Introduction
Gastric cancer (GC) is a serious public health
concern, and is one of the most common malignancies in the world
(1). Despite reductions in
incidence and mortality rates over the past few decades, it remains
the fifth most frequent type of cancer and the fourth leading cause
of cancer-related mortality worldwide, with an estimated 1,089,103
new cases per year, and 768,793 GC-related deaths recorded in
2020(2).
The occurrence and progression of GC have been
attributed to a number of factors; these factors may involve an
infection of the gastric epithelium by Helicobacter pylori
(H. pylori), a bacteria found in half of the population
worldwide (3); or by Epstein-Barr
virus (EBV), a double-stranded DNA virus that has recently been
associated with GC (4).
Researchers investigating this type of tumor believe that these two
agents can lead to prolonged and irreversible infection in the
epithelial tissue of the stomach, which eventually leads to stomach
cancer (5).
Recently, microRNAs (miRNAs/miRs) have attracted
increasing interest in tumor biology due to their differential
expression patterns observed in numerous types of cancer (6-8).
miRNAs are small non-coding RNAs (21-25 nucleotides in length) that
play a critical role in regulating genre expression at the
post-transcriptional level by regulating the stability and
translation of protein-coding mRNAs, typically through
sequence-specific binding to the 3'-untranslated region (3'-UTR) of
a target messenger RNAs (mRNAs) (9). The miRNA-mRNA interaction usually
causes a translocation and/or cleavage repression of the mRNA, thus
reducing the production of the final protein (10,11).
Several studies have revealed that various miRNAs play critical
roles in cell growth, differentiation, apoptosis and carcinogenesis
(12-16).
miR-21 has been found to be upregulated in certain types of solid
cancers of the breast, colon, lung, pancreas, prostate and stomach
(17). In addition, the
overexpression of miR-223 and miR-19 has been reported in GC
tissues (18). Thus, it can be
assumed from previous studies that miR-21, miR-223 and miR-19 may
function as oncogenes in GC. However, the targets of these miRNAs
have not yet been identified, and their association with the
clinicopathological characteristics of GC has also not been
illustrated, at least to the best of our knowledge. In recent
decades, among the >2,000 mature miRNAs discovered to date, only
120 miRNAs have been studied in GC or GC cell lines (19). Extensive research has been
conducted individually on the differential expression of miRNAs and
H. pylori/EBV infections, and the association of these
miRNAs with infections in GC specimens is also being increasingly
reported (20-23).
Recent studies have suggested that H. pylori and EBV
infections contribute to the dysregulation of these miRNAs
(24-26).
Thus, the detection of miRNAs will not only provide
potential biomarkers for the early detection and evaluation of GC,
but may also guide future functional analyses of miRNAs in order to
better understand their role in gastric carcinogenesis. The present
study aimed to investigate the differential expression of miR-21,
miR-223 and miR-19 in tissues from patients with GC and their
association with H. pylori and EBV infections.
Patients and methods
Study population and samples
Clinical data and 70 gastric tissue specimens (of
which 35 were tumor tissues and 35 were corresponding adjacent
normal tissues) were collected during the period June 2020, to
March, 2021 from patients, who found to have gastric adenocarcinoma
by an endoscopic biopsy, and who underwent gastric resection at the
Department of Surgery of Ibn Rochd University Hospital Center,
Casablanca, Morocco. Patients who simultaneously had another type
of cancer were excluded from the study.
The Biomedical Research Ethics Committee of the
Faculty of Medicine and Pharmacy of Casablanca, Morocco (Reference
no. 13/19 and committee reference no. 02/DRC/00) approved the
study, and each patient signed an informed consent form.
DNA extraction of H. pylori and
EBV
The collected tissue samples (both tumor and
adjacent normal) were then immediately frozen and stored at -80˚C
until use. DNA extraction from the biopsies was performed using the
pure link Invitrogen® Genomic DNA mini kit (Thermo
Fisher Scientific, Inc.), following the manufacturer's
instructions. The quality and quantity of the DNA extracted were
evaluated using a NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.).
Detection of H. pylori and
cytotoxin-associated gene A (cagA)
H. pylori was detected in biopsies using PCR
with glmM primers as previously described (27). To enhance the detection
sensitivity, cagA was examined using the primers indicated in
Table SI and as previously
described (28). For each
reaction, DNA from H. pylori CagA+ strain and a tube
containing water in place of DNA were assayed as positive and
negative controls used for comparisons.
