Introduction
Breast cancer is known to have a complex,
multi-factorial etiology, with contributions from genetic and
environmental factors (1–2). Certain low-penetrance genes have been
reported to be involved in breast cancer carcinogenesis (3).
Mitochondria are the major source of Reactive Oxygen
Species (ROS) (4), and are
important in cell energy production (5), by playing active roles in cell death
and cell proliferation (6).
Mitochondria may act individually or in combination with other
mitochondrial DNA variations or through interaction with nuclear
genes or environmental factors to modify cancer risk (7–9). The
10398 nucleotide position (np) in the human mitochondrial genome is
highly polymorphic (3). A10398G
polymorphism (A to G transition) results in an amino acid change
from threonine to alanine. Additionally, the A10398G polymorphism
in the nicotinamide adenine dinucleotide (NADH)
dehydrogenase subunit 3 (ND3) gene was shown to
modify the risk of certain aging-associated diseases (10–13).
The local ROS produced by redox cycling metabolites
within breast cells with an increase in cellular oxidative stress
was suggested to lead to carcinogenesis (4). The association between ND3
A10389G polymorphism and breast cancer risk has been previously
investigated (3,5,7),
however, the results obtained in this study were not always
consistent. A meta-analysis on eligible case-control studies was
conducted herein to estimate the association between A10389G
polymorphism and the risk of breast cancer.
Materials and methods
Search strategy
Searches were conducted using PubMed, Embase and the
Cochrane Library in English and VIP, CNKI, and Sinomed (up to
November 15, 2011) in Chinese. The key words and subject terms used
were: ‘ND3’ or ‘mitochondrial A10389G polymorphism’ and
‘breast cancer’. We also used the search terms: ‘ND3’ and
‘breast cancer’ and ‘genetic association’ in HuGE Navigator.
References of received articles were also searched. Articles
included in this meta-analysis were defined as: i) case-control
study (including nested case-control study); ii) not family-based
study; iii) evaluation of the relationship between A10389G
polymorphism of the ND3 gene and risk of breast cancer; and
iv) utilizing the previous study if multiple studies were based on
the same population. We excluded review articles, case reports,
editorials and information articles for patients only. Any studies
without sufficient information about the ND3 A10389G
polymorphism were also excluded.
Data extraction
Two investigators (Q.X. Mao and L.G. Gao) searched
the studies, and screened them for inclusion and appraisal.
Discrepancies were discussed with all the reviewers. Agreements
were reached after discussions. Data including author, year of
publication, country, ethnicity, study design, sample size,
resources of controls, and the information of the ND3 gene
A10398G polymorphism was collected from each publication. The
Newcastle-Ottawa-Scale (NOS) was used to quantify study quality
(14).
Statistical analysis
Unadjusted odds ratio (OR) with corresponding 95%
confidence interval (CI) of each selected study was first
calculated. The pooled OR was examined using the Z-test.
Heterogeneity among studies was measured by the Q-statistic and
I-square statistic tests. Random-effect models busing DerSimonian
and Laird method were used in the meta-analysis. Hardy-Weinberg
(H-W) equilibrium was assessed using Pearson’s Chi-square test for
the controls in each study.
Stratified analysis was performed according to
ethnicity and source of controls. Sensitivity analysis was
performed to assess the stability of the results. Potential
publication bias was assessed by Begg’s funnel plot and Egger’s
linear regression. Analyses were performed using software Stata,
version 8.0. P<0.05 was considered to indicate a statistically
significant difference. Tests were two-sided.
Results
Study characteristics and meta-analysis
database
Forty-five potentially related studies were searched
based on the search terms from the databases of PubMed, the HuGE
Navigator, Embase and the Cochrane Library in English. No related
study in Chinese was identified. One animal-related study, two
reviews, and 34 unrelated studies were excluded. Another study
concerning the ND3 genotypes was also excluded. Two studies
were based on the same population, the second (15) of which was also excluded. Therefore,
six studies (3,5,7,16–18)
including 11 study populations were included in this meta-analysis,
with a total of 5,580 patients and 5,749 controls for the
ND3 A10398G polymorphism.
