Introduction
Small interfering RNAs (siRNAs) are intermediate
products in RNA interference (RNAi) pathways that can cause target
gene silencing via RNAi pathways (1). Traditional siRNA delivery vehicles
fail to address the instability and poor bioavailability of siRNAs
and are associated with low delivery efficiency, poor uptake by
target cells and immunotoxicity for protein carriers. Effective
siRNA delivery to target systems without causing damage to normal
cells or tissue remains the biggest challenge for in vivo
studies (2). Aptamers are
oligonucleotides (single-stranded DNA or RNA) that bind to target
molecules with a high affinity and good specificity and can be
obtained using the systematic evolution of ligands by exponential
enrichment (SELEX) system (3). This
technique adopts random oligonucleotide libraries of
1013–1015 molecules and exponentially
enriches oligonucleotides that bind to the target molecule
specifically via polymerase chain reaction amplification. This
technique, when used to select aptamers by targeting living cells,
is known as cell-SELEX (4), which
offers more advantages over protein-based SELEX. Cell-SELEX
obviates the requirement of protein purification and
immobilization, allows target molecules to maintain their natural
spatial structure and keeps protein glycosylation sites on the cell
surface in a natural state. Aptamers obtained in this way can bind
target molecules in their natural state and can be directly used
for cell identification and detection.
Glioblastoma multiforme (GBM) is the most common and
aggressive human malignant brain tumor, accounting for 50% of solid
brain tumors. A common feature of patients with GBM is the presence
of an increased expression and mutation of human epidermal growth
factor receptor (EGFR), with the most common mutant being EGFR
variant III (EGFRvIII). EGFRvIII is produced from the deletion of
801 base pairs in exons 2–7 of the extracellular domain of EGFR and
is closely associated with the progression of GBM and its
resistance to radiotherapy and chemotherapy. Varying degrees of a
high EGFRvIII expression are found in other malignancies, including
non-small cell lung cancer and colorectal cancer. However, EGFRvIII
is not expressed in normal tissue (5). Therefore, EGFRvIII can be harnessed as
a specific target for tumor therapy. c-Met is encoded by
proto-oncogene c-Met and is a tyrosine kinase receptor of
hepatocyte growth factor (HGF). In GBM patients, overexpression of
c-Met often leads to poor prognosis, and the expression of c-Met is
significantly higher in recurrent GBM compared with primary GBM
(6).
Thus far, aptamers that have been designed for siRNA
delivery are mainly aimed at prostate cancer (7–9), AIDS
(10–12) and breast cancer (13). We have previously studied the
selection of high-affinity and high-specificity aptamers targeting
a U87MG glioma cell line with EGFRvIII overexpression
(U87-EGFRvIII). One of these aptamers, designated as 32, was
internalized and located in the nucleus of U87-EGFRvIII cells
(14). In the present study,
aptamer 32 was biotinylated and coupled to biotin-labeled c-Met
siRNA via streptavidin, and this delivered c-Met siRNA into
U87-EGFRvIII cells in a targeted manner. Subsequently, significant
changes in the c-Met protein expression, apoptosis and
proliferation of U87-EGFRvIII cells were observed.
Materials and methods
Aptamer and siRNA
The 5′-biotin-labeled aptamer 32, termed BA, was
synthesized by Invitrogen Life Technologies (Carlsbad, CA, USA).
The sequence was as follows: biotin-5′-GCAATG
GTACGGTACTTCCTGAATGTTGTTTTTTCTCTTT
TCTCTATAGTACAAAAGTGCACGCTACTTTGCTAA-3′. The c-Met siRNA sequence
(synthesized by Ribo Biotech Co. Ltd., Guangzhou, Guangdong, China)
was 5′-AGC CAAUUUAUCAGGAGGUTT-3′ (sense); and 5′-ACCUCC
UGAUAAAUUGGCUTT-3′ (antisense). The negative control siRNA was
Cy3-labeled siR-Ribo™ Transfection Control (siN05815122144).