EBV detection was performed using nested PCR, which
consists of two successive PCRs in such a way that the product of
the first PCR serves as a matrix for the second, and the sequence
amplified by the second PCR is located inside the first fragment.
The primers for the EBV (Table
SI) were as previously described (29).
Briefly, the PCR reaction was carried out in a 25 µl
reaction mixture containing genomic DNA (8 ng), 2X Taq PCR Master
Mix (Qiagen, Inc.), 10 µmol forward and reverse primers. PCR
amplification was performed using a PerkinElmer 2400 GeneAmp PCR
System 2400 Thermal Cycler® (Thermo Fisher Scientific,
Inc.). The primers used are presented in Table SI. The cycling conditions were as
follows: Denaturation at 94˚C for 3 min, followed by 35 cycles of
denaturation at 94˚C for 1 min, annealing at the specific
temperature for 1 min, extension at 72˚C for 1 min, and the
reaction was terminated by a 10-min extension at 72˚C.
miRNA extraction and the detection of
mature miRNAs using reverse transcription-quantitative PCR
(RT-qPCR)
miRNAs were extracted and purified from cancerous
tissues and adjacent normal tissues using the mirVanaTM miRNA
isolation kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. The reverse transcription process (RT)
of the miRNA samples into cDNA was performed using a
TaqManTM MicroRNA assay (Applied Biosystems; Thermo
Fisher Scientific, Inc.; details are presented in Table SII), and reagents from the
TaqManTM MicroRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). For the qPCR process,
the PCR amplification was performed using TaqMan microRNA assays,
and TaqManTM Universal Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.).
The expression level of the three candidate miRNAs
relative to the internal control, small nuclear U6 RNA, was
determined using the following equation: 2-ΔΔCq, where
ΔΔCq=(Cq target-Cq U6 RNA)cancer-(Cq target-Cq U6 RNA)normal
adjacent (30). The
quantification cycle (Cq) is defined as the intersection between
the amplification plot of the PCR product and the threshold line.
The Cq value is relative to the PCR product concentration.
Statistical analysis
The statistical analysis of the results was
performed using SPSS 23.0 statistical software (IBM Corp.), and the
association between the miRNA expression levels, and the various
clinicopathological parameters were analyzed using a Student's
t-test (paired t-test), or a Chi-squared test or Fisher exact test.
A P-value <0.05 was considered to indicate a statistically
significant difference.
Receiver operating characteristic (ROC) curve
analysis was performed on the three miRNAs examined to investigate
their performance as a discriminatory tool for classifying tumor
and normal tissues. It is a plot of the true positive rate
(sensitivity) in function of the false positive rate
(100-specificity) for different cut-off points of miRNA expression.
Cut-off values were determined using the Youden index, and P-values
<0.05 for any parameters were considered to indicate
statistically significant differences. Sensitivity indicates the
ability of a ROC curve to correctly identify cancerous tissues
(true positive). Specificity is the ability of ROC curve to
correctly identify non-cancerous tissues (true negative). The area
under the ROC curve (AUC) indicates the effectiveness of a miRNA to
distinguish between cancerous tissues and non-cancerous
tissues.
Results
Detection of H. pylori and EBV
Overall, the results indicated the presence of H.
pylori DNA in 31.4% of the GC samples. Additionally, all H.
pylori-positive tissues were cagA-positive. EBV DNA found was
in 40% of the GC tissues, and co-infection was found in 11.4% of
the GC samples.
