Introduction
Obesity has increased epidemically and is currently
an important global health problem, since numerous individuals may
be classified as overweight or obese (1). Obesity is commonly assessed by
calculating the body mass index (BMI) [weight/(height)2
in kg/m2] (2). According
to the World Health Organization guidelines regarding BMI revised
for Asian populations, individuals with BMI≥23 kg/m2 are
classified as overweight, whereas those with BMI≥25
kg/m2 are defined as obese (3). Multiple genes, environmental factors
and gene-environment interactions play crucial roles in obesity and
in the tendency to gain weight. Although the maintenance of body
weight is under genetic control, mutations in a single gene rarely
result in severe obesity (4).
Previous studies reported associations between BMI or obesity and
genetic variants, suggesting that polymorphisms in the genes linked
to various pathways may contribute to the development of obesity
(5–7).
The annexin A5 gene (ANXA5), mapped to the
chromosome 4q28-q32, is 1.6-kb long and contains 13 exons that code
for a polypeptide chain of 320 amino acids (8,9). ANXA5
has also been described as placental anticoagulant protein I,
vascular anticoagulant-α, endonexin II, lipocortin V, placental
protein 4 and anchorin CII. ANXA5 is a member of the annexin family
of calcium-dependent phospholipid-binding proteins (10). In addition, a previous study by
Dennis et al (11) reported
that ANXA5 may affect triglyceride metabolism. These results
suggested that an association between ANXA5 and lipid metabolism
may be an important aspect of obesity. However, the genetic role of
ANXA5 in obesity has not been fully elucidated. To the best of our
knowledge, there is only one available association study on
ANXA5 single-nucleotide polymorphism (SNP), which reported
that the intronic SNPs (iSNPs) rs4833229 and rs6830321 were
associated with restenosis in patients who had undergone
percutaneous coronary intervention (PCI) (12). Taking into consideration that the
role of ANXA5 in coagulation is linked to triglyceride biosynthesis
(11), we hypothesized that
ANXA5 SNPs may affect the development of obesity.
To the best of our knowledge, the association
between ANXA5 polymorphisms and obesity has not been
previously investigated. The aim of this study was to investigate
whether ANXA5 SNPs were associated with obesity in a Korean
population.
Subjects and methods
Subjects
A total of 213 overweight/obese (BMI≥23
kg/m2) and 159 control (18.0<BMI<23
kg/m2) subjects were recruited from Kyung Hee University
Medical Center and Keimyung University Dongsan Medical Center. All
the subjects were ethnic Korean. Their demographic and biochemical
characteristics are shown in Table
I. DNA was isolated from peripheral blood using the G-DEX™ IIb
Genomic DNA Extraction kit (iNtRON Biotechnology, Seongnam, Korea).
Collection of the subjects was performed according to the
Declaration of Helsinki guidelines. All the subjects provided
written informed consent prior to enrollment, and informed consent
was obtained by legal guardians of the patients if they were of
minor age. This study was approved by the the ethics review
committee of the Medical Research Institute, School of Medicine,
Kyung Hee University and the Institutional Review Board of Kyung
Hee University Medical Center, Seoul, Korea.
 | Table I.Demographic and clinical
characteristics of study subjects. |
Table I.
Demographic and clinical
characteristics of study subjects.
| Variables | Control (n=159) | Overweight/obese
(n=213) | P-value |
|---|
| Age (years) | 43.43±6.08 | 44.79±6.40 | 0.042 |
| BMI
(kg/m2) | 21.15±1.21 | 25.59±2.03 | <0.001 |
| SBP (mmHg) | 115.53±16.17 | 123.89±17.57 | <0.001 |
| DBP (mmHg) | 71.88±10.37 | 77.66±11.19 | <0.001 |
| Fasting plasma
glucose (mg/dl) | 90.10±11.62 | 93.81±14.86 | <0.001 |
| HbA1C
(%) | 5.34±0.41 | 5.47±0.66 | 0.037 |
| TG (mg/dl) | 97.79±56.88 | 140.39±118.28 | <0.001 |
| TC (70 mg/dl) | 186.10±29.92 | 196.24±33.66 | 0.003 |
| LDL-C (mg/dl) | 109.08±28.92 | 118.31±31.43 | 0.004 |
| HDL-C (mg/dl) | 56.83±13.25 | 49.85±11.37 | <0.001 |
SNP selection and genotyping
Five iSNPs within the ANXA5 gene were
selected as follows: SNP tagging was performed using the tagging
option of the Tagger program (http://www.broad.mit.edu/mpg/tagger/) with known
heterozygosity and minor allele frequency >0.05 (http://www.hapmap.org). Genotyping was performed with
the Affymetrix Targeted Genotyping Chip array (Affymetrix, Santa
Clara, CA, USA) according to the manufacturer’s instructions. Each
genotyping was analyzed using GCOS software (Affymetrix).
