |
|
| LN metastasis |
|
|
| 0.562 |
|
|
| 0.267 |
|
Positive | 86 | 70 (81.4) | 16 (18.6) |
| 86 | 76 (88.4) | 10 (11.6) |
|
|
Negative | 116 | 98 (84.5) | 18 (15.5) |
| 116 | 96 (82.8) | 20 (17.2) |
|
| TNM stage |
|
|
| 0.193 |
|
|
| 1.000 |
| I | 184 | 155 (84.2) | 29 (15.8) |
| 186 | 158 (84.9) | 28 (15.1) |
|
|
II/III/IV | 18 | 13 (72.2) | 5 (27.8) |
| 16 | 14 (87.5) | 2 (12.5) |
|
Levels of TNIK and p-TNIK in PTC
Based on the data above, there was no significant
difference in the levels of TNIK and p-TNIK between benign thyroid
tumors A and benign thyroid tumors B, as well as between PTC
adjacent tissue and normal tissues. In the following analysis,
benign thyroid tumors B were selected as benign tumors, and in
addition, normal tissues were used for further investigation.
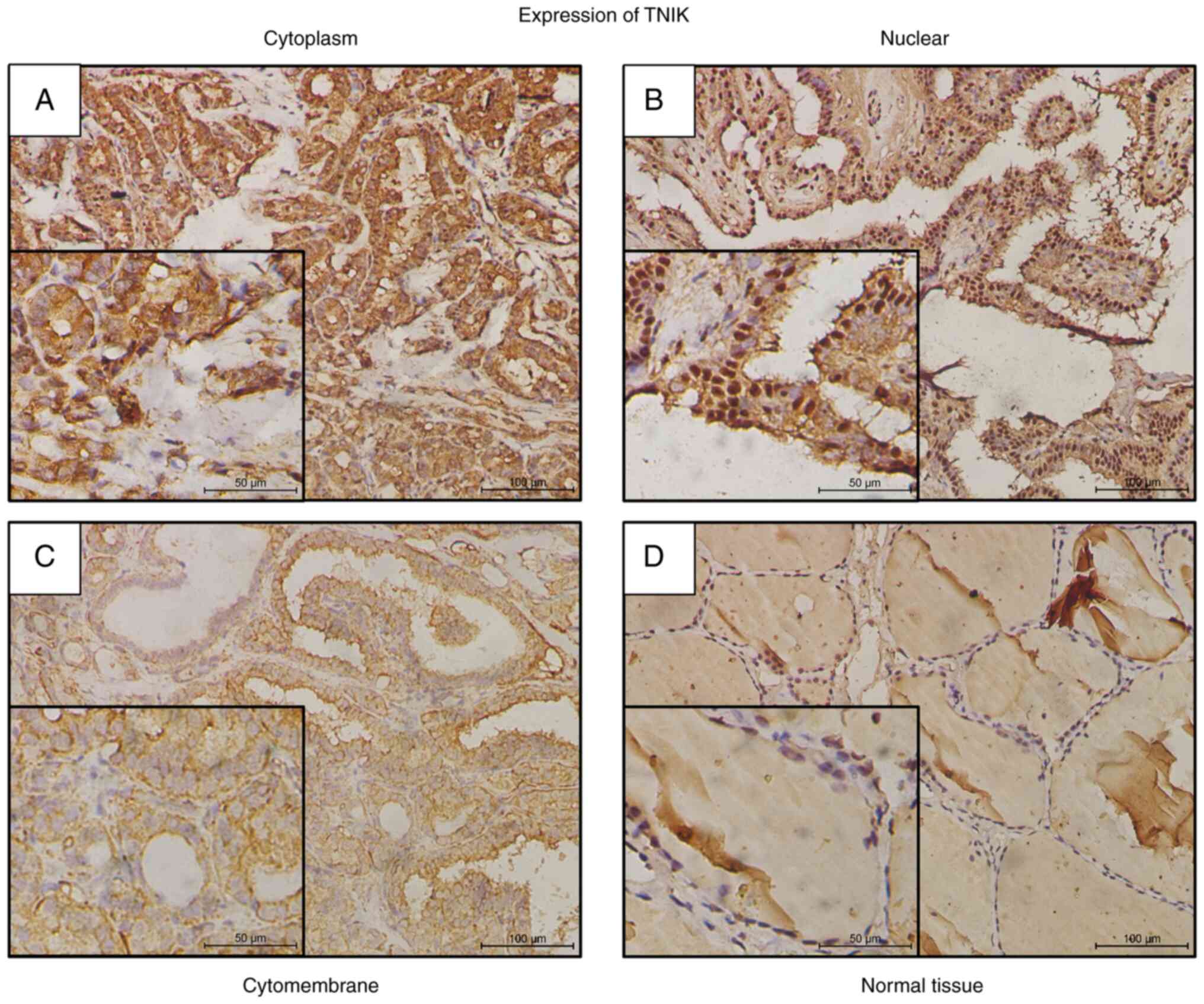
TNIK-positive staining was observed in 187 out of 202 (92.6%) cases
in the cytoplasm, nuclei or cytomembrane of PTC cells. In the 187
positive cases, cytoplasm expression was identified in 162 cases
(86.6%), nuclear expression in 17 cases (9.1%) and cytomembrane
expression in 8 cases (4.3%) (Table
II, Fig. 2). There was no
significant difference in expression location distribution among
PTC tissue, benign thyroid tumors and normal tissue (Table II).