A dataset was established based on the extracted
information from each included study (Table I). Cases of breast cancer were
confirmed by medical records and clinical examinations. Information
on the ND3 A10398G polymorphism was obtained via sequencing,
Taqman, allele-specific polymerase chain reaction (PCR) and
restriction fragment length polymorphism (RFLP). Quality assessment
for the eligible studies according to the NOS is shown in Table II.
 | Table ICharacteristics of studies included in
this analysis. |
Table I
Characteristics of studies included in
this analysis.
| ID | First author | Year (study) | Country | Ethnicity | Genotyping
method | Source of
controls | Sample size
(case/control) | Allele distribution
| Refs. |
|---|
| (case/control) |
|---|
| A | G |
|---|
| 1 | Fang | 2010 | China | Asian | Sequence | HB | 104/114 | 28/45 | 76/69 | 16 |
| 2 | Pezzotti | 2009 (1) | USA | Mixed | Taqman | PB | 1561/2209 | 1242/2162 | 319/447 | 17 |
| 2009 (2) | USA | Mixed | Taqman | PB | 678/669 | 529/518 | 149/151 |
| 3 | Czarnecka | 2009 | Poland | European | PCR and RFLP | PB | 44/100 | 34/97 | 10/3 | 3 |
| 4 | Setiawan | 2008 (1) | USA | African-American | Taqman | PB | 541/282 | 38/11 | 493/266 | 18 |