miRNA expression
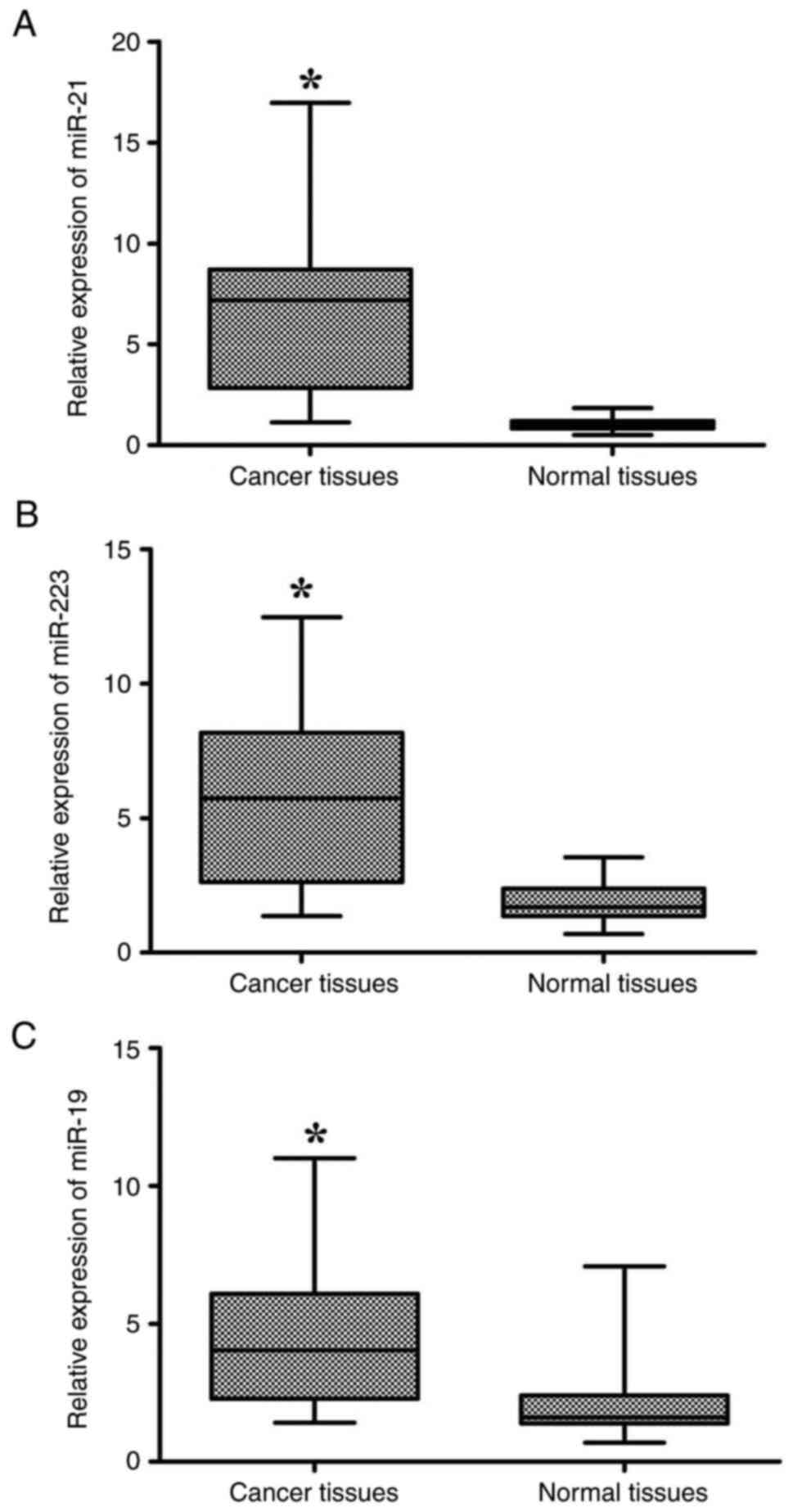
RT-qPCR detection revealed that the miR-21, miR-223
and miR-19 expression levels, normalized to U6 RNA, were
significantly increased in cancerous tissues compared with normal
tissues (mean ± SD: 6.5±3.7 vs. 1.5±0.4, P<0.0001; 5.73±3.09 vs.
1.84±0.75, P<0.0001; 4.37±2.55 vs. 1.93±1.12, P<0.0001,
respectively; Fig. 1). Using the
average miRNA expression levels (2-ΔΔCq) in GC tissues
as a maximum, all patients were divided into two groups as follows:
A group with a high miRNA expression, and a group with a low miRNA
expression. The analysis of the association between the
clinicopathological parameters of the patients, and the miRNA
expression level revealed that miR-21, miR-223 and miR-19
expression levels were not associated with age, sex, or toxic
habits (e.g., smoking or alcohol consumption; P>0.05); however,
they were associated with histological type, tumor size, lymphatic
vessel invasion, the presence of lymph node metastasis and tumor
stage (P<0.05; Table I).