 | Table II.Positive expression percentage of
TNIK in cytoplasm, nuclei and cytomembrane of different
samples. |
Table II.
Positive expression percentage of
TNIK in cytoplasm, nuclei and cytomembrane of different
samples.
|
|
| TNIK
expression |
|
|---|
| Sample type | n | Cytoplasm | Nuclear | Cytomembrane | P-value |
|---|
| PTC | 187 | 162 (86.6) | 17 (9.1) | 8 (4.3) | 0.883 |
| Benign tumor | 89 | 76 (85.4) | 9 (10.1) | 4 (4.5) |
|
| Normal | 83 | 75 (90.4) | 6 (7.2) | 2 (2.4) |
|
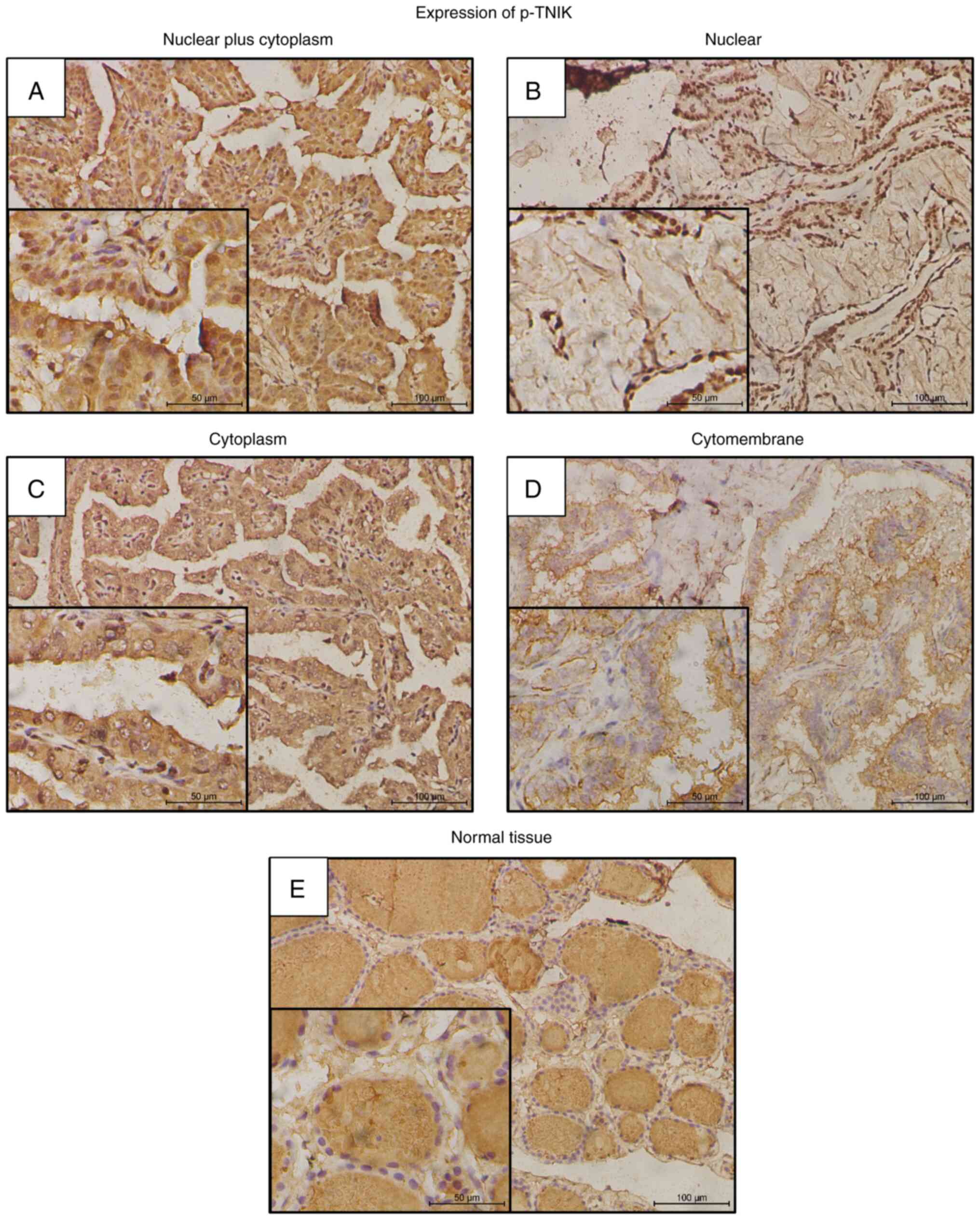
p-TNIK-positive staining was observed in 179 out of
202 (88.6%) cases in the nuclei, cytoplasm or cytomembrane of PTC
cells. In the 179 p-TNIK-positive cases, nuclear plus cytoplasm
expression was identified in 142 cases (79.3%), nuclear expression
in 9 cases (5.0%), cytoplasm expression in 21 cases (11.7%) and
cytomembrane expression in 7 cases (3.9%) (Table III, Fig. 3). Specifically, the most positive
p-TNIK expression pattern was strong staining in nuclei plus weak
or strong staining in the cytoplasm. In comparison, in benign
thyroid tumors and normal tissue, the main positive expression was
located in the nuclei (Table
III).
 | Table III.Percentage of positivity for p-TNIK
in nuclei, cytoplasm and cytomembrane of different samples. |
Table III.
Percentage of positivity for p-TNIK
in nuclei, cytoplasm and cytomembrane of different samples.
|
|
| Positivity for
p-TNIK |
|
|---|
|
|
|
|
|
|---|
| Sample type | n | Nuclear plus
cytoplasm | Nuclear | Cytoplasm | Cytomembrane | P-value |