| 2008 (2) | USA | African-American | Taqman | PB | 391/460 | 26/29 | 391/460 |
| 2008 (3) | USA | African-American | Taqman | PB | 524/236 | 29/16 | 479/219 |
| 5 | Bai | 2007 | USA | European | AS-PCR | Not mentioned | 156/260 | 50/54 | 106/206 | 7 |
| 6 | Canter | 2005 (1) | USA | African-American | Taqman | HB | 48/54 | 7/3 | 41/51 | 5 |
| 2005 (2) | USA |
African-American | Taqman | PB | 654/605 | 84/52 | 570/553 |
| 2005 (3) | USA | European | Taqman | PB | 879/760 | 700/601 | 179/159 |
 | Table IIQuality assessment for the eligible
studies according to NOS. |
Table II
Quality assessment for the eligible
studies according to NOS.
| ID | First author | Selection
(stars) | Comparability
(stars) | Exposure
(stars) | Refs. |
|---|
| 1 | Fang | 3 | 2 | 1 | 16 |
| 2 | Pezzotti | 4 | 2 | 1 | 17 |
| 3 | Czarnecka | 4 | 2 | 1 | 3 |
| 4 | Setiawan | 4 | 2 | 1 | 18 |
| 5 | Bai | 3 | 2 | 1 | 7 |
| 6 | Canter | 3 | 2 | 1 | 5 |
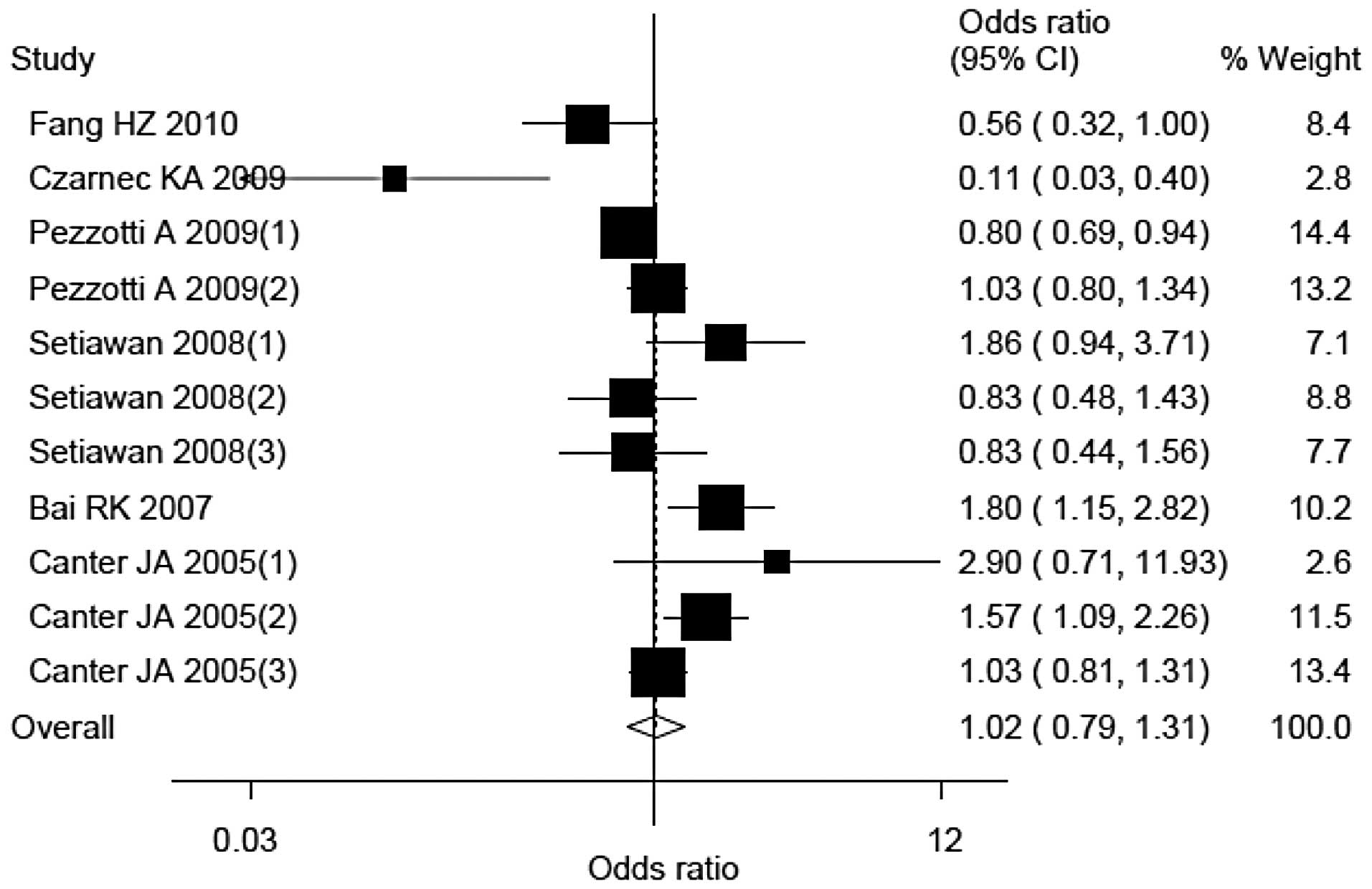
Quantitative synthesis
Findings obtained from the eligible studies
indicated that compared with individuals carrying the G allele,
individuals carrying the A allele did not exhibit an increased
breast cancer risk. The OR (95% CI) and Pheterogeneity
value were 1.02 (0.79–1.31) and <0.01, respectively (Fig. 1).
