1. Introduction
Cell death, survival, proliferation and
differentiation represent fundamental processes of life. Cell death
plays a pivotal role in embryonic development, maintaining the
homeostasis of the organism and eliminating damaged cells. Cell
death was initially divided into three types (1): Type I cell death (apoptosis), type
II cell death (autophagy) and type III cell death (necrosis). In
recent years, multiple novel cell death modalities have been
identified and characterized concerning their corresponding
stimuli, molecular mechanisms and morphologies. Some of these
modalities share overlapping, but not identical signal pathways and
fail to be incorporated into the type I-III categories. In 2018,
the Nomenclature Committee on Cell Death listed multiple cell death
types in a molecule-oriented manner (2). Tang et al also provided
historical origins of items used during cell death research
development and a brief summary of molecular machinery involved in
regulated cell death (3).
However, the hierarchical association among different cell death
types remained vague and the molecular interplays led to further
confusion. Therefore, the present review article aims to provide a
simpler classification system and key features of different cell
death modalities are abstracted.
Cell death entities can be categorized into
programmed or non-programmed cell death based on their signal
dependency (Fig. 1). Programmed
cell death (PCD) is driven by tightly regulated intracellular
signal transduction pathways. By contrast, accidental cell death is
referred to as non-PCD as a result of unexpected cell injury. Given
the morphological characteristics and molecular mechanisms, PCD can
be further categorized into apoptotic cell death and non-apoptotic
cell death. Apoptosis retains cell membrane integrity and occurs in
a caspase-dependent manner. By contrast, non-apoptotic cell death
is mostly characterized by membrane rupture and
caspase-independency. For simplicity, the present review article
focuses on the key features of the diverse cell death modes and
their assessment methods commonly utilized in research (Table I), and refers the reader to
specialized recent review articles describing the processes of each
cell death mode in further detail (4-15).
 | Table ICell death modalities, their features
and common detection methods. |
Table I
Cell death modalities, their features
and common detection methods.
| Classification | Cell death
modality | Key molecules | Key morphology | Detection
methods |
|---|
| Non-PCD | Necrosis | None | Cell swelling;
membrane rupture; loss of organelle | Lactate
dehydrogenase activity detection; visualizing membrane integrity
loss by cell-impermeable DNA binding dye |
| PCD-apoptotic |
Apoptosis/anoikis | DRs and their
ligands, Bax, Bak, AIF, caspase-8, caspase-3, caspase-9 | Cell shrinkage;
membrane blebbing; loss of positional organization of organelles in
the cytoplasm; DNA condensation and | Chromosome
condensation detection; TUNEL assay; Annexin V assay; caspase
assay; PARP cleavage assay; applying apoptosis inhibitors
fragmentation; nuclear membrane rupture |
| PCD-vacuole
presenting | Autophagy | UKL1, PI3KIII,
ATGs, LC3 | Large intracellular
vesicles; membrane blebbing; enlarged organelles; depletion of
cytoplasmic organelles | Turnover of
long-lived proteins; LDH sequestration; western blot analysis with
autophagy specific antibodies |
| | Entosis | RhoA, ROCKI/II,
E-cadherin, α-catenin, actomyosin, LC3, ATGs | Cell-in-cell
formation | Morphology
observation with fluorescence imaging and electron microscopy |
| | Methuosis | Ras, Rac1, Arf6,
LAMP1, Rab7 | Accumulation of
large fluid-filled single membrane vacuoles; cell swelling;
membrane rupture | Morphology
observation with electron microscopy |
| | Paraptosis | Unclear | Accumulation of
large fluid-filled single membrane vacuoles; dilation of ER or
mitochondria | Morphology
observation with electron microscopy |
|
PCD-mitochondriadependent | Mitoptosis | Bax, Bak,
TIMM8a(DDP), Drp1 | Mitochondria
disappearance; decomposition of the mitochondrial reticulum to
small spherical organelles | Morphology
observation with fluorescence microscopy and electron microscopy;
western blot analysis with mitoptosis-specific antibodies |
| | Parthanatos | PARP, AIF | Membrane rupture;
mitochondrial outer membrane permeabilization; chromatin
condensation; DNA large-scale fragmentation | Western blot
analysis with parthanatos specific antibodies; Mitochondrial
depolarization detection with fluorescent probe |
| PCD-iron
dependent | Ferroptosis | System XC−, GPX4,
Lipid ROS | Diminutive
mitochondria with decreased cristae and collapsed and ruptured
membrane | Applying
ferroptosis inhibitors; measuring lipid peroxides e.g.