|---|
| PTC | 179 | 142 (79.3) | 9 (5.0) | 21 (11.8) | 7 (3.9) |
|
| Benign tumor | 81 | 0 (0) | 69 (85.2) | 7 (8.6) | 5 (6.2) | <0.001 |
| Normal | 85 | 0 (0) | 73 (85.9) | 9 (10.6) | 3 (3.5) |
|
Comparison of sensitivity and
specificity among BRAFV600E mutation, TNIK expression
and p-TNIK levels in PTC diagnosis
The sensitivity and specificity of
BRAFV600E mutation (routinely detected by the Molecular
Cell Diagnostic Center of our institution) in the patients enrolled
in the present study were 68.6 and 89.7%, respectively (Table IV), which indicated a higher
sensitivity and a lower specificity compared to prior studies
(7,8). As presented in Tables V and VI, the sensitivity and specificity of
TNIK and p-TNIK in the diagnosis of PTC were 92.6 and 11.0%,
respectively, as well as 88.6 and 19.0%, respectively, affirming
that both TNIK and p-TNIK have high sensitivity and poor
specificity. In spite of the desirable sensitivity compared to
BRAFV600E mutation, in view of the unsatisfactory
specificity, TNIK and p-TNIK may be regarded as oncogenes, but not
be used as indicators for PTC diagnosis.
 | Table IV.Specificity and sensitivity of
BRAFV600E mutation in PTC diagnosis. |
Table IV.
Specificity and sensitivity of
BRAFV600E mutation in PTC diagnosis.
|
| Pathologic
diagnosis of PTC |
|---|
|
|
|
|---|
|
BRAFV600E | + | - |
|---|
| + | 107 (68.6)
(sensitivity) | 7 (10.3) |
| - | 49 (31.4) | 61 (89.7)
(specificity) |
 | Table V.Specificity and sensitivity of TNIK
expression in PTC diagnosis. |
Table V.