Cell culture
U87MG cells were cultured in Dulbecco’s modified
Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 100
U/ml penicillin and 100 μg/ml streptomycin (P/S). The U87-EGFRvIII
cell line (15) (obtained from Dr
Webster Cavenee, Ludwig Cancer Institute, San Diego, CA, USA) was
grown in DMEM supplemented with 10% FBS, 1% P/S and 100 mg/ml
geneticin. The cells were cultured at 37°C in a 5%
CO2/95% air environment.
Lipofectamine 2000 transfection and
BA-mediated delivery
The U87-EGFRvIII cells were seeded in 12-well plates
at a density of 1×105 cells/well. The following day, the
cells were harvested for transfection at 30–50% confluence. The
media were switched to 1 ml of serum-free, antibiotic-free media
prior to transfection. A total of 5 μl/well of 20 μM BA and 5
μl/well of 20 μM biotin-labeled c-Met siRNA were added into 100 μl
of Opti-MEM (Gibco-BRL, Carlsbad, CA, USA) respectively, gently
pipetted 3–5 times and allowed to stand for 5 min. Subsequently,
the two solutions were mixed with 2.5 μl/well of 20 μM streptavidin
(Sigma-Aldrich, St. Louis, MO, USA) and left to stand for 20 min
prior to being added to the culture media. The transfection system
was 200 μl, and the final concentration of BA and siRNA was 100 nM.
A total of 5 μl/well of liposome (Invitrogen Life Technologies) and
5 μl/well of c-Met siRNA plus 2.5 μl/well of streptavidin served as
a positive control and underwent the same aforementioned
procedures. BA + control siRNA + streptavidin and liposome +
control siRNA + streptavidin were used as negative controls for BA
and liposome, respectively. Separate groups exposed to BA or
streptavidin alone were provided to exclude the interference from
BA or streptavidin, respectively. Following transfection, the cells
were cultured in an incubator (37°C, 5% CO2) for 6 h,
following which the cells were cultured for another 72 h in DMEM
containing 10% FBS.
Western blotting
Cells were washed with phosphate-buffered saline
(PBS) 72 h after transfection and lysed in cell lysis buffer
containing 1% protease inhibitor. The protein content was measured
using the bicinchoninic acid method. Following denaturation, the
samples (10 μg/well) were subjected to 8% SDS-PAGE, transferred to
a polyvinylidene fluoride membrane, and incubated with rabbit
anti-human c-Met (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and rabbit anti-human GAPDH antibodies (Santa Cruz
Biotechnology, Inc.) at 4°C overnight prior to exposure to
horseradish peroxidase-labeled secondary antibody at room
temperature for 1 h. Target proteins were visualized using enhanced
chemiluminescence detection and analyzed with Fluorchem SP imaging
system (Alpha Innotech Corporation, San Leandro, CA, USA).
Annexin V-fluorescein
isothiocyanate/propidium iodine (PI) assay
The cells were washed with PBS and digested with
0.25% EDTA-free trypsin 72 h after transfection. Following
termination of the digestion, the cell suspension was transferred
to a 1.5 ml Eppendorf tube and centrifuged at 100 × g for 5 min.
The supernatant was discarded, while the cell pellets were
resuspended in PBS (500 μl/tube) and centrifuged as aforementioned.
Following the addition of 200 μl detection buffer to each tube, 2
μl of Annexin V and 2 μl of PI (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, Jiangsu, China) were added in sequence. The resultant
mixture was incubated on ice in the dark for 10 min before
performing flow cytometry to detect the percentage of apoptotic
cells. To prove the targeting specificity of BA for U87-EGFRvIII
cells, U87MG cells were transfected as control cells in this
experiment and for the MTT assays.