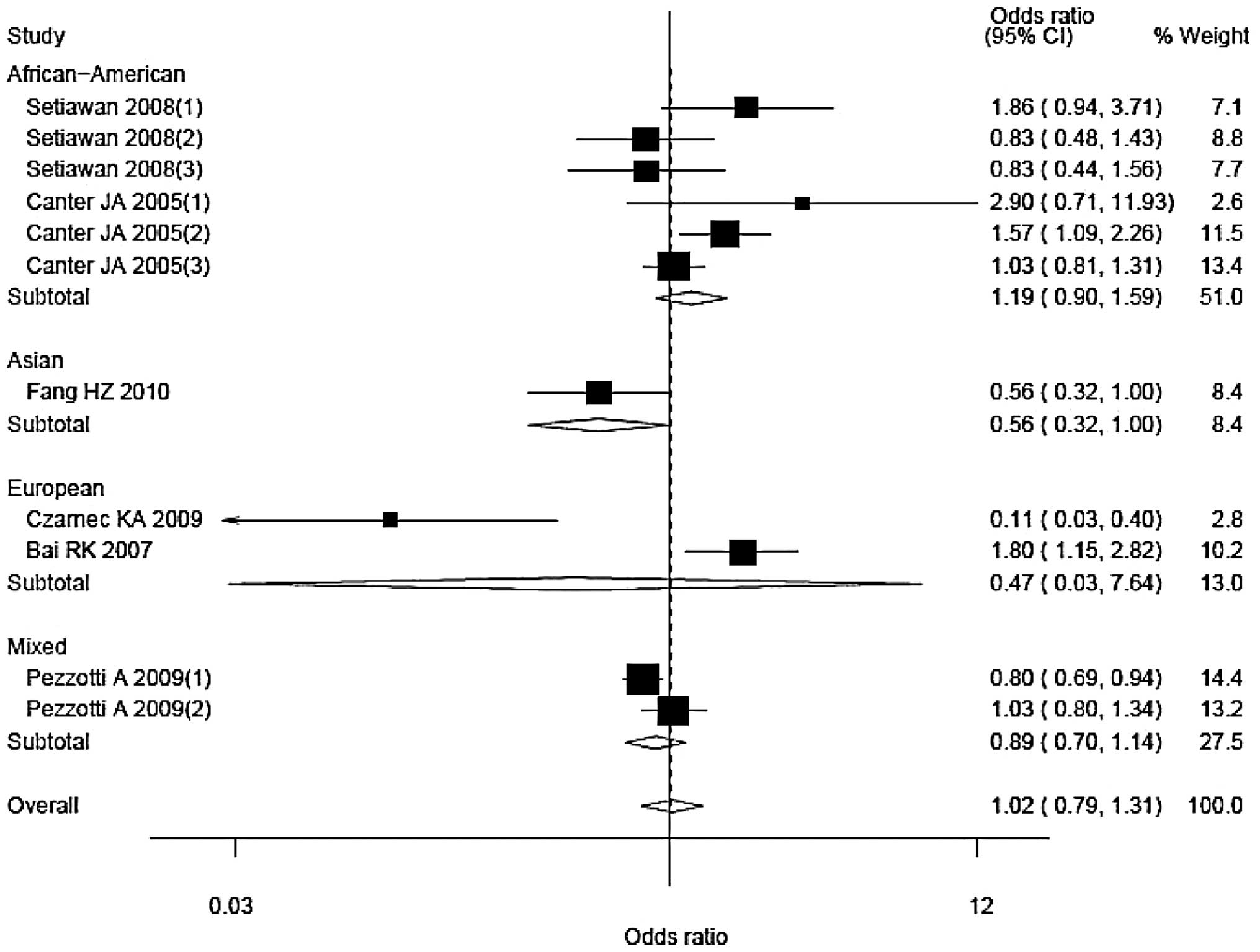
Stratified analyses were performed as per ethnicity
and source of controls (Figs. 3 and
4). The corresponding ORs (95% CIs)
and Pheterogeneity values were 1.19 (0.90–1.59), 0.09
for African-American; 0.47 (0.03–7.64), <0.01 for European; and
0.89 (0.70–1.14), 0.10 for mixed populations, respectively. There
was only one Asian study and the OR (95% CI) was 0.56 (0.32–1.00).