Specificity and sensitivity of TNIK
expression in PTC diagnosis.
|
| Pathologic
diagnosis of PTC |
|---|
| TNIK
expression |
|
|---|
| + | - |
|---|
| + | 187 (92.6)
(sensitivity) | 89 (89.0) |
| - | 15 (7.4) | 11 (11.0)
(specificity) |
 | Table VI.Specificity and sensitivity of p-TNIK
positivity in PTC diagnosis. |
Table VI.
Specificity and sensitivity of p-TNIK
positivity in PTC diagnosis.
|
| Pathologic
diagnosis of PTC |
|---|
| p-TNIK status |
|
|---|
| + | - |
|---|
| + | 179 (88.6)
(sensitivity) | 81 (81.0) |
| - | 23 (11.4) | 19 (19.0)
(specificity) |
Discussion
The present study was the first, to the best of our
knowledge, to determine the protein levels of TNIK and p-TNIK in
PTC clinical tissue samples. The results indicated that TNIK and
p-TNIK were significantly elevated in PTC tissues compared to
benign thyroid tumors and normal tissues, and p-TNIK was
significantly associated with extrathyroidal extension, while the
expression of TNIK did not exhibit any association with any of the
clinicopathological parameters of the patients with PTC, which
explained that p-TNIK, as an active form of TNIK, may have a
crucial role in regulating transcriptional activity in PTC. IHC
analysis then indicated that the expression of TNIK was mainly
located in the cytoplasm, while the location of p-TNIK was in the
nuclei and cytoplasm. Strikingly, to sum up, the present study
suggested that p-TNIK/TNIK may be considered an oncogene to
participate in the carcinogenesis of PTC.
Multifocality in PTC is common and has been
considered a significant risk factor for disease progression and
risk of recurrence in PTC (29).
Therefore, in the present study, to verify whether benign thyroid
tumors are implicated in the accompanied PTC, the relative
expression of TNIK and levels of p-TNIK were detected and compared
in benign thyroid tumors accompanied by PTC and benign thyroid
tumors not accompanied by PTC. The results indicated that there was
no significant difference in TNIK and p-TNIK expression between
benign thyroid tumors from the two different groups, which
suggested that the elevated TNIK and p-TNIK expression in PTC
tissue did not include the adjacent benign thyroid tumors.
Similarly, there was no significant difference between PTC adjacent
tissue and normal tissues, indicating that the upregulated TNIK and
p-TNIK in PTC tissues did not extend to the adjacent tissues.
The phosphorylation and dephosphorylation of
proteins on serine residues are essential for regulating a broad
range of cellular functions in eukaryotes, including cell division,
homeostasis and apoptosis (30,31).
The serine 764 (S764) residue of human TNIK has been identified as
a phosphorylation site by liquid chromatography tandem mass
spectrometry-based random sequencing of protein kinases (32). P-TNIK then translocates into the
nucleus and augments the transcriptional activity of TCF4. A study
reported that TNIK-positive staining was detected in 92.7% of
hepatocellular carcinomas in the cytoplasm and p-TNIK expression
was identified in the cytoplasm of 55.6% and nuclei of 7.9% of
samples (17). A study on
colorectal cancer suggested that TNIK protein was observed in the
cytoplasm of cancer cells (18) and
TNIK protein was distributed along the filamentous cytoskeleton,
whereas p-TNIK was detected mainly in the nuclei and colocalized
with the TCF4/β-catenin complex (16). This was almost coincident with our
finding that TNIK-positive staining was mainly located in the
cytoplasm and p-TNIK-positive staining was present in the nuclei
plus cytoplasm. A noteworthy finding was that the most positive
p-TNIK expression pattern in PTC was strong staining in nuclear
plus weak or strong staining in the cytoplasm. However, in benign
thyroid tumors and normal tissue, the positive expression of p-TNIK
was mainly located in the nuclei. As is well known, human cancers
are a heterogeneous disease and multiple cancer genes consist of
all genetic alterations that modify the normal DNA/mRNA sequences
triggering a cataract of molecular reactions (33). Therefore, it was speculated that in
the tumorigenesis of PTC, one or more genes hinder the transfer of
p-TNIK from the cytoplasm to the nucleus, and this key finding may
help uncover the mechanism of PTC. Furthermore, at least, in the
present study, this different location of p-TNIK may help
distinguish PTC from benign tumors.