 | Table IAssociation of miRNA expression with
various clinicopathological features of patients with gastric
cancer. |
Table I
Association of miRNA expression with
various clinicopathological features of patients with gastric
cancer.
| | miR-21
expression | | miR-223
expression | | miR-19
expression | |
|---|
| Clinicopathological
features | No. of cases | Low | High | P-value | Low | High | P-value | Low | High | P-value |
|---|
| Age group | | | | | | | | | | |
|
<59
years | 17 | 6 | 11 | 0.38 | 9 | 8 | 0.86 | 7 | 10 | 0.84 |
|
≥59
years | 18 | 9 | 9 | | 9 | 9 | | 8 | 10 | |
| Sex | | | | | | | | | | |
|
Male | 20 | 9 | 11 | 0.77 | 12 | 8 | 0.24 | 10 | 10 | 0.32 |
|
Female | 15 | 6 | 9 | | 6 | 9 | | 5 | 10 | |
| Smoking status | | | | | | | | | | |
|
Smoker | 17 | 7 | 10 | 0.85 | 9 | 8 | 0.86 | 7 | 10 | 0.84 |
|
Non-smoker | 18 | 8 | 10 | | 9 | 9 | | 8 | 10 | |
| Alcoholic
status | | | | | | | | | | |
|
Alcoholic | 6 | 1 | 5 | 0.15 | 3 | 3 | 0.93 | 1 | 5 | 0.20 |
|
Non-alcoholic | 29 | 14 | 15 | | 15 | 14 | | 14 | 15 | |
| Tumor size | | | | | | | | | | |
|
<5
cm | 7 | 6 | 1 | 0.027a | 6 | 1 | 0.042a | 6 | 1 | 0.02a |
|
≥5 cm | 28 | 9 | 19 | | 12 | 16 | | 9 | 19 | |
| Histopathological
differentiation | | | | | | | | | | |
|
Well | 8 | 7 | 1 | 0.011a | 7 | 1 | 0.041a | 7 | 1 | 0.01a |
|
Moderate/poor | 27 | 8 | 19 | | 11 | 16 | | 8 | 19 | |
| Lauren's
classification | | | | | | | | | | |
|
Intestinal
type | 16 | 11 | 5 | 0.005a | 11 | 5 | 0.02a | 11 | 5 | 0.005a |
|
Diffuse
type | 19 | 4 | 15 | | 6 | 13 | | 4 | 15 | |
| Lymph node
metastasis | | | | | | | | | | |
|
Negative | 10 | 8 | 2 | 0.008a | 8 | 2 | 0.032a | 8 | 2 | 0.008a |
|
Positive | 25 | 7 | 18 | | 10 | 15 | | 7 | 18 | |
| Lymphatic duct
vessel invasion | | | | | | | | | | |
|
Negative | 11 | 9 | 2 | 0.003a | 9 | 2 | 0.015a | 9 | 2 | 0.003a |
|
Positive | 24 | 6 | 18 | | 9 | 15 | | 6 | 18 | |
| Tumor stage | | | | | | | | | | |
|
Low (I and
II) | 11 | 10 | 1 | 0.0001a | 10 | 1 | 0.002a | 10 | 1 | 0.0001a |
|
High (III
and IV) | 24 | 5 | 19 | | 8 | 16 | | 5 | 19 | |
Moreover, a significant association between
infections and miRNA overexpression was observed. It was found that
patients who had one infection or co-infection with H.
pylori/EBV had a higher expression level of all three miRNAs
compared to those with no infections (P<0.05; Table II).
 | Table IIAssociation between miRNA expression
and H. pylori and EBV infections in patients with gastric
cancer. |
Table II
Association between miRNA expression
and H. pylori and EBV infections in patients with gastric
cancer.