malondialhyde and 4-hydroxynonenal quantification |
| PCD-immune
reactive | Pyroptosis | NLRs, ALRs,
caspase-1, caspase-11 | Cell swelling;
membrane rupture; DNA condensation and fragmentation | Quantification of
cytoplasmic LDH; visualizing membrane integrity loss by
fluorescence microscopy; western blot analysis with
pyroptosis-specific antibodies |
| | NETosis | NOX4, PAD4 | Chromatin
decondensation; membrane rupture | Morphology
observation with fluorescence microscopy; free-cell DNA and
DNA-neutrophil derived protein complex detection with fluorescent
probe and immunoblot |
| Other type | Necroptosis | DRs, TLRs, TCR,
RIPKs, MLMK | Cell swelling;
membrane rupture; loss of organelle; mitochondria swelling | Visualizing
membrane integrity loss; mitochondrial depolarization detection;
applying necroptosis specific inhibitors; western blot analysis
with necroptosis-specific antibodies |
2. Non-programmed cell death
Non-programmed necrosis
Non-programmed necrosis is stimulated by a number of
external factors, e.g., infection, toxins and physical injury,
which lead to morphological alterations, such as cytoplasmic
swelling [oncosis, pre-lethal phase caused by the disruption of
ionic pumps such as Ca+ influx (16)], plasma membrane rupture and the
subsequent loss of intracellular organelles without severe
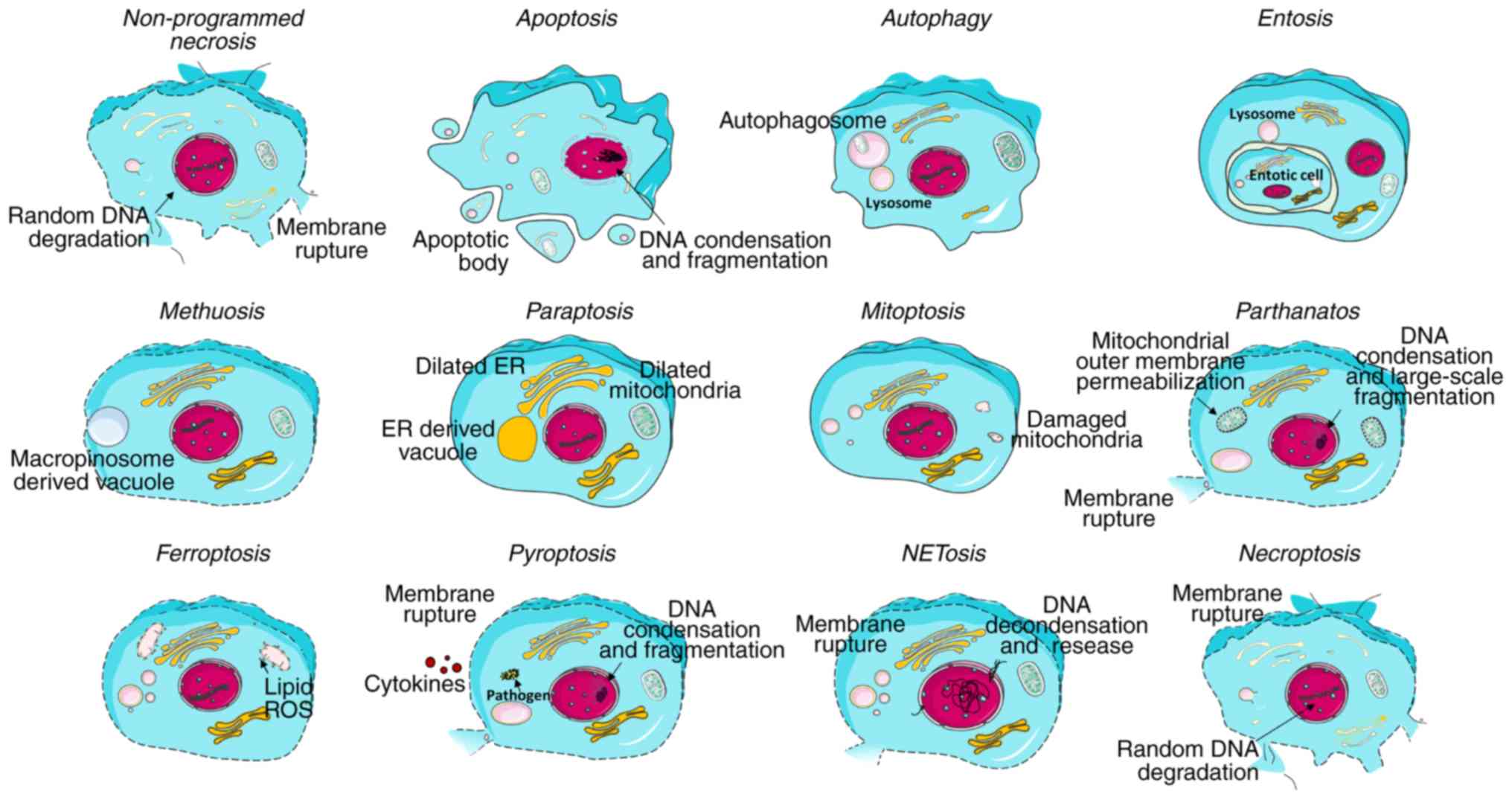
chromatin condensation, but randomly degraded DNA (17) (Fig.
2). Non-programmed necrosis is often observed in ischemia,
trauma and possibly some forms of neurodegeneration. It is commonly
considered as a passive process, which does not require de
novo macromolecular synthesis, but minimal energy (4).
Based on the morphological features of necrosis, a
number of methods, including lactate dehydrogenase (LDH) activity
detection and cell-impermeable DNA binding dye, are commonly used
to certify the cellular leakage and membrane permeability (Table I).
3. Programmed apoptotic cell death
Apoptosis
Apoptosis involves a series of tightly controlled
events and is characterized by cell shrinkage, membrane blebbing,
positional organelle loss, DNA condensation and fragmentation
(Fig. 2). Three signaling
pathways are known to trigger apoptotic cell death: The extrinsic
(death receptors) pathway, the intrinsic (mitochondrial) pathway
and the perforin/granzyme pathway (Fig. 3) (5).
 | Figure 3Synopsis of cell death processes. Ten
cell death modalities (apoptosis, autophagy, entosis, methuosis,
paraptosis, mitoptosis, parthanatos, ferroptosis, pyroptosis and
necroptosis) are presented. Anoikis shares identical signaling
pathways as apoptosis, apart from the fact that it is stimulated by
inadequate or inappropriate cell-matrix interactions. The cell
death modalities (necrosis and NETosis) without elucidative
mechanism were not included. Grey color indicates non-functional
molecules. Arrow direction indicates the causal association. RIPK,
receptor-interacting protein kinase; MLKL, mixed lineage kinase
domain-like protein; NLRs, NOD-like receptors; MOMP, mitochondrial
outer membrane permeabilization; LC3, microtubule-associated
protein light chain 3; ROCK, Rho associated coiled-coil containing
protein kinase; GPX4, glutathione peroxidase 4; ROS, reactive
oxygen species; UKL complex, UKL1 in a complex with FIP200, ATG13
and ATG101. |
Anoikis is a particular type of apoptosis, which
essentially shares identical pathways as with apoptosis; however,
is triggered by inadequate or inappropriate cell-matrix
interactions (18) (Fig. 3). The architectural state of the
cytoskeleton is expected to interfere with the function of
integrin, a pro-survival effector (6). However, the connection between cell
architecture alteration and apoptosis remains poorly identified. It
has recently been indicated that c-JUN NH2-terminal kinase (JNK)
signaling is required for efficient anoikis through a
BAK/BAX-dependent manner by increasing BCL2-like 11 (BIM)
expression and BCL-2 modifying factor (BMF) phosphorylation
(19).