Emerging evidence suggested that TNIK as an oncogene
is involved in the progression of gastric cancer, lung cancer,
pancreatic cancer, colorectal cancer, synovial sarcoma,
osteosarcoma, prostate cancer and breast cancer (11,12,19,21–26),
while p-TNIK expression was observed to be increased in
hepatocellular carcinoma and ERG-positive prostate cancer and
associated with poor prognosis (17,25).
Based on this, it was speculated that TNIK may be an oncogene that
participates in PTC tumorigenesis. In the current first study of
TNIK expression in PTC, both TNIK and p-TNIK were found to be
upregulated in PTC and p-TNIK was more frequently observed in
extrathyroidal extension, while the expression of TNIK did not
exhibit any association with any clinicopathological parameters of
patients with PTC. When TNIK is activated, the resulting p-TNIK is
upregulated and the tumor cells detach and disseminate, leading to
metastasis (17). It was indicated
that high levels of p-TNIK were coincident with tumor progression,
which suggests that p-TNIK may serve as a tumor activator in PTC.
Taken together, the present study suggests that high levels of
p-TNIK may function as an oncogene and have an important role in
the progression of PTC.
BRAFV600E mutation, as one of the most
common mutations, is frequently present in thyroid cancer (34,35).
Of note, the BRAFV600E mutation in thyroid cancer occurs
in ~50% of PTC and PTC-derived anaplastic TC cases, but rarely
occurs in follicular TC or other types of thyroid tumor (36). The data of the present study
indicated a higher sensitivity and a lower specificity of
BRAFV600E mutation compared to prior studies, which
perhaps resulted from the limited sample number. In spite of the
desirable sensitivity compared to BRAFV600E mutation,
TNIK and p-TNIK were impractical to be adopted as diagnostic
indicators for PTC due to the poor specificity. However, the
present work provides important insight into TNIK or p-TNIK serving
as a novel biomarker, and the distinct differences in expression
location of p-TNIK between PTC and benign tumor may contribute to
PTC diagnosis.
Besides as an oncogene, another research focus on
TNIK is its utility as a target molecule for anti-cancer treatment.
TNIK has recently been considered a first-in-class anti-cancer
target molecule to regulate the Wnt signaling pathway. Previous
studies have proved that the small-molecule TNIK inhibitor NCB-0846
suppressed tumorigenesis, epithelial-to-mesenchymal transition,
cell viability, colony formation and apoptotic cell death in
vitro and induced regression of xenografts or abolished cancer
metastasis in in vivo models (21–25).
108600, as a novel TNIK inhibitor, was confirmed to suppress the
growth and colony- and mammosphere-forming capacity of breast
cancer stem cell-like cells, induce apoptosis and overcome
chemotherapy resistance in mice bearing triple-negative tumors
(26).
In conclusion, both TNIK and p-TNIK were upregulated
in PTC tissues, p-TNIK was significantly associated with
extrathyroidal extension and both TNIK and p-TNIK may function as
an oncogene to participate in the carcinogenesis and progression of
PTC. Subsequent work will follow up the patients of the present
study to explore the prognostic significance of TNIK and p-TNIK.
Furthermore, in vitro and in vivo studies will be
performed to elucidate the biological role and potential mechanism
of TNIK, with the purpose of clarifying whether TNIK affects the
biological behavior of PTC. In addition, whether TNIK functions as
a critical oncogene in PTC through Wnt/β-catenin or other
biochemical pathways deserves further investigation and further
experiments should be performed to determine whether TNIK
inhibition may serve as a promising therapeutic approach for
patients with PTC.
Acknowledgements
The authors would like to thank Professor Yueping
Liu and Ms. Fei Lv (Pathology Department, The Fourth Hospital of
Hebei Medical University, Shijiazhuang, China) for their technical
assistance with the preparation of paraffin-embedded samples.
Funding
The present study was supported by the Beijing-Tianjin-Hebei
Integration Project (grant no. J200004).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL: Execution of the majority of the experiments and
manuscript writing. LL, YX and SL: Obtaining the tissue samples
from the patients. ML, GH and ZW: Collection of clinicopathological
data of patients. GW, YZ and JS: Statistical analysis of data. JW
and YS: Extraction of RNA and execution of qPCR. RZ: Conception and
design of the study, final review and supervision. GW and YZ
confirm the authenticity of all the raw data. All authors have read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the Fourth Hospital of Hebei
Medical University (Shijiazhuang, China) approved this study
(approval no. 2018MEC106).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Stat Facts, . Thyroid Cancer
website. Available. https://seer.cancer.gov/statfacts/html/thyro.htmlSeptember
20–2018
|
|
3
|
Fagin JA and Wells SA Jr: Biologic and
clinical perspectives on thyroid cancer. N Engl J Med.