| | miR-21
expression | | miR-223
expression | | miR-19
expression | |
|---|
| Infections
status | No. of cases | Low | High | P-value | Low | High | P-value | Low | High | P-value |
|---|
| H. pylori
status | | | | | | | | | | |
|
Infected | 11 (31.4%) | 0 | 11 | 0.001a | 1 | 10 | 0.001a | 0 | 11 | 0.001a |
|
Uninfected | 24 (68.6%) | 15 | 9 | | 17 | 7 | | 15 | 9 | |
| EBV status | | | | | | | | | | |
|
Infected | 14 (40%) | 2 | 12 | 0.005a | 4 | 10 | 0.027a | 1 | 13 | 0.001a |
|
Uninfected | 21 (60%) | 13 | 8 | | 14 | 7 | | 14 | 7 | |
| Co-infection | | | | | | | | | | |
|
Co-infected | 4 (11.4%) | 0 | 4 | 0.002a | 1 | 3 | 0.005a | 0 | 4 | 0.0003a |
|
Uninfected | 14 (40%) | 13 | 1 | | 14 | 0 | | 14 | 0 | |
miRNA expression profiles for
differentiating between GC and normal tissues
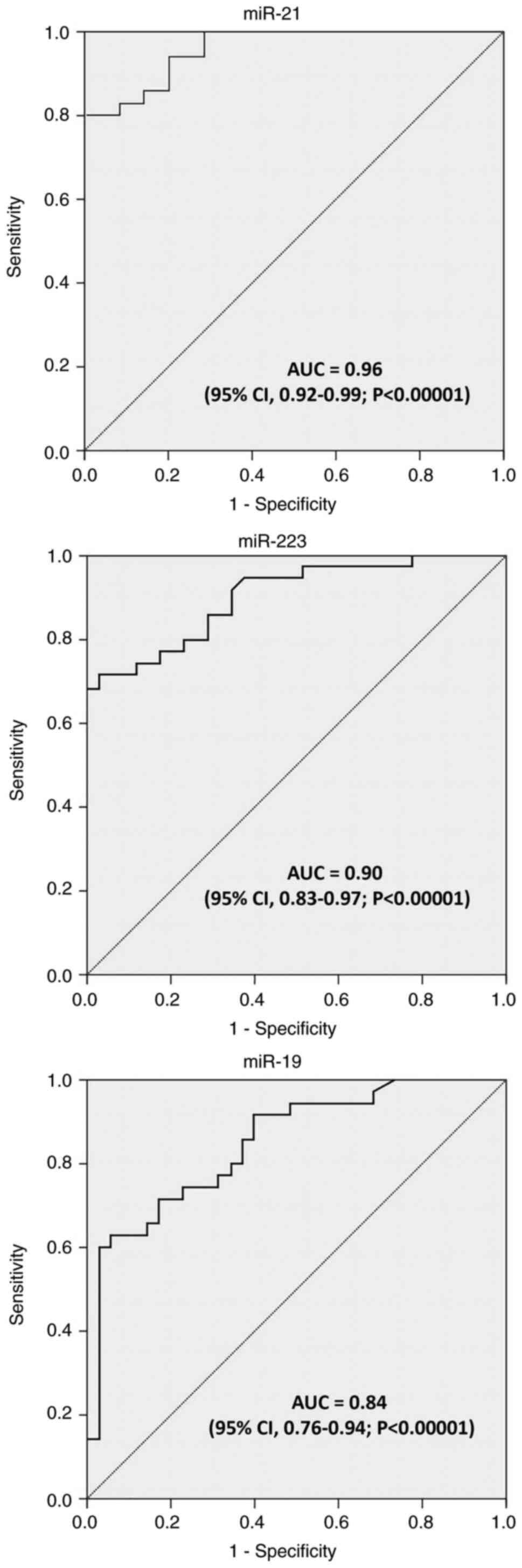
ROC curve analysis was performed on the three miRNAs
to investigate their performance as discriminatory tools for tumor
and normal tissue classification. As demonstrated in Fig. 2 and Table III, the ROC curves for the three
miRNAs indicated that these miRNAs were able to discriminate
between cancerous and non-cancerous tissues with a relatively high
sensitivity and specificity. Moreover, the sensitivity and
specificity of miR-21 were detected at 82.9 and 85.7%,
respectively, with an AU of 0.96 (95% CI, 0.92-0.99; P<0.00001),
based on the reported data (Fig.