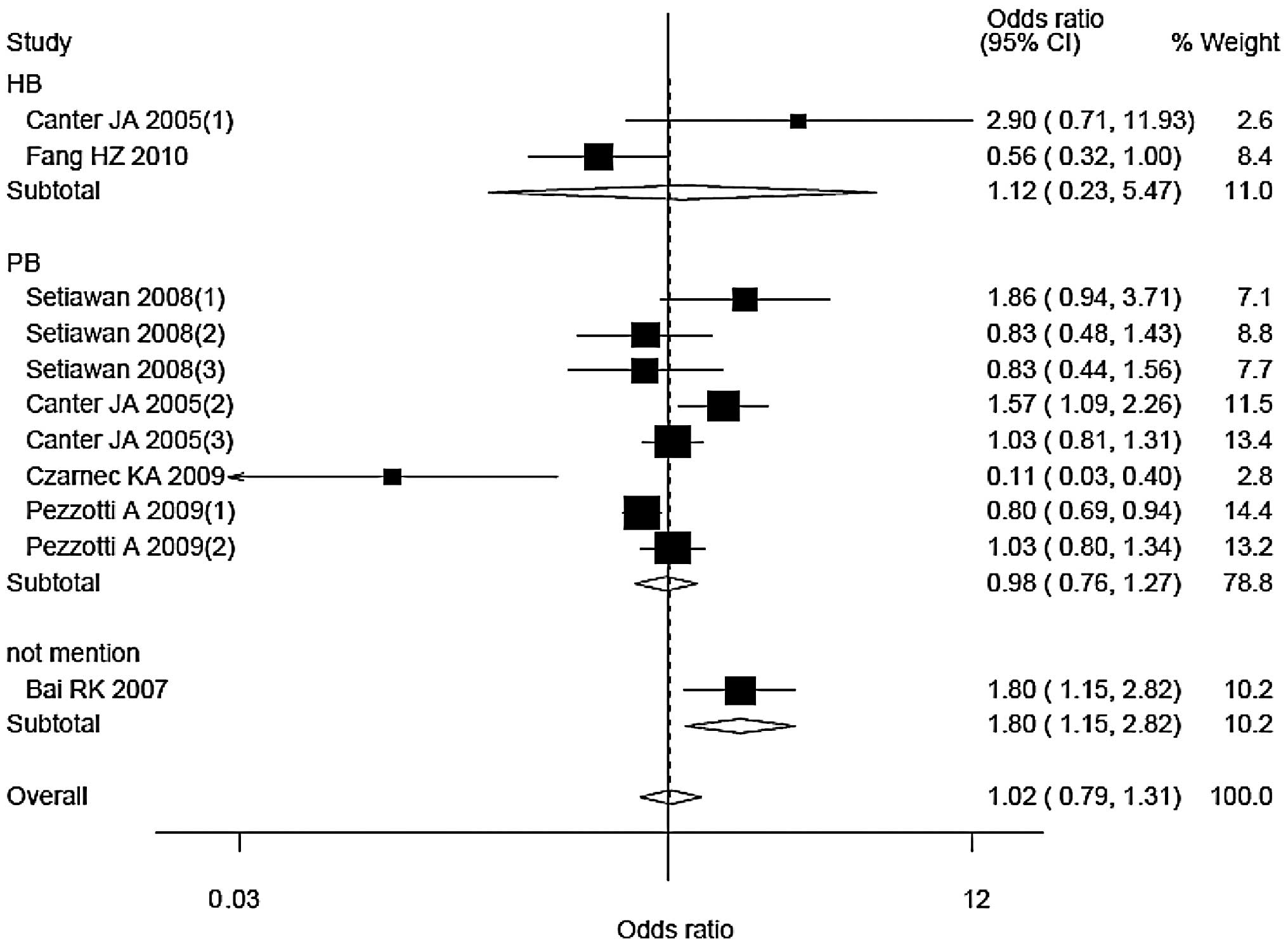
The corresponding ORs (95% CIs) and Pheterogeneity
values were 1.12 (0.23–5.47), 0.04 for hospital-based studies; and
0.98 (0.76–1.27), <0.01 for population-based studies. Only one
study did not mention the source of control, with an OR (95% CI) of
1.80 (1.15–2.82).