Apoptosis assessment methods have been rapidly
developed over the past years (Table
I). Terminal deoxynucleotidyl transferase dUPT nick-end
labeling (TUNEL) assay and comet assay are able to detect the
presence of fragmented DNA. Annexin V in combination with
cell-impermeable DNA staining dye is used to detect the outwards
exposed phosphatidylserine on cell membrane and cellular integrity.
Alternatively, some assays evaluate the intermediate modulators,
e.g., caspase assay and poly-ADP ribose polymerase (PARP)
cleavage assay (20).
Furthermore, specific apoptosis inhibitors, such as the pan-caspase
inhibitor, zVAD-fmk, can also shed some light on the presence of
apoptosis.
4. Programmed non-apoptotic cell death
Vacuole-presenting cell death
Autophagy
Autophagic cell death is characterized by the
appearance of large intracellular vesicles, plasma membrane
blebbing, enlarged organelles and the depletion of cytoplasmic
organelles in the absence of chromatin condensation (21) (Fig.
2). Noticeably, it functions as a lever in the cell process.
Autophagy is initiated upon cellular stress as a protective
response. Once the cellular stress is irreversible, the cell will
be committed to death also through excessive levels of autophagy.
There are three forms of autophagy: Macro-autophagy (Fig. 3), micro-autophagy and
chaperone-mediated autophagy (7).
The macro-autophagic process has been well documented (22-24)
(Fig. 3). In micro-autophagy, the
cytoplasmic components are directly sequestrated into the
lysosomes, where acidic hydrolases further mediate the degradation.
Chaperone-mediated autophagy selectively targets KFERQ motif
(Lys-Phe-Glu-Arg-Gln)-containing proteins. These proteins can be
recognized by chaperones, are subsequently hijacked into lysosomes
and eventually degraded (25).
The specific degradation of the mitochondria is referred to as
mitophagy. The selective autophagy of foreign pathogens is coined
as xenophagy. There are also some other selective autophagy forms,
such as lipophagy, aggrephagy and lysophagy (26).
The detection methods are mostly developed for
macro-autophagy embodying direct measurement of autophagic activity
(e.g., turnover of long-lived proteins and LDH sequestration) and
indirect analysis with autophagy specific antibodies through
western blot-based assay, fluorescence microscopy-based assay and
flow cytometry-based assay (27)
(Table I).
Entosis. Entosis (or cannibalism) is
characterized by cell-in-cell formation (Fig. 2). Upon internalization, the
entotic cells remain viable for a short period of time. This
process is frequently followed by lysosome-mediated degradation and
non-apoptotic cell death, while a fraction of the internalized
cells can also extricate themselves or are expelled from the host
cell (28). Entosis is believed
to be triggered by integrin-extracellular matrix (ECM) detachment
(29). Unlike phagocytosis, the
engulfment of entotic cells represents a self-control process
through RhoA and the Rho-associated coiled-coil containing protein
kinases (ROCK). The entotic cell and the host cell interact with
each other through the E-cadherin and α-catenin cell junction
interface. RhoA and ROCK in entotic cells lead to specific
accumulation of actin and myosin complex (actomyosin) at the cell
cortex opposite to the junctional interface, which generates the
unbalanced contractile force driving cell-in-cell formation.
However, entosis is also observed in matrix-attached epithelial
cells. Wan et al proposed that the overactivation of myosin
or unbalanced myosin activation through regulatory polarity
proteins between the contacting cells acted as the driving force
for entosis in matrix-attached epithelial cells (30). The engulfment is followed by
lysosome-mediated degradation, which differs from autophagic cell
death (31). The autophagic
protein, microtubule-associated protein light chain 3 (LC3), does
not participate to form the autophagosome. Instead, LC3 is directed
to the single-membrane vacuole in the host cell that harbors the
engulfed cell through lipidation with the help of autophagy-related
protein (ATG)5, ATG7 and Vps34, and promotes lysosome fusion
followed by lysosome-mediated degradation (8) (Fig.
3).
However, there is as yet no specific assay available
for the detection of entosis, at least to the best of our
knowledge. The presence of entosis is deduced from its typical
cell-in-cell structure, as detected by fluorescence imaging and
electron microscopy (32,33) (Table
I).
Methuosis. Methuosis represents a type of
cell death characterized by the presence of the massive
accumulation of large fluid-filled single membrane vacuoles derived
from macropinosomes, which is specifically accompanied with Ras
hyper-activation and apoptosis impairment. Intriguingly, methuosis
is not associated with the conventional Ras-Raf-MEK-ERK axis or
class III phosphoinositide 3-kinase (PI3K) signaling (34). The consequent morphology resembles
necrosis in the manner of cell swelling and plasma membrane
integrity loss. In methuosis, activated Ras stimulates
micropinocytosis through the downstream activation of Rac family
small GTPase 1 (Rac1). Coincidently, the reduction of ADP
ribosylation factor 6-GTP (Arf6-GTP) impedes macropinosome
recycling (35). The abnormal
coalescence of nascent macropinosomes gives rise to massive
cytoplasmic vacuolization. The vacuoles formed in the early stages
of methuosis are decorated with late endosomal markers [e.g.,
lysosomal-associated membrane protein 1 (LAMP1) and Rab7] (9). The massive vacuoles, which are not
able to be recycled or merged with lysosomes, will finally lead to
cell death. Methuosis with its typical morphology, is often
assessed by electron microscopy in research (36-38)
(Table I).
Paraptosis. The hallmark of paraptosis is the
extensive cytoplasmic vacuolization derived from the dilated
endoplasmic reticulum (ER) or the mitochondria (39) (Fig.
2). It has been reported that the activation of insulin-like
growth factor 1 receptor (IGF1R) and its downstream signaling
incorporating mitogen-activated protein kinases (MAPKs) and JNK
pathways can induce paraptosis, despite the fact that IGF1R is
commonly considered as a pro-survival modulator (40). A number of studies have indicated
that paraptosis is associated with reactive oxygen species (ROS)
generation and the accumulation of misfolded proteins in the ER, as
well as mitochondrial Ca2+ overload (10,41-43),
which exert an osmotic force to distend the ER lumen and
mitochondria for vacuolization. In spite of the current available
evidence, the molecular mechanisms underlying paraptosis have not
yet been fully addressed.