2). As regards miR-223, it achieved a suitable diagnostic
accuracy for distinguishing GC tissues from non-cancerous tissues,
with a sensitivity of 80%, a specificity of 74.3%, and an AUC of
0.90 (95% CI, 0.83-0.97; P<0.00001). In addition, it was found
that miR-19 expression profiles had a sensitivity of 71.4% and a
specificity of 82.9% in distinguishing GC tissues from normal
non-cancerous, with an AUC of 0.84 (95% CI, 0.76-0.94;
P<0.00001) (Fig. 2).
 | Table IIIROC curves tested for specificity and
sensitivity and the AUC of miR-21, miR-223 and miR-19. |
Table III
ROC curves tested for specificity and
sensitivity and the AUC of miR-21, miR-223 and miR-19.
| miRNA | AUC | Sensitivity | Specificity | P-value | Youden index | Cut-off (ΔΔCq) |
|---|
| miR-21 | 0.96 | 82.9 | 85.7 | 0.0001 | 0.69 | 1.46 |
| miR-223 | 0.90 | 80 | 74.3 | 0.0001 | 0.54 | 2.35 |
| miR-19 | 0.84 | 71.4 | 82.9 | 0.0001 | 0.54 | 2.50 |
Discussion
Despite advancements made in the early diagnosis and
surgical techniques, the majority of patients with GC continue to
have a relatively poor prognosis. The molecular biomarkers for both
early diagnosis and prognosis prediction are urgently required.
Research has been conducted in an attempt to identify biomarkers
with diagnostic and prognostic implications for GC. miRNAs can be
critical regulators of carcinogenesis and tumor progression
(31,32). The present study evaluated three
miRNAs, including miR-21, miR-223 and miR-19 using RT-qPCR.
The results of the present study revealed that all
three miRNAs were overexpressed in tumor tissues relative to their
corresponding normal tissues, indicating that their levels of
expression may be used as suitable potential biomarkers for
detecting and distinguishing cancer from non-cancerous tissues.
Previous research has highlighted the potential role of miR-21 as
an appropriate biomarker for the early diagnosis of GC. Wang et
al (33) suggested miR-21 to
be a potential biomarker, and significant differences in expression
values were found in GC tissues. Emami et al (34) found that the miR-21 expression
levels in patients with GC were considerably higher than those in
normal subjects (P<0.001).
Previous research has also demonstrated that the
upregulation of miR-21 can alter the biological process of cancer
cells, including proliferation, apoptosis and cell invasion,
possibly by targeting reversion-inducing-cysteine-rich protein with
kazal motifs (RECK) and phosphatase and tensin homolog (PTEN) as
major tumor suppressor genes. For example, the study by Zhang et
al (35) found an inverse
association between the levels of RECK expression and miR-21, where
the increased levels of expression in miR-21 were indicative of a
decrease in the expression of RECK. It has been suggested that the
downregulation of RECK plays an essential role in GC progression
(36,37).
Other studies have revealed that F-box and WD repeat
domain containing 7 (FBXW7, also known as hCdc4), one of the major
target genes of miR-223 functions as a general tumor suppressor in
human cancer. Li et al (38) demonstrated that miR-223 functions
as an oncogene in GC by mediating the downregulation of
FBXW7/hCdc4. Another study demonstrated that miR-19 was upregulated
in GC (39). The function of
miR-19 in GC has not yet been fully defined; however, it has been
suggested that miR-19 upregulation may play a significant role in
the progression of GC (40). The
present study confirmed these previous findings, demonstrating that
miR-19 expression was upregulated in GC tissues. Notably, the
results of the present study indicated that the overexpression of
miR-21, miR-223 and miR-19 was associated with histological type,
tumor size, lymphatic invasion, the presence of lymph node
metastasis and tumor stage (P<0.05), which are the main
prognostic factors for GC. Their expression was also associated
with H. pylori and EBV infection (P<0.05).
In the study by Chang et al (41), miRNAs exhibited a differential
expression signature between H. pylori-infected and
non-H. pylori-infected gastric tissues. Riley et al
(42) found that EBV infection
affected cellular miRNA expression. Given the role of H.
pylori and EBV in the development of GC, this difference in
miRNA expression signatures may be used as a prognostic factor.
According to recent studies, miRNA expression has a specific
association with lymph node metastasis, and is a prognostic factor
for patients with GC (43,44).
In conclusion, considering the significant
differences in the expression of the three tested miRNAs (miR-21,
miR-223 and miR-19) in cancer tissues compared with normal tissues,
as well as their association with H. pylori and EBV
infections and patient clinicopathological parameters, it is
suggested that they play a crucial role in GC initiation and
progression. These miRNAs may thus be considered as effective
biomarkers for the accurate early diagnosis of GC. However, these
results are only at a preliminary stage; therefore, further studies
are warranted to extend these findings and identify their specific
roles in GC diagnosis.