375:1054–1067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao L, Pang P, Zang L, Luo Y, Wang F,
Yang G, Du J, Wang X, Lyu Z, Dou J and Mu Y: Features and trends of
thyroid cancer in patients with thyroidectomies in Beijing, China
between 1994 and 2015: A retrospective study. BMJ Open.
9:e0233342019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haugen BR, Alexander EK, Bible KC, Doherty
GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM,
Schlumberger M, et al: 2015 American thyroid association management
guidelines for adult patients with thyroid nodules and
differentiated thyroid cancer: The American thyroid association
guidelines task force on thyroid nodules and differentiated thyroid
cancer. Thyroid. 26:1–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cibas ES and Ali SZ: The 2017 bethesda
system for reporting thyroid cytopathology. Thyroid. 27:1341–1346.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jinih M, Foley N, Osho O, Houlihan L, Toor
AA, Khan JZ, Achakzai AA and Redmond HP: BRAFV600E
mutation as a predictor of thyroid malignancy in indeterminate
nodules: A systematic review and meta-analysis. Eur J Surg Oncol.
43:1219–1227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jia Y, Yu Y, Li X, Wei S, Zheng X, Yang X,
Zhao J, Xia T and Gao M: Diagnostic value of B-RAF(V600E) in
difficult-to-diagnose thyroid nodules using fine-needle aspiration:
Systematic review and meta-analysis. Diagn Cytopathol. 42:94–101.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ito Y, Miyauchi A, Inoue H, Fukushima M,
Kihara M, Higashiyama T, Tomoda C, Takamura Y, Kobayashi K and Miya
A: An observational trial for papillary thyroid microcarcinoma in
Japanese patients. World J Surg. 34:28–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dong W, Horiuchi K, Tokumitsu H, Sakamoto
A, Noguchi E, Ueda Y and Okamoto T: Time-varying pattern of
mortality and recurrence from papillary thyroid cancer: Lessons
from a long-term follow-up. Thyroid. 29:802–808. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu DH, Zhang X, Wang H, Zhang L, Chen H,
Hu M, Dong Z, Zhu G, Qian Z, Fan J, et al: The essential role of
TNIK gene amplification in gastric cancer growth. Oncogenesis.
2:e892014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Torres-Ayuso P, An E, Nyswaner KM, Bensen
RC, Ritt DA, Specht SI, Das S, Andresson T, Cachau RE, Liang RJ, et
al: TNIK is a therapeutic target in lung squamous cell carcinoma
and regulates FAK activation through merlin. Cancer Discov.
11:1411–1423. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mahmoudi T, Li VS, Ng SS, Taouatas N,
Vries RG, Mohammed S, Heck AJ and Clevers H: The kinase TNIK is an
essential activator of Wnt target genes. EMBO J. 28:3329–3340.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shitashige M, Satow R, Honda K, Ono M,
Hirohashi S and Yamada T: Regulation of Wnt signaling by the
nuclear pore complex. Gastroenterology. 134:1961–1971. 1971.e1–e4.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gui J, Yang B, Wu J and Zhou X: Enormous
influence of TNIK knockdown on intracellular signals and cell
survival. Hum Cell. 24:121–126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shitashige M, Satow R, Jigami T, Aoki K,
Honda K, Shibata T, Ono M, Hirohashi S and Yamada T: Traf2- and
Nck-interacting kinase is essential for Wnt signaling and
colorectal cancer growth. Cancer Res. 70:5024–5033. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin J, Jung HY, Wang Y, Xie J, Yeom YI,
Jang JJ and Lee KB: Nuclear expression of phosphorylated TRAF2- and
NCK-interacting kinase in hepatocellular carcinoma is associated
with poor prognosis. Pathol Res Pract. 210:621–627. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takahashi H, Ishikawa T, Ishiguro M,
Okazaki S, Mogushi K, Kobayashi H, Iida S, Mizushima H, Tanaka H,
Uetake H and Sugihara K: Prognostic significance of Traf2- and
Nck-interacting kinase (TNIK) in colorectal cancer. BMC Cancer.