Similar to entosis and methuosis, there is no
specific assay available for the detection of paraptosis, at least
to the best of our knowledge. It is mostly defined by the
appearance of multiple single-membraned cytoplasmic vacuoles, as
detected by electron microscopy (44) (Table
I).
Mitochondrial-dependent cell death
Mitoptosis
Unlike mitophagy (autophagic degradation of
mitochondria), mitoptosis, also known as mitochondrial suicide,
represents a process of programmed fission and fusion of the
mitochondria with the concomitant disruption of the adenosine
triphosphate (ATP) supply. As a consequence, mitoptosis can be
associated with both apoptosis (45) and autophagy (46). The degraded mitochondria either
become autophagosomes or mitoptotic bodies, which are extruded from
the cell. In this sense, mitoptosis itself is not a cell death
pathway, but a mitochondrial death pathway. However, the extensive
mitochondrial fragmentation through elevated fission finally leads
to cell death (47). Mechanically
speaking, mitochondrial outer membrane permeabilization (MOMP)
induced by BAX/BAK triggers the release of a mitochondrial
intermembrane space protein termed translocase of inner
mitochondrial membrane 8a (TIMM8a/DDP). DDP subsequently binds to
DRP1 in the cytoplasm. The interaction between DDP and DRP1 leads
to the recruitment of DRP1 and retention in the mitochondria, which
induces mitochondrial fission and finally, mitoptosis (48). Nevertheless, the process remains
poorly understood and is described mostly by its morphological
features.
As a manner of mitochondrial suicide, the
visualization of fragmented mitochondria with mitochondria-specific
dyes (e.g., MitoTracker Green®) by utilizing
fluorescence microscopy and a close observation with electron
microscopy provide certain clues on the presence of mitoptosis
(45). Moreover, specific
antibodies against cytochrome c and TIMM8a/DDP are also
utilized in research (48)
(Table I).
Parthanatos. Parthanatos represents a
mitochondrial-linked, but caspase-independent cell death and is
characterized by the hyperactivation of PARP. PARP mediates the
synthesis of poly(ADP-ribose) (PAR), which further shuttles from
the nucleus to the cytoplasm and binds to specific mitochondrial
proteins followed by apoptosis-inducing factor (AIF) release. Free
AIF is translocated from the mitochondria into the nucleus. In the
nucleus, AIF induces chromatin condensation and DNA breakage
(49). Compared to the apoptotic
process, intact PARP and its activation is required, rather than
PARP cleavage. Moreover, parthanatos cannot be inhibited by
broad-spectrum caspase inhibitors (50), which proves its independency of
caspases. Parthanatos does not involve the formation of apoptotic
bodies. Furthermore, the DNA fragmentation is large-scale rather
than small-to-moderate scale, as typically observed in apoptosis
(11) (Fig. 2).
PAR accumulation, PARP-1 activation and nuclear AIF
are practically used as biomarkers of parthanatos. The process can
be further confirmed with mitochondrial depolarization, as detected
with fluorescent probe staining (Table I).
Iron-dependent cell death
Ferroptosis
Ferroptosis is normally associated with a
normal-appearing morphology, with an intact cell membrane without
blebbing and normal-sized nucleus free of chromatin condensation,
although with diminutive mitochondria with decreased cristae and
collapsed and ruptured membranes (51) (Fig.
2). It is initiated by the failure of the glutathione-dependent
antioxidant defense through defects in system
XC− or glutathione peroxidase 4 (GPX4)
(12). System
XC− transports extracellular cystine into the
cell, which is then transformed into cysteine for glutathione (GSH)
synthesis. GPX4 can directly catalyze the reaction between
glutathione and lipid hydroperoxides to reduce the cellular level
of lipid peroxidation. Either the depletion of GSH or the
inhibition of GPX4 results in lipid hydroperoxide accumulation.
Free iron interacts with lipid hydroperoxides through the Fenton
reaction and forms lipid ROS (Fig.
3). Excessive lipid ROS generation finally leads to the cell
death.
The induction of ferroptosis can be confirmed by
applying ferroptosis inhibitors (e.g., ferrostatin-1 and
liproxstatin-1) and by measuring lipid peroxides (e.g.,
malondialhyde quantification and 4-hydroxynonenal quantification)
(Table I).
Immune-reactive cell death
Pyroptosis
Pyroptosis is an inflammatory form of programmed
cell death that commonly occurs upon the recognition of
intracellular pathogens in immune cells. The inflammation sensors
[e.g., NOD-like receptors (NLRs)] of infected macrophages recognize
the flagellin components of pathogens and initiate the formation of
multi-protein complex inflammasomes, which subsequently activate
caspase-1(13) (Fig. 3). Upon activation, caspase-1
mediates the membrane pore formation through the cleavage of
gasdermin D, allowing the rupture of the cell membrane (52). The process is also accompanied by
DNA condensation and fragmentation (Fig. 2). Moreover, caspase-11 can be
directly activated by bacterial lipopolysaccharide (LPS) and
induces pyroptosis (53).
Pyroptosis can be evaluated through the
quantification of released cytoplasmic LDH, the visualization of
membrane integrity loss by fluorescence microscopy, the detection
of interleukin (IL)-1β, caspase activation and gasdermin D cleavage
by western blot analysis (54)
(Table I).
Neutrophil extracellular trap-associated cell
death (NETosis). NETosis, a unique form of cell death, is
initiated by the presence of pathogens or their components and
mostly occurs in immune cells, particularly neutrophils. Upon the
recognition of pathogens within neutrophils, the cells undergo
histone modification, chromatin decondensation and neutrophil
extracellular trap [NET, comprising chromatin and antimicrobial
components including myeloperoxidase, neutrophil elastase,
cathepsin G, lysozyme and defensins (55)] release and this eventually leads
to cell death. The process is promoted through superoxide generated
by NADPH oxidase 4 (NOX4), autophagy and peptidylarginine deiminase
4 (PAD4)-dependent histone citrullination (56,57). However, further research is
expected to provide a clear molecular elucidation.