Supplementary Material
Primer sequences and PCR conditions
for the detection of Helicobacter pylori and Epstein-Barr
virus infections.
TaqMan™ MicroRNA assay ID
details.
Acknowledgements
The authors would like to deeply thank the
researchers, Professor Hajri Amal, Dr Lafkih Oussama, Dr Elyamine
Othmane, Dr Bonkoukou Harouna, Dr Hajjaji Reda and Dr Boussat
Zineb, digestive cancers surgery service members in Ibn Rochd
Hospital Center of Casablanca for their contribution to this study,
for their assistance during the collection of the biopsies.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FER was involved in the conceptualization and
methodology of the study, as well as in the formal analysis and
investigation, and in the writing, review and editing of the
manuscript. DE was involved in the conceptualization of the study,
as well as in the provision of resources and in the reviewing of
the manuscript. BA was involved in the conceptualization and
methodology of the study, as well as in the reviewing and editing
of the manuscript. HC was involved in the formal analysis and
investigative aspects of the study, and in the reviewing and
editing of the manuscript. FC was involved in the conceptualization
of the study, as well as in the provision of resources and in the
reviewing of the manuscript. MME was involved in the
conceptualization and methodology of the study, as well as in the
reviewing and editing of the manuscript, and in study supervision.
FER and FC confirm the authenticity of all raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The Biomedical Research Ethics Committee of the
Faculty of Medicine and Pharmacy of Casablanca, Morocco (Reference
no. 13/19 and committee reference no. 02/DRC/00) approved the
study, and each patient signed an informed consent form.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Konturek PC, Konturek SJ and Brzozowski T:
Helicobacter pylori infection in gastric cancerogenesis. J
Physiol Pharmacol. 60:3–21. 2009.PubMed/NCBI
|
|
4
|
Thompson MP and Kurzrock R: Epstein-Barr
virus and cancer. Clin Cancer Res. 10:803–821. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fuccio L, Eusebi LH and Bazzoli F: Gastric
cancer, Helicobacter pylori infection and other risk
factors. World J Gastrointest Oncol. 2:342–347. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ali Syeda Z, Langden SSS, Munkhzul C, Lee
M and Song SJ: Regulatory mechanism of microRNA expression in
cancer. Int J Mol Sci. 21(1723)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hussen BM, Hidayat HJ, Salihi A, Sabir DK,
Taheri M and Ghafouri-Fard S: MicroRNA: A signature for cancer
progression. Biomed Pharmacother. 138(111528)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Arif KMT, Elliott EK, Haupt LM and
Griffiths LR: Regulatory mechanisms of epigenetic miRNA
relationships in human cancer and potential as therapeutic targets.
Cancers (Basel). 12(2922)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lytle JR, Yario TA and Steitz JA: Target
mRNAs are repressed as efficiently by microRNA-binding sites in the
5'UTR as in the 3'UTR. Proc Natl Acad Sci USA. 104:9667–9672.
2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Behm-Ansmant I, Rehwinkel J, Doerks T,
Stark A, Bork P and Izaurralde E: mRNA degradation by miRNAs and
GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping
complexes. Genes Dev. 20:1885–1898. 2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Catalanotto C, Cogoni C and Zardo G:
MicroRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17(1712)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Miska EA: How microRNAs control cell
division, differentiation and death. Curr Opin Genet Dev.
15:563–568. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261.
2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Petrocca F, Visone R, Onelli MR, Shah MH,
Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini
M, et al: E2F1-regulated microRNAs impair TGFbeta-dependent
cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell.
13:272–286. 2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kozomara A, Birgaoanu M and
Griffiths-Jones S: miRBase: From microRNA sequences to function.
Nucleic Acids Res. 47:D155–D162. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yuan C, Zhang Y, Tu W and Guo Y:
Integrated miRNA profiling and bioinformatics analyses reveal
upregulated miRNAs in gastric cancer. Oncol Lett. 18:1979–1988.
2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Irmak-Yazicioglu MB: Mechanisms of
microRNA deregulation and microRNA targets in gastric cancer. Oncol
Res Treatment. 39:136–139. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu L, Zhao Y, Fan G, Shuai T, Li B and Li
Y: Helicobacter pylori infection enhances heparanase leading
to cell proliferation via mitogen-activated protein kinase
signalling in human gastric cancer cells. Mol Med Rep.