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) was assessed
for each of the five selected SNPs using the SNPStats software
(13). The linkage disequilibrium
(LD) block of the five selected SNPs was assessed using Haploview
software version 4.1 (14).
Multiple logistic regression models were performed for the odds
ratios (ORs), 95% confidence intervals (CIs) and corresponding
P-values, controlling for age and gender as covariables (15). The Student’s unpaired t-test was
used for assessing statistical differences in age, BMI, systolic
blood pressure (SBP), diastolic blood pressure (DBP), fasting
plasma glucose, glycosylated hemoglobin (HbA1c),
triglycerides (TG), total cholesterol (TC), low-density lipoprotein
cholesterol (LDL-C) and high-density lipoprotein cholesterol
(HDL-C) between the control and the overweight/obese groups. SPSS
software version 18.0 (SPSS, Inc., Chicago, IL, USA) was used to
analyze statistical significance.
Results
Demographic and clinical characteristics
of the study subjects
The demographic and clinical characteristics, i.e.,
age, BMI, SBP, DBP, fasting plasma glucose, HbA1c, TG,
TC, LDL-C and HDL-C, were significantly different between the
control and overweight/obese subjects (P<0.05, Table I), with some of the clinical
characteristics, including BMI, SBP, DBP, fasting plasma glucose,
TG and HDL-C, exhibiting more statistically significant differences
(P<0.001) (Table I). The
genotype distributions of the five selected SNPs were in HWE
(P>0.05, data not shown).
Genotype and allele frequencies of ANXA5
SNPs
The genotype and allele frequencies of the
investigated SNPs (rs12510548, rs4240260, rs3756281, rs13136094 and
rs6534313) are provided in Table
II. The genotype frequencies of the five selected SNPs
exhibited protective effects on the development of
overweight/obesity in the codominant 2 model (rs12510548; OR=0.25,
95% CI: 0.10–0.64; P=0.004) (rs4240260; OR=0.19, 95% CI: 0.07–0.55;
P=0.002; Fisher’s exact P=0.0006) (rs3756281; OR=0.35, 95% CI:
0.15–0.82; P=0.016) (rs13136094; OR=0.21, 95% CI: 0.07–0.59;
P=0.0030; Fisher’s exact P=0.0011) (rs6534313; OR=0.17, 95% CI:
0.06–0.50; P=0.0010; Fisher’s exact P=0.0003) and recessive model
(rs12510548; OR=0.26, 95% CI: 0.11–0.64; P=0.0019) (rs4240260;
OR=0.20, 95% CI: 0.07–0.56; P=0.0007; Fisher’s exact P=0.0007)
(rs3756281; OR=0.35, 95% CI: 0.15–0.79; P=0.0094) (rs13136094;
OR=0.21, 95% CI: 0.08–0.59; P=0.0012; Fisher’s exact P=0.0013)
(rs6534313; OR=0.18, 95% CI: 0.07–0.51; P=0.0003; Fisher’s exact
P=0.0003). Four out of the five investigated SNPs (rs12510548,
rs4240260, rs13136094 and rs6534313) exhibited protective effects
in the log-additive model (rs12510548; OR=0.68, 95% CI: 0.48–0.96;
P=0.027) (rs4240260; OR=0.66, 95% CI: 0.47–0.94; P=0.020; Fisher’s
exact P=0.0019) (rs13136094; OR=0.69, 95% CI: 0.49–0.97; P=0.034;
Fisher’s exact P=0.0035) (rs6534313; OR=0.62, 95% CI: 0.44–0.88;
P=0.0075; Fisher’s exact P=0.0010) (Table II).
 | Table II.Genotype and allele frequencies of
ANXA5 SNPs in the control and overweight/obese groups. |
Table II.