The staining of co-localized neutrophil-derived
proteins and extracellular DNA, as well as citrullinated histones
is utilized to evaluate NETosis. Moreover, cell-free DNA and
DNA-neutrophil derived protein complexes can be detected by
PicoGreen® and ELISA. Both morphology and cell-appendant
NETosis components can be detected through flow cytometry (58) (Table
I).
Other types Necroptosis
Necroptosis, also known as programmed necrosis, is
characterized by the activation of receptor-interacting protein
kinases (RIPKs) through several signaling pathways (15). RIPKs are activated upon
recruitment to macromolecular complexes from various cell-surface
receptors: Death receptors (DRs), Toll-like receptors (TLRs), and
the T-cell receptor (TCR) (Fig.
3) (59,60). RIPK1 and RIPK3 function as the key
components of necrosome (61).
RIPK3 further activates downstream molecule mixed lineage kinase
domain-like protein (MLKL) through phosphorylation (62,63), which leads to MLKL
oligomerization. The oligomerized MLKL inserts into and
permeabilizes cellular membrane, which finally gives rise to cell
death (64). Moreover,
RIP3-dependent necroptosis is also triggered by the cytosolic DNA
sensor, DNA-dependent activator of interferon (DAI) regulatory
factors, following viral infection or the presence of
double-stranded viral DNA (65).
Necroptosis reveals the necrotic morphology with membrane rupture
and loss of organelles (Fig.
2).
Necroptosis can be assessed by the loss of plasma
membrane integrity by utilizing cell-impermeable DNA binding dyes,
the release of cellular contents, including LDH, high mobility
group box 1 protein (HMGB1) and cyclophilin A by western blot
analysis, mitochondrial potential by fluorescent probes and
morphology by electron microscopy. The utilization of necroptosis
specific inhibitors, such as necrostatin-1 and measuring key
proteins in the pathway represent alternative strategies (66) (Table
I).
5. Implications of cell death in human
diseases
The dysregulation of cell death processes is highly
relevant to tumorigenesis, as well as to the pathogenesis of a
number of other diseases, such as degenerative, cardiovascular and
autoimmune diseases. The association between cell death and cancer
is complex. The complexity is attributed to several factors: On the
one hand, there is more than one type of cell death endogenously
engaged in cancer. On the other hand, some types of cell death have
dual and even opposing effects on tumorigenesis. Firstly, apoptosis
is involved in cancer. Cancerous cells can evade apoptosis by
downregulating or blocking apoptosis signaling (67). Unexpectedly, apoptosis can also
drive tumor formation by promoting cell proliferation as a
compensation for cell loss (68).
Secondly, necrosis is commonly observed in tumors due to hypoxic
microenvironments (67). Thirdly,
cancerous cells with defects in apoptosis tend to utilize autophagy
as a pro-survival mechanism. Paradoxically, impeded autophagy is
also associated with tumorigenesis (69). Fourthly, entosis represents tumor
suppressive activity in pancreatic cancer, whereas it promotes
tumor progression in most other situations (70,71). Although the other cell death types
are much less endogenously involved in cancer development, they are
mostly utilized as anti-cancer defense strategies of the body and
defects in their signaling plays an important role in drug
resistance and clinical failures.
As for neurodegenerative diseases, the initial phase
of cell death in ischemia represents necrotic cell death, while
delayed cell death is apoptotic in nature due to the fact that the
ischemic core tends to be necrotic and the penumbra region
apoptotic (72). Autophagic cell
death and parthanatos are linked to ischemia (11,73). In Parkinson's disease, apoptosis
contributes to the loss of nigral neurons due to the fact that
almost every Lewy body-containing neuron (as a pathological feature
of Parkinson's disease) is positive for pro-apoptotic modulator
staining (74). Another study
demonstrated that necrostatin-1, an inhibitor of necroptosis,
ameliorated neuronal loss in a model of Parkinson's disease
(75), indicating that
necroptosis may also play a role in Parkinson's disease. There is
also evidence suggesting the role of apoptosis in Huntington's
disease. However, its role in Alzheimer's disease remains under
debate (76).
Cell death modes, such as apoptosis, necrosis and
autophagy in cardiac myocytes have been frequently reported to
affect a variety of cardiovascular diseases, including myocardial
infarction, diabetic cardiomyopathy, ischemic cardiomyocyte and
congestive heart failure (77-79).
In addition, ferroptosis, pyroptosis, as well as parthanatos are
also documented to contribute to ischemia/reperfusion injury
(80). The other cell death types
have been studied to a much lesser extent as compared to
cardiovascular diseases. Likewise, apoptosis and secondary necrosis
are considered as major modes of cell death in systemic autoimmune
diseases. Recent evidence indicates that NETosis accounts for
certain immunological features in systemic lupus erythematosus
(81).
6. Conclusions and perspectives
The cell death modes presented in the present review
article are mostly distinguished by stimuli, molecules and
morphologies. Apart from non-programmed necrosis, the other cell
death modes are regulated in a signal-dependent manner, despite the
fact that a number of the pathways have not yet been fully
addressed. Some cell death modes are intensively interacting with
others. For instance, the activation of tumor necrosis factor
receptor (TNFR) can stimulate both apoptosis and necroptosis;
however, compromised apoptosis can shift the downstream pathway to
necroptosis (82) and vice versa
(83). Some processes during cell
death are connected; for instance, the occurrence of mitoptosis can
turn out as autophagic cell death or apoptotic cell death. In
general, necrosis-like cell death is associated with membrane
rupture. The consequent release of intracellular inflammatory
factors can give rise to inflammation as observed in necrosis,
necroptosis, NETosis and pyroptosis. By contrast, apoptotic cells
do not stimulate inflammation, since they are rapidly eliminated by
phagocytes. However, if apoptotic cells are not properly processed,
they can develop secondary necrosis. These mutual connections
indicate that different cell death types are not isolated from each
other. The molecular links await to be unveiled in greater detail.