18:5733–5741. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ding N, Zou Z, Sha H, Su S, Qian H, Meng
F, Chen F, Du S, Zhou S, Chen H, et al: iRGD synergizes with PD-1
knockout immunotherapy by enhancing lymphocyte infiltration in
gastric cancer. Nat Commun. 10(1336)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Prinz C, Mese K and Weber D: MicroRNA
Changes in gastric carcinogenesis: Differential dysregulation
during Helicobacter pylori and EBV Infection. Genes.
12(597)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Săsăran MO, Meliț LE and Dobru ED:
Microrna modulation of host immune response and inflammation
triggered by Helicobacter pylori. Int J Mol Sci.
22(1406)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Iizasa H, Kim H, Kartika AV, Kanehiro Y
and Yoshiyama H: Role of viral and host microRNAs in immune
regulation of Epstein-Barr virus-associated diseases. Front
Immunol. 11(367)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q,
Chong SK and Lee CH: Comparison of five PCR methods for detection
of Helicobacter pylori DNA in gastric tissues. J Clin
Microbiol. 37:772–774. 1999.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ortiz-Princz D, Guariglia-Oropeza V, Avila
M, Correnti M, Perrone M, Gutierrez B, Torres J, Megraud F and
Cavazza ME: Helicobacter pylori cagA and vacA genotypes in
Cuban and Venezuelan populations. Mem Inst Oswaldo Cruz.
105:331–335. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lu H, Zhu C, Li F, Xu W, Tao D and Feng X:
Putative periodontopathic bacteria and herpesviruses in pregnant
women: A case-control study. Sci Rep. 6(27796)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ebert MP and Röcken C: Molecular screening
of gastric cancer by proteome analysis. Eur J Gastroenterol
Hepatol. 18:847–853. 2006.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liu X, Cai H and Wang Y: Prognostic
significance of tumour markers in Chinese patients with gastric
cancer. ANZ J Surg. 84:448–453. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang D, Fan Z, Liu F and Zuo J: Hsa-miR-21
and Hsa-miR-29 in tissue as potential diagnostic and prognostic
biomarkers for gastric cancer. Cell Physiol Biochem. 37:1454–1462.
2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Emami SS, Nekouian R, Akbari A, Faraji A,
Abbasi V and Agah S: Evaluation of circulating miR-21 and miR-222
as diagnostic biomarkers for gastric cancer. J Cancer Res Ther.
15:115–119. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu
W, Xiao S and Lu H: miR-21 plays a pivotal role in gastric cancer
pathogenesis and progression. Lab Invest. 88:1358–1366.
2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Du YY, Dai DQ and Yang Z: Role of RECK
methylation in gastric cancer and its clinical significance. World
J Gastroenterol. 16:904–908. 2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhao H, Wang Y, Yang L, Jiang R and Li W:
MiR-25 promotes gastric cancer cells growth and motility by
targeting RECK. Mol Cell Biochem. 385:207–213. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Li J, Guo Y, Liang X, Sun M, Wang G, De W
and Wu W: MicroRNA-223 functions as an oncogene in human gastric
cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol.
138:763–774. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Qin S, Ai F, Ji WF, Rao W, Zhang HC and
Yao WJ: miR-19a promotes cell growth and tumorigenesis through
targeting SOCS1 in gastric cancer. Asian Pac J Cancer Prev.
14:835–840. 2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Chang H, Kim N, Park JH, Nam RH, Choi YJ,
Lee HS, Yoon H, Shin CM, Park YS, Kim JM and Lee DH: Different
microRNA expression levels in gastric cancer depending on
Helicobacter pylori infection. Gut Liver. 9:188–196.
2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Riley KJ, Rabinowitz GS, Yario TA, Luna
JM, Darnell RB and Steitz JA: EBV and human microRNAs co-target
oncogenic and apoptotic viral and human genes during latency. EMBO
J. 31:2207–2221. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zheng Q, Chen C, Guan H, Kang W and Yu C:
Prognostic role of microRNAs in human gastrointestinal cancer: A
systematic review and meta-analysis. Oncotarget. 8:46611–46623.
2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Sun X, Zhang K and Li D: Prognostic
potential of miR-21-3p in gastric cancer. J BUON. 25:2678–2682.
2020.PubMed/NCBI
|