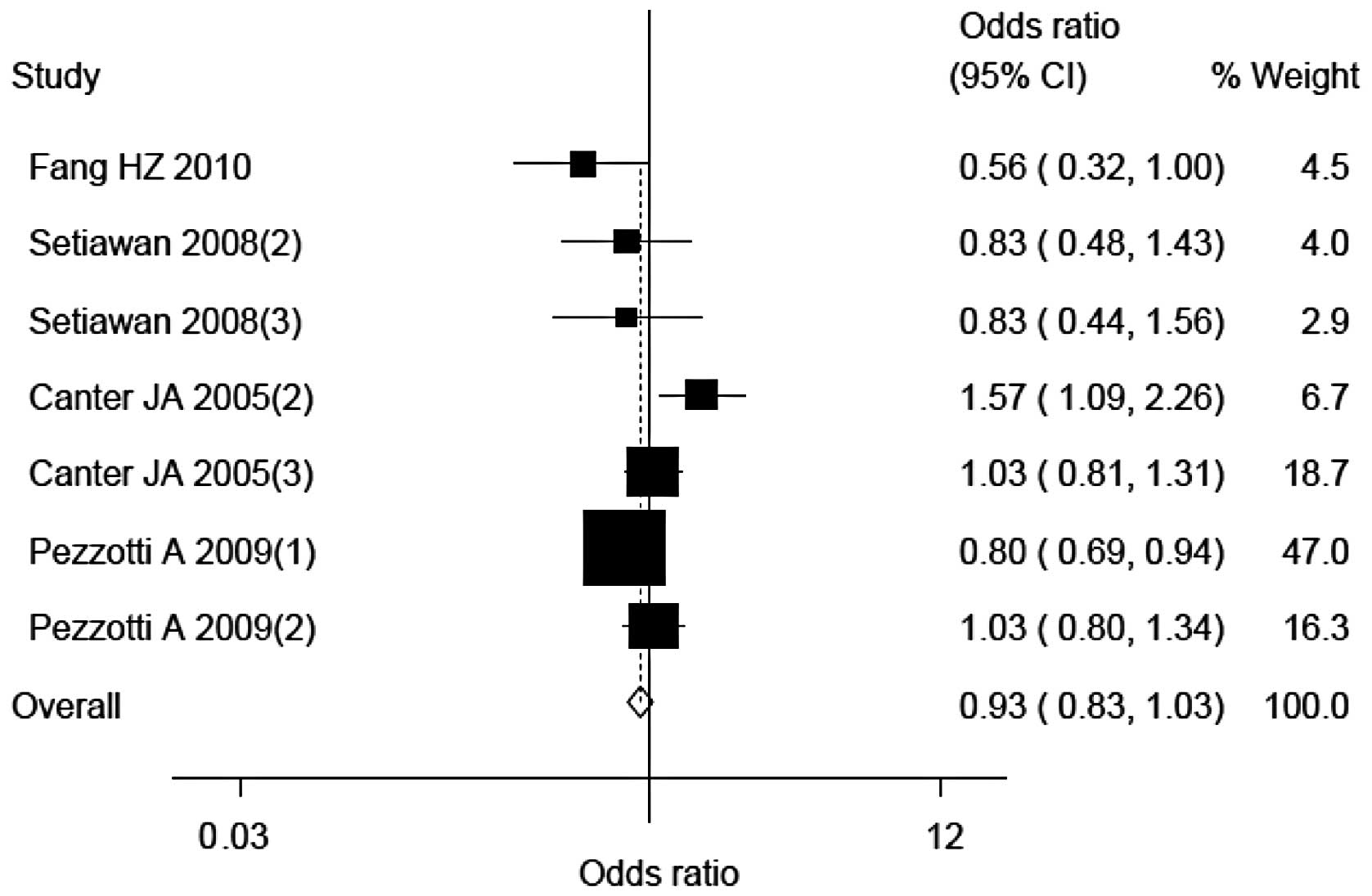
Sensitivity analysis
Four study populations [by Czarnecka et al
(3), Setiawan et al (one
study; 18), Bai et al (7)
and Canter et al (one study; 5)] were the main origin of
heterogeneity of this meta-analysis. The heterogeneity was
decreased following the exclusion of the four studies, with
P-values based on the heterogeneity test of 0.02. The corresponding
OR (95% CI) were altered to 0.93 (0.83–1.03), respectively
(Fig. 5).