Their implications on diverse diseases are expected to be unraveled
in the near future, since current studies on cell death modes
involved in diseases are mostly confined to the more classical cell
death categories. Green (84)
also addressed five quite interesting and inspiring questions about
the balance and context of cell death. In fact, much is still
unknown. Noticeably, this review article has primarily focused on
the features of pathological cell death and is limited to the
animal kingdom. However, there also exist physiologic cell death
such as cornification (85) to
form termination differentiation and some cell death types are also
similarly present in the plant kingdom (e.g., apoptosis-like cell
death) (86).
Acknowledgements
Not applicable.
Funding
The authors are grateful to PhD stipends given to GY
(by the Chinese Scholarship Council) and to ME (by the German
Academic Exchange Service, DAAD).
Availability of data and materials
Not applicable.
Authors' contributions
GY was responsible for the drafting of the
manuscript and cell death information collection. ME was
responsible for information presentesst and figure construction. TE
was responsible for the initial conception of the study and for the
revision of the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Green DR and Llambi F: Cell death
signaling. Cold Spring Harb Perspect Biol. 7:
pii(a006080)2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Galluzzi L, Vitale I, Aaronson SA, Abrams
JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews
DW, et al: Molecular mechanisms of cell death: Recommendations of
the nomenclature committee on cell death 2018. Cell Death Differ.
25:486–541. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tang D, Kang R, Berghe TV, Vandenabeele P
and Kroemer G: The molecular machinery of regulated cell death.
Cell Res. 29:347–364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Syntichaki P and Tavernarakis N: Death by
necrosis. Uncontrollable catastrophe, or is there order behind the
chaos? EMBO Rep. 3:604–609. 2002.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Elmore S: Apoptosis: A review of
programmed cell death. Toxicol Pathol. 35:495–516. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Paoli P, Giannoni E and Chiarugi P:
Anoikis molecular pathways and its role in cancer progression.
Biochim Biophys Acta. 1833:3481–3498. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ravanan P, Srikumar IF and Talwar P:
Autophagy. The spotlight for cellular stress responses. Life Sci.
188:53–67. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Krishna S and Overholtzer M: Mechanisms
and consequences of entosis. Cell Mol Life Sci. 73:2379–2386.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Maltese WA and Overmeyer JH: Methuosis.
Nonapoptotic cell death associated with vacuolization of
macropinosome and endosome compartments. Am J Pathol.
184:1630–1642. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lee D, Kim IY, Saha S and Choi KS:
Paraptosis in the anti-cancer arsenal of natural products.
Pharmacol Ther. 162:120–133. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fatokun AA, Dawson VL and Dawson TM:
Parthanatos: Mitochondrial-linked mechanisms and therapeutic
opportunities. Br J Pharmacol. 171:2000–2016. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yu H, Guo P, Xie X, Wang Y and Chen G:
Ferroptosis, a new form of cell death, and its relationships with
tumourous diseases. J Cell Mol Med. 21:648–657. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bergsbaken T, Fink SL and Cookson BT:
Pyroptosis. Host cell death and inflammation. Nat Rev Microbiol.
7:99–109. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Neubert E, Meyer D, Rocca F, Günay G,
Kwaczala-Tessmann A, Grandke J, Senger-Sander S, Geisler C, Egner
A, Schön MP, et al: Chromatin swelling drives neutrophil
extracellular trap release. Nat Commun. 9(3767)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Vanlangenakker N, Vanden Berghe T and
Vandenabeele P: Many stimuli pull the necrotic trigger, an
overview. Cell Death Differ. 19:75–86. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Won SJ, Kim DY and Gwag BJ: Cellular and
molecular pathways of ischemic neuronal death. J Biochem Mol Biol.
35:67–86. 2002.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Weerasinghe P and Buja LM: Oncosis. An
important non-apoptotic mode of cell death. Exp Mol Pathol.
93:302–308. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Frisch SM and Screaton RA: Anoikis
mechanisms. Curr Opin Cell Biol. 13:555–562. 2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Girnius N and Davis RJ: JNK promotes
epithelial cell anoikis by transcriptional and post-translational
regulation of BH3-only proteins. Cell Rep. 21:1910–1921.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Muganda PM (ed): Apoptosis methods in
toxicology. Humana Press, New York, NY, 2016.
|
|
21
|
Liu Y and Levine B: Autosis and autophagic
cell death: The dark side of autophagy. Cell Death Differ.
22:367–376. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pajares M, Jiménez-Moreno N, García-Yagüe
ÁJ, Escoll M, de Ceballos ML, van Leuven F, Rábano A, Yamamoto M,
Rojo AI and Cuadrado A: Transcription factor NFE2L2/NRF2 is a
regulator of macroautophagy genes. Autophagy. 12:1902–1916.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mercer CA, Kaliappan A and Dennis PB: A
novel, human Atg13 binding protein, Atg101, interacts with ULK1 and
is essential for macroautophagy. Autophagy. 5:649–662.
2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen Y and Klionsky DJ: The regulation of
autophagy-unanswered questions. J Cell Sci. 124:161–170.
2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mizushima N: A brief history of autophagy
from cell biology to physiology and disease. Nat Cell Biol.
20:521–527. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hansen M, Rubinsztein DC and Walker DW:
Autophagy as a promoter of longevity: Insights from model
organisms. Nat Rev Mol Cell Biol. 19:579–593. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Orhon I and Reggiori F: Assays to monitor
autophagy progression in cell cultures. Cells. 6:
pii(E20)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
White E: Entosis: It's a cell-eat-cell
world. Cell. 131:840–842. 2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ishikawa F, Ushida K, Mori K and Shibanuma
M: Loss of anchorage primarily induces non-apoptotic cell death in
a human mammary epithelial cell line under atypical focal adhesion
kinase signaling. Cell Death Dis. 6(e1619)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wan Q, Liu J, Zheng Z, Zhu H, Chu X, Dong
Z, Huang S and Du Q: Regulation of myosin activation during
cell-cell contact formation by Par3-Lgl antagonism: Entosis without
matrix detachment. Mol Biol Cell. 23:2076–2091. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Garanina AS, Kisurina-Evgenieva OP,
Erokhina MV, Smirnova EA, Factor VM and Onishchenko GE: Consecutive
entosis stages in human substrate-dependent cultured cells. Sci
Rep. 7(12555)2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sun Q and Overholtzer M: Methods for the
study of entosis. Methods Mol Biol. 1004:59–66. 2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Huang H, Chen A, Wang T, Wang M, Ning X,
He M, Hu Y, Yuan L, Li S, Wang Q, et al: Detecting cell-in-cell
structures in human tumor samples by E-cadherin/CD68/CD45 triple
staining. Oncotarget. 6:20278–20287. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kaul A, Overmeyer JH and Maltese WA:
Activated Ras induces cytoplasmic vacuolation and non-apoptotic
death in glioblastoma cells via novel effector pathways. Cell
Signal. 19:1034–1043. 2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bhanot H, Young AM, Overmeyer JH and
Maltese WA: Induction of nonapoptotic cell death by activated Ras
requires inverse regulation of Rac1 and Arf6. Mol Cancer Res.