Genotype and allele frequencies of
ANXA5 SNPs in the control and overweight/obese groups.
| SNP | Type | Control [n (%)] | Overweight/obese [n
(%)] | Model | OR (95% CI) | P-value | Fisher's exact P |
|---|
rs12510548
intron | G/G | 71 (44.9) | 107 (51.4) | Codominant 1 | 0.93 (0.60–1.44) | 0.74 | |
| C/G | 68 (43.0) | 94 (45.2) | Codominant 2 | 0.25 (0.10–0.64) | 0.004 | |
| C/C | 19 (12.0) | 7 (3.4) | Dominant | 0.78 (0.51–1.19) | 0.25 | |
| | | Recessive | 0.26 (0.11–0.64) | 0.0019 | |
| | | Log-additive | 0.68 (0.48–0.96) | 0.027 | |
| G | 210 (66.5) | 308 (74.0) | | | | |
| C | 106 (33.5) | 108 (26.0) | Allele | 0.69 (0.50–0.96) | 0.026 | |
rs4240260
intron | G/G | 71 (44.9) | 108 (51.7) | Codominant 1 | 0.92
(0.59–1.43) | 0.71 | |
| A/G | 69 (43.7) | 96 (45.9) | Codominant 2 | 0.19
(0.07–0.55) | 0.002 | 0.0006 |
| A/A | 18 (11.4) | 5 (2.4) | Dominant | 0.77
(0.51–1.18) | 0.23 | |
| | | Recessive | 0.20
(0.07–0.56) | 0.0007 | 0.0007 |
| | | Log-additive | 0.66
(0.47–0.94) | 0.020 | 0.0019 |
| G | 211 (66.8) | 312 (74.6) | | | | |
| A | 105 (33.2) | 106 (25.4) | Allele | 0.68
(0.50–0.94) | 0.020 | |
rs3756281
intron | A/A | 67 (43.2) | 90 (46.4) | Codominant 1 | 1.01
(0.64–1.58) | 0.98 | |
| A/G | 69 (44.5) | 95 (49.0) | Codominant 2 | 0.35
(0.15–0.82) | 0.016 | |
| G/G | 19 (12.3) | 9 (4.6) | Dominant | 0.86
(0.56–1.33) | 0.51 | |
| | | Recessive | 0.35
(0.15–0.79) | 0.0094 | |
| | | Log-additive | 0.75
(0.53–1.06) | 0.098 | |
| A | 203 (65.5) | 275 (70.9) | | | | |
| G | 107 (34.5) | 113 (29.1) | Allele | 0.78
(0.57–1.07) | 0.13 | |
rs13136094
intron | T/T | 79 (50.0) | 119 (56.4) | Codominant 1 | 0.96
(0.61–1.48) | 0.84 | |
| C/T | 62 (39.2) | 87 (41.2) | Codominant 2 | 0.21
(0.07–0.59) | 0.0030 | 0.0011 |
| C/C | 17 (10.8) | 5 (2.4) | Dominant | 0.80
(0.52–1.21) | 0.29 | |
| | | Recessive | 0.21
(0.08–0.59) | 0.0012 | 0.0013 |
| | | Log-additive | 0.69
(0.49–0.97) | 0.034 | 0.0035 |
| T | 220 (69.6) | 325 (77.0) | | | | |
| C | 96 (30.4) | 97 (23.0) | Allele | 0.68
(0.49–0.95) | 0.024 | |
rs6534313
intron | G/G | 74 (49.0) | 121 (58.2) | Codominant 1 | 0.88
(0.56–1.37) | 0.57 | |
| C/G | 59 (39.1) | 82 (39.4) | Codominant 2 | 0.17
(0.06–0.50) | 0.0010 | 0.0003 |
| C/C | 18 (11.9) | 5 (2.4) | Dominant | 0.71
(0.46–1.09) | 0.12 | |
| | | Recessive | 0.18
(0.07–0.51) | 0.0003 | 0.0003 |
| | | Log-additive | 0.62
(0.44–0.88) | 0.0075 | 0.0010 |
| G | 207 (68.5) | 324 (77.9) | | | | |
| C | 95 (31.5) | 92 (22.1) | Allele | 0.62
(0.44–0.87) | 0.005 | |
The allele frequencies of four SNPs (rs12510548,
rs4240260, rs13136094 and rs6534313) were significantly different
between the control and overweight/obese subjects (rs12510548;
OR=0.69; 95% CI: 0.50–0.96) (rs4240260; OR=0.68, 95% CI: 0.50–0.94)
(rs13136094; OR=0.68, 95% CI: 0.49–0.95) (rs6534313; OR=0.62, 95%
CI: 0.44–0.87) (Table II). Their
minor allele frequencies were ~1.3–1.4-fold lower in the
overweight/obese group compared to the control group and they
exerted protective effects on the development of
overweight/obesity.