Publication bias
Begg’s funnel plots and Egger’s tests were conducted
to determine publication bias. No publication bias was identified
for A10389G polymorphism: P=0.94 (Fig.
2).
Discussion
This meta-analysis included a total of 5,580
patients and 5,749 controls from six eligible studies (11 study
populations).
The 10398 np is highly polymorphic in the human
mitochondrial genome (3). A10398G
is probably one of the most thoroughly studied mitochondrial single
nucleotide polymorphisms, particularly for its potential effect on
tumorigenesis, and it serves as a breast cancer predisposition
factor (16,19). In a study based on a regional
Chinese population, with a total of 104 cases and 114 controls, G
allele was found to increase the risk of breast cancer, with an OR
(95% CI) of 1.77 (1.00–3.14) (16).
In the study by Bai et al (7), the 10398G allele was also shown to
increase risk of breast cancer in European-American women (7). However, these results were not always
consistent. Canter et al (5)
detected an inverse association between A10398G polymorphism and
the risk of breast cancer (5).
African-American women carrying the 10398A allele had a
significantly increased risk of breast cancer (OR, 1.60; 95% CI,
1.10–2.31). However, no statistically significant correlation was
identified in Caucasian women. Of note, such a statistically
significant association between A10398G polymorphism and the risk
of breast cancer was not always duplicated in other studies.
Pezzotti et al (17)
reported that the frequencies of the G allele among cases and
controls from both the Nurses’ Health Study (NHS) population
(including 1,561 cases and 2,209 controls) and Women’s Health Study
(including 678 cases and 669 controls) were similar to each other.
Findings of that study also demonstrated that alcohol consumers
carrying the 10398G allele had a higher risk of breast cancer as
compared to non-drinkers among the NHS population. However,
Setiawan et al (18)
observed that G10398A was not significantly associated with breast
cancer risk among African-American women. Similarly, no association
was detected in three study populations including the San Francisco
Bay Area case-control study comprising 541 cases and 282 controls,
a nested case-control study based on the Multiethnic Cohort Study
comprising 391 cases and 460 controls and a case-control study
based on the Los Angeles component of the Women’s Contraceptive and
Respective Experiences Study and the LIFE study comprising 524
cases and 236 controls. For our meta-analysis, the results also
support that individuals carrying A allele did not exhibit an
increased risk of breast cancer as per the eligible studies, when
compared with individuals carrying the G allele. Even in stratified
analyses, no statistically significant relationship between A10398G
genotypes and breast cancer risk was detected.
There are limitations to our meta-analysis that
should be considered when interpreting the results. First, since
the original data of the reviewed studies were lacking, only
unadjusted estimates were assessed and the potential effect on
breast cancer risk from other possible variables including age and
family history could not be evaluated. At the same time, our
evaluation of potential interactions of gene-gene, gene-environment
or different polymorphic loci in the same gene that may affect
breast cancer risk was limited. Second, the number of studies
included in this analysis were not sufficiently large for a
comprehensive analysis. Despite the limitations, however, our
meta-analysis has important advantages. First, the substantial
number of cases and controls included in this meta-analysis
significantly increased the statistical power of the analysis as
compared to any individual study. Second, the quality of the
case-control studies included in the current pooling analysis was
satisfactory and met the inclusion criterion.
In conclusion, our meta-analysis shows that
ND3 gene A10398G polymorphism might not statistically
modulate breast cancer risk. Although a systematic investigation of
the relationship between A10398G polymorphisms and the risk of
breast cancer could not be conducted because of the aforementioned
limitations, it is important to gain a better understanding of the
effect of A10398G polymorphisms on breast cancer risk. At the same
time, we assume that studies using standardized unbiased methods,
enrolling precisely defined breast cancer cases and healthy
controls, with more detailed individual data are needed.
Furthermore, more and larger studies, particularly studies
stratified by gene-gene and gene-environmental interactions, should
be performed to clarify the possible roles of the ND3
A10398G polymorphism in the etiology of breast cancer.