8:1358–1374. 2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Overmeyer JH, Young AM, Bhanot H and
Maltese WA: A chalcone-related small molecule that induces
methuosis, a novel form of non-apoptotic cell death, in
glioblastoma cells. Mol Cancer. 10(69)2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Trabbic CJ, Dietsch HM, Alexander EM, Nagy
PI, Robinson MW, Overmeyer JH, Maltese WA and Erhardt PW:
Differential induction of cytoplasmic vacuolization and methuosis
by novel 2-indolyl-substituted pyridinylpropenones. ACS Med Chem
Lett. 5:73–77. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Silva-Pavez E, Villar P, Trigo C, Caamaño
E, Niechi I, Pérez P, Muñoz JP, Aguayo F, Burzio VA, Varas-Godoy M,
et al: CK2 inhibition with silmitasertib promotes methuosis-like
cell death associated to catastrophic massive vacuolization of
colorectal cancer cells. Cell Death Dis. 10(73)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Sperandio S, de Belle I and Bredesen DE:
An alternative, nonapoptotic form of programmed cell death. Proc
Natl Acad Sci USA. 97:14376–14381. 2000.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sperandio S, Poksay K, de Belle I,
Lafuente MJ, Liu B, Nasir J and Bredesen DE: Paraptosis: Mediation
by MAP kinases and inhibition by AIP-1/Alix. Cell Death Differ.
11:1066–1075. 2004.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yoon MJ, Lee AR, Jeong SA, Kim YS, Kim JY,
Kwon YJ and Choi KS: Release of Ca2+ from the endoplasmic reticulum
and its subsequent influx into mitochondria trigger
celastrol-induced paraptosis in cancer cells. Oncotarget.
5:6816–6831. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gandin V, Pellei M, Tisato F, Porchia M,
Santini C and Marzano C: A novel copper complex induces paraptosis
in colon cancer cells via the activation of ER stress signalling. J
Cell Mol Med. 16:142–151. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ghosh K, De S, Das S, Mukherjee S and
Sengupta Bandyopadhyay S: Withaferin a induces ROS-mediated
paraptosis in human breast cancer cell-lines MCF-7 and MDA-MB-231.
PLoS One. 11(e0168488)2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kessel D: Apoptosis, paraptosis and
autophagy: Death and survival pathways associated with photodynamic
therapy. Photochem Photobiol. 95:119–125. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lyamzaev KG, Nepryakhina OK, Saprunova VB,
Bakeeva LE, Pletjushkina OY, Chernyak BV and Skulachev VP: Novel
mechanism of elimination of malfunctioning mitochondria
(mitoptosis). Formation of mitoptotic bodies and extrusion of
mitochondrial material from the cell. Biochim Biophys Acta.
1777:817–825. 2008.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jangamreddy JR and Los MJ: Mitoptosis, a
novel mitochondrial death mechanism leading predominantly to
activation of autophagy. Hepat Mon. 12(e6159)2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Youle RJ and Karbowski M: Mitochondrial
fission in apoptosis. Nat Rev Mol Cell Biol. 6:657–663.
2005.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Arnoult D, Rismanchi N, Grodet A, Roberts
RG, Seeburg DP, Estaquier J, Sheng M and Blackstone C:
Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated
mitochondrial fission and mitoptosis during programmed cell death.
Curr Biol. 15:2112–2118. 2005.PubMed/NCBI View Article : Google Scholar
|
|
49
|
David KK, Andrabi SA, Dawson TM and Dawson
VL: Parthanatos, a messenger of death. Front Biosci (Landmark Ed).
14:1116–1128. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
50
|
Yu SW, Wang H, Poitras MF, Coombs C,
Bowers WJ, Federoff HJ, Poirier GG, Dawson TM and Dawson VL:
Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by
apoptosis-inducing factor. Science. 297:259–263. 2002.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Latunde-Dada GO: Ferroptosis. Role of
lipid peroxidation, iron and ferritinophagy. Biochim Biophys Acta
Gen Subj. 1861:1893–1900. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liu X, Zhang Z, Ruan J, Pan Y, Magupalli
VG, Wu H and Lieberman J: Inflammasome-activated gasdermin D causes
pyroptosis by forming membrane pores. Nature. 535:153–158.
2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Lacey CA, Mitchell WJ, Dadelahi AS and
Skyberg JA: Caspase-1 and caspase-11 mediate pyroptosis,
inflammation, and control of brucella joint infection. Infect
Immun. 86: pii(e00361-18)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
den Hartigh AB and Fink SL: Pyroptosis
induction and detection. Curr Protoc Immunol: Jul 20, 2018 (Epub
ahead of print).
|
|
55
|
Branzk N and Papayannopoulos V: Molecular
mechanisms regulating NETosis in infection and disease. Semin
Immunopathol. 35:513–530. 2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Remijsen Q, Vanden Berghe T, Wirawan E,
Asselbergh B, Parthoens E, De Rycke R, Noppen S, Delforge M,
Willems J and Vandenabeele P: Neutrophil extracellular trap cell
death requires both autophagy and superoxide generation. Cell Res.