Linkage disequilibrium and
haplotypes
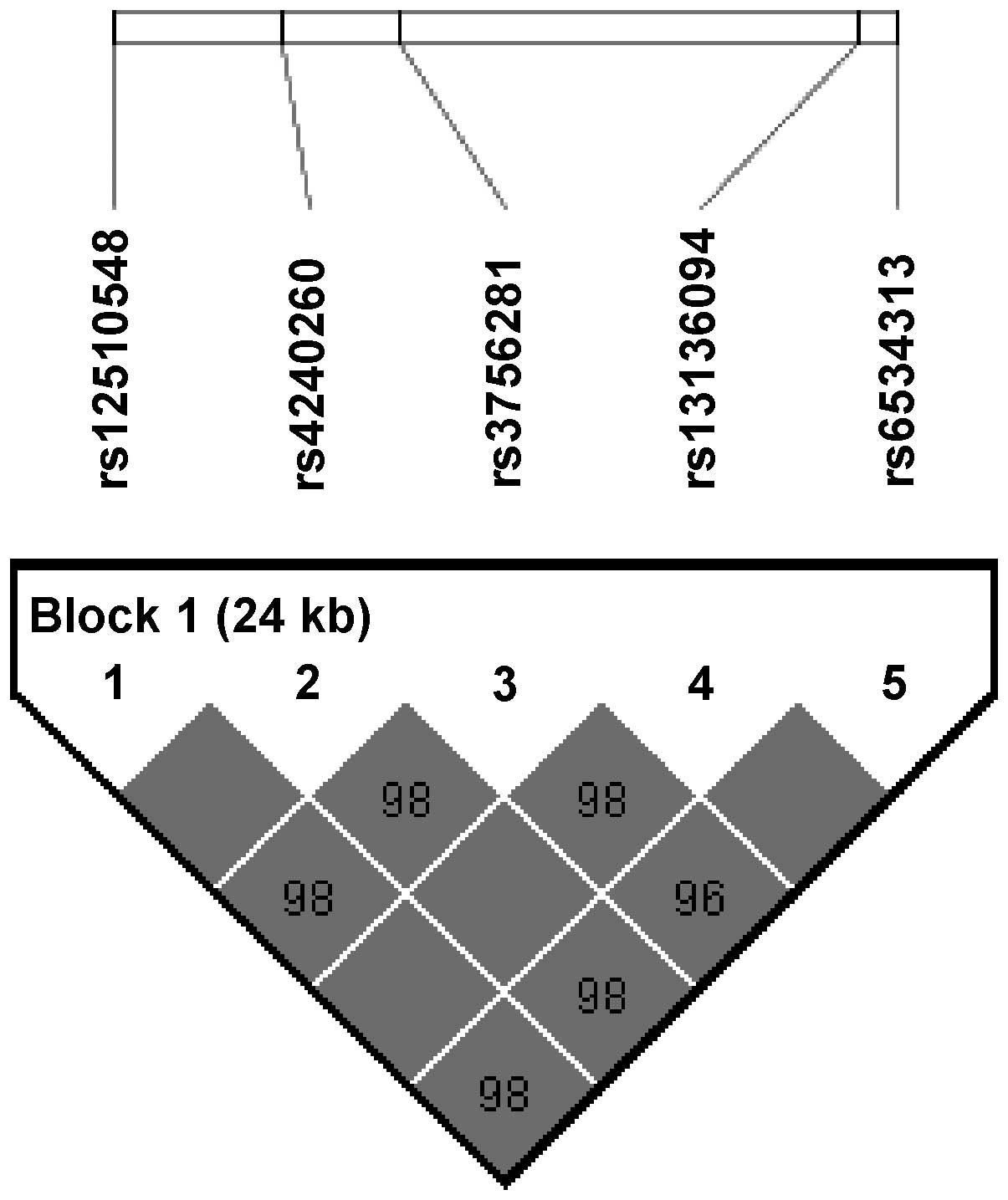
Fig. 1 shows an LD
block consisting of the five consecutive SNPs (rs12510548,
rs4240260, rs3756281, rs13136094 and rs6534313). In the LD block,
three haplotypes were formed (GGATG, CAGCC and CAGTG). The
frequencies of the GGATG, CAGCC and CAGTG haplotypes were 70.2,
25.7 and 3.6%, respectively, and the results indicated that the
GGATG (P=0.037) and CAGCC (P=0.020) haplotypes were weakly
associated with the development of obesity (Table III).