Acknowledgements
The study was supported by the
National Nature Science Foundation for Young Scientists of China
(grant no. 81102142), the National Research Institute for Family
Planning (grant no. 2010GJSSJKA10) and the General Financial Grant
from the China Postdoctoral Science Foundation (grant no. 2011M
500154).
References
|
1
|
Olopade OI, Grushko TA, Nanda R and Huo D:
Advances in breast cancer: pathways to personalized medicine. Clin
Cancer Res. 14:7988–7999. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rohan TE, Wong LJ, Wang T, et al: Do
alterations in mitochondrial DNA play a role in breast
carcinogenesis? J Oncol. 2010:6043042010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Czarnecka AM, Krawczyk T, Zdrozny M, et
al: Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism
(A10398G) and sporadic breast cancer in Poland. Breast Cancer Res
Treat. 121:511–518. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Darvishi K, Sharma S, Bhat AK, et al:
Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup
N a risk for breast and esophageal cancer. Cancer Lett.
249:249–255. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Canter JA, Kallianpur AR, Parl FF and
Millikan RC: Mitochondrial DNA G10398A polymorphism and invasive
breast cancer in African-American women. Cancer Res. 65:8028–8033.
2005.
|
|
6
|
Wallace DC: Mitochondria as chi. Genetics.
179:727–735. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bai RK, Leal SM, Covarrubias D, et al:
Mitochondrial genetic background modifies breast cancer risk.
Cancer Res. 67:4687–4694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aikhionbare FO, Khan M, Carey D, et al: Is
cumulative frequency of mitochondrial DNA variants a biomarker for
colorectal tumor progression? Mol Cancer. 3:302004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Datta S, Majumder M, Biswas NK, et al:
Increased risk of oral cancer in relation to common Indian
mitochondrial polymorphisms and autosomal GSTP1 locus. Cancer.
110:1991–1999. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van der Walt JM, Nicodemus KK, Martin ER,
et al: Mitochondrial polymorphisms significantly reduce the risk of
Parkinson disease. Am J Hum Genet. 72:804–811. 2003.PubMed/NCBI
|
|
11
|
van der Walt JM, Dementieva YA, Martin ER,
et al: Analysis of European mitochondrial haplogroups with
Alzheimer disease risk. Neurosci Lett. 365:28–32. 2004.PubMed/NCBI
|
|
12
|
Mancuso M, Conforti FL, Rocchi A, et al:
Could mitochondrial haplogroups play a role in sporadic amyotrophic
lateral sclerosis? Neurosci Lett. 371:158–162. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giacchetti M, Monticelli A, De Biase I, et
al: Mitochondrial DNA haplogroups influence the Friedreich’s ataxia
phenotype. J Med Genet. 41:293–295. 2004.PubMed/NCBI
|
|
14
|
Wells GA, Shea B, O’Connell D, et al: The
Newcastle-Ottawa Scale (NOS) for assessing the quality of
nonrandomised studies in meta-analyses. Available from: URL:
http://www.ohri.ca/programs/clinical_epidemiology/oxford.aspuri.
Accessed on November 26, 2011.
|
|
15
|
Covarrubias D, Bai RK, Wong LJ and Leal
SM: Mitochondrial DNA variant interactions modify breast cancer
risk. J Hum Genet. 53:924–928. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang H, Shen L, Chen T, et al: Cancer
type-specific modulation of mitochondrial haplogroups in breast,
colorectal and thyroid cancer. BMC Cancer. 10:4212010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pezzotti A, Kraft P, Hankinson SE, et al:
The mitochondrial A10398G polymorphism, interaction with alcohol
consumption, and breast cancer risk. PLoS One. 4:e53562009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Setiawan VW, Chu LH, John EM, et al:
Mitochondrial DNA G10398A variant is not associated with breast
cancer in African-American women. Cancer Genet Cytogenet.
181:16–19. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brandon M, Baldi P and Wallace DC:
Mitochondrial mutations in cancer. Oncogene. 25:4647–4662. 2006.
View Article : Google Scholar
|