21:290–304. 2011.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wang Y, Li M, Stadler S, Correll S, Li P,
Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, et al: Histone
hypercitrullination mediates chromatin decondensation and
neutrophil extracellular trap formation. J Cell Biol. 184:205–213.
2009.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Masuda S, Nakazawa D, Shida H, Miyoshi A,
Kusunoki Y, Tomaru U and Ishizu A: NETosis markers: Quest for
specific, objective, and quantitative markers. Clin Chim Acta.
459:89–93. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Kaiser WJ, Sridharan H, Huang C, Mandal P,
Upton JW, Gough PJ, Sehon CA, Marquis RW, Bertin J and Mocarski ES:
Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. J
Biol Chem. 288:31268–31279. 2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Weinlich R, Oberst A, Beere HM and Green
DR: Necroptosis in development, inflammation and disease. Nat Rev
Mol Cell Biol. 18:127–136. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
He S, Wang L, Miao L, Wang T, Du F, Zhao L
and Wang X: Receptor interacting protein kinase-3 determines
cellular necrotic response to TNF-alpha. Cell. 137:1100–1111.
2009.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Sun L, Wang H, Wang Z, He S, Chen S, Liao
D, Wang L, Yan J, Liu W, Lei X and Wang X: Mixed lineage kinase
domain-like protein mediates necrosis signaling downstream of RIP3
kinase. Cell. 148:213–227. 2012.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wang H, Sun L, Su L, Rizo J, Liu L, Wang
LF, Wang FS and Wang X: Mixed lineage kinase domain-like protein
MLKL causes necrotic membrane disruption upon phosphorylation by
RIP3. Mol Cell. 54:133–146. 2014.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Cai Z, Jitkaew S, Zhao J, Chiang HC,
Choksi S, Liu J, Ward Y, Wu LG and Liu ZG: Plasma membrane
translocation of trimerized MLKL protein is required for
TNF-induced necroptosis. Nat Cell Biol. 16:55–65. 2014.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Maelfait J, Liverpool L, Bridgeman A,
Ragan KB, Upton JW and Rehwinkel J: Sensing of viral and endogenous
RNA by ZBP1/DAI induces necroptosis. EMBO J. 36:2529–2543.
2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Vanden Berghe T, Grootjans S, Goossens V,
Dondelinger Y, Krysko DV, Takahashi N and Vandenabeele P:
Determination of apoptotic and necrotic cell death in vitro and in
vivo. Methods. 61:117–129. 2013.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Messmer MN, Snyder AG and Oberst A:
Comparing the effects of different cell death programs in tumor
progression and immunotherapy. Cell Death Differ. 26:115–129.
2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Labi V and Erlacher M: How cell death
shapes cancer. Cell Death Dis. 6(e1675)2015.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967.
2007.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Durgan J and Florey O: Cancer cell
cannibalism: Multiple triggers emerge for entosis. Biochim Biophys
Acta Mol Cell Res. 1865:831–841. 2018.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Wang X, Li Y, Li J, Le Li Zhu H, Chen H,
Kong R, Wang G, Wang Y, Hu J and Sun B: Cell-in-cell phenomenon and
its relationship with tumor microenvironment and tumor progression:
A review. Front Cell Dev Biol. 7(311)2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Nitatori T, Sato N, Waguri S, Karasawa Y,
Araki H, Shibanai K, Kominami E and Uchiyama Y: Delayed neuronal
death in the CA1 pyramidal cell layer of the gerbil hippocampus
following transient ischemia is apoptosis. J Neurosci.
15:1001–1011. 1995.PubMed/NCBI
|
|
73
|
Uchiyama Y, Koike M and Shibata M:
Autophagic neuron death in neonatal brain ischemia/hypoxia.
Autophagy. 4:404–408. 2008.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Lev N, Melamed E and Offen D: Apoptosis
and Parkinson's disease. Prog Neuropsychopharmacol Biol Psychiatry.
27:245–250. 2003.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Iannielli A, Bido S, Folladori L, Segnali
A, Cancellieri C, Maresca A, Massimino L, Rubio A, Morabito G,
Caporali L, et al: Pharmacological inhibition of necroptosis
protects from dopaminergic neuronal cell death in parkinson's
disease models. Cell Rep. 22:2066–2079. 2018.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Chi H, Chang HY and Sang TK: Neuronal cell
death mechanisms in major neurodegenerative diseases. Int J Mol
Sci. 19: pii(E3082)2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Clarke M, Bennett M and Littlewood T: Cell
death in the cardiovascular system. Heart. 93:659–664.
2007.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Lee Y and Gustafsson AB: Role of apoptosis
in cardiovascular disease. Apoptosis. 14:536–548. 2009.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Chiong M, Wang ZV, Pedrozo Z, Cao DJ,
Troncoso R, Ibacache M, Criollo A, Nemchenko A, Hill JA and
Lavandero S: Cardiomyocyte death: Mechanisms and translational
implications. Cell Death Dis. 2(e244)2011.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Del Re DP, Amgalan D, Linkermann A, Liu Q
and Kitsis RN: Fundamental mechanisms of regulated cell death and
implications for heart disease. Physiol Rev. 99:1765–1817.
2019.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Darrah E and Andrade F: NETs: The missing
link between cell death and systemic autoimmune diseases? Front
Immunol. 3(428)2013.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Vanden Berghe T, Kaiser WJ, Bertrand MJ
and Vandenabeele P: Molecular crosstalk between apoptosis,
necroptosis, and survival signaling. Mol Cell Oncol.
2(e975093)2015.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Ali M and Mocarski ES: Proteasome
inhibition blocks necroptosis by attenuating death complex
aggregation. Cell Death Dis. 9(346)2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Green DR: The coming decade of cell death
research: Five riddles. Cell. 177:1094–1107. 2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Eckhart L, Lippens S, Tschachler E and
Declercq W: Cell death by cornification. Biochim Biophys Acta.
1833:3471–3480. 2013.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Emanuele S, Oddo E, D'Anneo A, Notaro A,
Calvaruso G, Lauricella M and Giuliano M: Routes to cell death in
animal and plant kingdoms. From classic apoptosis to alternative
ways to die-a review. Rend Lincei Sci Fis. 29:397–409. 2018.
|