 | Table III.Haplotypes of the ANXA5 SNPs
in the control and overweight/obese groups. |
Table III.
Haplotypes of the ANXA5 SNPs
in the control and overweight/obese groups.
| Haplotype | Frequency (%) | Control
| Overweight/obese
| χ2 | P-value |
|---|
| − | + | − | + |
|---|
| GGATG | 70.2 | 107 | 209 | 114 | 312 | 4.36 | 0.037 |
| CAGCC | 25.7 | 221 | 95 | 330 | 96 | 5.38 | 0.020 |
| CAGTG | 3.6 | 306 | 10 | 409 | 17 | 0.35 | 0.55 |
Discussion
ANXA5 is an intracellular protein that is abundantly
present in endothelial cells and platelets and exhibits high
affinity for anionic phospholipids in lipid membranes (16). Based on its affinity,
fluorescently-labeled ANXA5 is often used in flow cytometric assays
to detect cells undergoing apoptosis, during which the lipid
consistency of cell membranes rapidly changes (17). ANXA5 is a potent anticoagulant that
regulates exocytosis and syncytiotrophoblast membrane fusion
(17) and expression of ANXA5 in
cancer tissues is an important factor in tumor infiltration, which
is associated with cellular energy metabolism and membrane
regulating function (18). ANXA5
protects the lipid membrane barrier against damage due to
inflammatory mediators (19) and
there is an association between inflammatory molecule levels and
visceral obesity (20). These
results suggested that ANXA5 may play a role in lipid
metabolism.
As regards ANXA5 polymorphisms, there has
been only one study on the association of ANXA5 iSNPs
(rs4833229 and rs6830321) and the restenosis rate of PCI (12). It may be considered that the role of
ANAX5 in coagulation pathways affected the therapeutic consequences
of atherosclerotic disease (12).
However, that study demonstrated a significant effect exerted by
polymorphisms in the intron region of ANXA5, although the
two SNPs involved (rs4833229 and rs6830321) were not included in
our study.
Our results demonstrated that the five SNPs of
ANXA5 included in this study (rs12510548, rs4240260,
rs3756182, rs13136094 and rs6534313) were associated with the risk
of obesity. In our haplotype analysis results, GGATG and CAGCC were
significantly associated with the risk of overweight/obesity,
suggesting that ANXA5 may be involved in the development of
obesity (21). All the significant
SNPs were intronic and unlikely to be directly protein-modifying
polymorphisms. However, iSNPs may interfere with the mRNA splicing
process and gene expression levels (22). The minor allele frequencies of
rs12510548, rs4240260, rs3756281, rs13136094 and rs6534313 in our
study population were similar to those in Japanese subjects (0.29
vs. 0.21, 0.29 vs. 0.23, 0.32 vs. 0.23, 0.26 vs. 0.22 and 0.26 vs.
0.22, respectively) in the dbSNP Build 137 of the NCBI database
(http://www.ncbi.nlm.nih.gov/SNP/).
In summary, our results suggest that there is an
association between the five iSNPs (rs12510548, rs4240260,
rs3756182, rs13136094 and rs6534313) of ANXA5 and the
development of obesity in a Korean population. To the best of our
knowledge, this is the first study demonstrating the association of
ANXA5 SNPs with the susceptibility to overweight and
obesity. Further studies are required to elucidate whether
additional ANXA5 SNPs are associated with obesity and
determine the precise role of ANXA5 in obesity in different
populations.
Acknowledgements
This study was supported by a grant
from the Kyung Hee University (KHU-20090641).
References
|
1.
|
Haslam DW and James WP: Obesity. Lancet.