15:7942015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Jiang H, Qin M, Su X, Cao Z and
Wang J: TNIK serves as a novel biomarker associated with poor
prognosis in patients with pancreatic cancer. Tumour Biol.
37:1035–1040. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fleuren EDG, Zhang L, Wu J and Daly RJ:
The kinome ‘at large’ in cancer. Nat Rev Cancer. 16:83–98. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Masuda M, Uno Y, Ohbayashi N, Ohata H,
Mimata A, Kukimoto-Niino M, Moriyama H, Kashimoto S, Inoue T, Goto
N, et al: TNIK inhibition abrogates colorectal cancer stemness. Nat
Commun. 7:125862016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sekita T, Yamada T, Kobayashi E, Yoshida
A, Hirozane T, Kawai A, Uno Y, Moriyama H, Sawa M, Nagakawa Y, et
al: Feasibility of targeting Traf2-and-Nck-interacting kinase in
synovial sarcoma. Cancers (Basel). 12:12582020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sugano T, Masuda M, Takeshita F, Motoi N,
Hirozane T, Goto N, Kashimoto S, Uno Y, Moriyama H, Sawa M, et al:
Pharmacological blockage of transforming growth factor-β signalling
by a Traf2- and Nck-interacting kinase inhibitor, NCB-0846. Br J
Cancer. 124:228–236. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirozane T, Masuda M, Sugano T, Sekita T,
Goto N, Aoyama T, Sakagami T, Uno Y, Moriyama H, Sawa M, et al:
Direct conversion of osteosarcoma to adipocytes by targeting TNIK.
JCI Insight. 6:e1372452021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee RS, Zhang L, Berger A, Lawrence MG,
Song J, Niranjan B, Davies RG, Lister NL, Sandhu SK, Rubin MA, et
al: Characterization of the ERG-regulated kinome in prostate cancer
identifies TNIK as a potential therapeutic target. Neoplasia.
21:389–400. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sato K, Padgaonkar AA, Baker SJ, Cosenza
SC, Rechkoblit O, Subbaiah DRCV, Domingo-Domenech J, Bartkowski A,
Port ER, Aggarwal AK, et al: Simultaneous CK2/TNIK/DYRK1 inhibition
by 108600 suppresses triple negative breast cancer stem cells and
chemotherapy-resistant disease. Nat Commun. 12:46712021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yokoyama Y, Sato S, Futagami M, Fukushi Y,
Sakamoto T, Umemoto M and Saito Y: Prognostic significance of
vascular endothelial growth factor and its receptors in endometrial
carcinoma. Gynecol Oncol. 77:413–418. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Joseph KR, Edirimanne S and Eslick GD:
Multifocality as a prognostic factor in thyroid cancer: A
meta-analysis. Int J Surg. 50:121–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hanks SK, Quinn AM and Hunter T: The
protein kinase family: Conserved features and deduced phylogeny of
the catalytic domains. Science. 241:42–52. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hunter T: Protein kinase classification.
Methods Enzymol. 200:3–37. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wissing J, Jänsch L, Nimtz M, Dieterich G,
Hornberger R, Kéri G, Wehland J and Daub H: Proteomics analysis of
protein kinases by target class-selective prefractionation and
tandem mass spectrometry. Mol Cell Proteomics. 6:537–547. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu R, Gao Q, Foltz SM, Fowles JS, Yao L,
Wang JT, Cao S, Sun H, Wendl MC, Sethuraman S, et al: Co-evolution
of tumor and immune cells during progression of multiple myeloma.
Nat Commun. 12:25592021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fukushima T, Suzuki S, Mashiko M, Ohtake
T, Endo Y, Takebayashi Y, Sekikawa K, Hagiwara K and Takenoshita S:
BRAF mutations in papillary carcinomas of the thyroid. Oncogene.
22:6455–6457. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kimura ET, Nikiforova MN, Zhu Z, Knauf JA,
Nikiforov YE and Fagin JA: High prevalence of BRAF mutations in
thyroid cancer: Genetic evidence for constitutive activation of the
RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma.
Cancer Res. 63:1454–1457. 2003.PubMed/NCBI
|
|
36
|
Xing M: BRAF mutation in thyroid cancer.
Endocr Relat Cancer. 12:245–262. 2005. View Article : Google Scholar : PubMed/NCBI
|