366:1197–1209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Sachdev HS, Fall CH, Osmond C, et al:
Anthropometric indicators of body composition in young adults:
relation to size at birth and serial measurements of body mass
index in childhood in the New Delhi birth cohort. Am J Clin Nutr.
82:456–466. 2005.PubMed/NCBI
|
|
3.
|
WHO Expert Consultation: Appropriate
body-mass index for Asian populations and its implications for
policy and intervention strategies. Lancet. 363:157–163. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Gazzerro P, Caruso MG, Notarnicola M, et
al: Association between cannabinoid type-1 receptor polymorphism
and body mass index in a southern Italian population. Int J Obes
(Lond). 31:908–912. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Dahlman I and Arner P: Obesity and
polymorphisms in genes regulating human adipose tissue. Int J Obes
(Lond). 31:1629–1641. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Duarte SF, Francischetti EA, Genelhu-Abreu
V, et al: p.Q223R leptin receptor polymorphism associated with
obesity in Brazilian multiethnic subjects. Am J Hum Biol.
18:448–453. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Rankinen T, Zuberi A, Chagnon YC, et al:
The human obesity gene map: the 2005 update. Obesity (Silver
Spring). 14:529–644. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Martin SJ, Finucane DM, Amarante-Mendes
GP, O’Brien GA and Green DR: Phosphatidylserine externalization
during CD95-induced apoptosis of cells and cytoplasts requires
ICE/CED-3 protease activity. J Biol Chem. 271:28753–28756. 1996.
View Article : Google Scholar
|
|
9.
|
Bratton DL, Fadok VA, Richter DA, et al:
Appearance of phosphatidylserine on apoptotic cells requires
calcium-mediated nonspecific flip-flop and is enhanced by loss of
the amino-phospholipid translocase. J Biol Chem. 272:26159–26165.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Reutelingsperger CP, Hornstra G and Hemker
HC: Isolation and partial purification of a novel anticoagulant
from arteries of human umbilical cord. Eur J Biochem. 151:625–629.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Dennis MW, Downey C, Brufatto N, et al:
Prothrombinase enhancement through quantitative and qualitative
changes affecting very low density lipoprotein in complex with
C-reactive protein. Thromb Haemost. 91:522–530. 2004.PubMed/NCBI
|
|
12.
|
Ewing MM, Karper JC, Sampietro ML, et al:
Annexin A5 prevents post-interventional accelerated atherosclerosis
development in a dose-dependent fashion in mice. Atherosclerosis.
221:333–340. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Sole X, Guino E, Valls J, Iniesta R and
Moreno V: SNPStats: a web tool for the analysis of association
studies. Bioinformatics. 22:1928–1929. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Cho AR, Lee SM, Kang WS, et al: Assessment
between dopamine receptor D2 (DRD2) polymorphisms and schizophrenia
in Korean population. Clin Psychopharmacol Neurosci. 10:88–93.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Willems GM, Janssen MP, Comfurius P, et
al: Competition of annexin V and anticardiolipin antibodies for
binding to phosphatidylserine containing membranes. Biochemistry.
39:1982–1989. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Arai T, Matsubayashi H, Sugi T, et al:
Anti-annexin A5 antibodies in reproductive failures in relation to
antiphospholipid antibodies and phosphatidylserine. Am J Reprod
Immunol. 50:202–208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Deng S, Wang J, Hou L, et al: Annexin A1,
A2, A4 and A5 play important roles in breast cancer, pancreatic
cancer and laryngeal carcinoma, alone and/or synergistically. Oncol
Lett. 5:107–112. 2013.PubMed/NCBI
|
|
19.
|
Creutz CE, Hira JK, Gee VE and Eaton JM:
Protection of the membrane permeability barrier by annexins.
Biochemistry. 51:9966–9983. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Tchernof A and Despres JP: Pathophysiology
of human visceral obesity: an update. Physiol Rev. 93:359–404.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Zhu X, Zhang S, Kan D and Cooper R:
Haplotype block definition and its application. Pac Symp Biocomput.
152–163. 2004.
|
|
22.
|
Barrett LW, Fletcher S and Wilton SD:
Regulation of eukaryotic gene expression by the untranslated gene
regions and other non-coding elements. Cell Mol Life Sci.
69:3613–3634. 2012. View Article : Google Scholar : PubMed/NCBI
|