MTT colorimetric assay
The cells were seeded in 24-well plates at a density
of 5×104 cells/well, with each well containing 500 μl of
cell suspension. The cells were switched to serum-free medium, 72 h
after transfection. Subsequently, 50 μl of MTT (5 mg/ml) was added
to each well and was incubated at 37°C for 4 h, the supernatant was
discarded, while the cell pellets were exposed to 500 μl of
dimethyl sulfoxide and incubated at 37°C for 15 min. Optical
absorbance (OD) at 570 nm was measured on a microplate reader.
Statistical analysis
Data were presented as the means ± SD. One-way
analysis of variance was performed using SPSS 13.0 statistical
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
BA-mediated delivery of c-Met siRNA into
U87-EGFRvIII cells downregulates c-Met protein expression
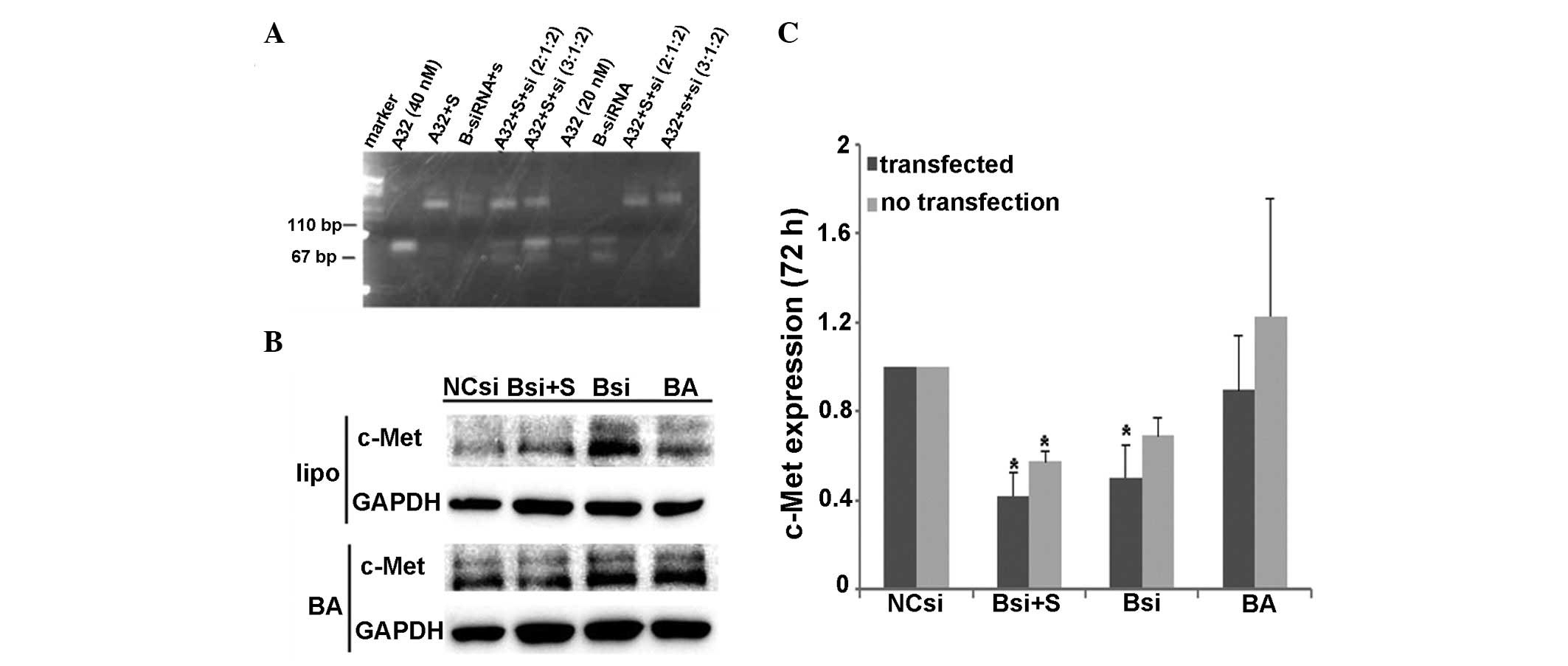
An agarose gel electrophoresis analysis was
performed to identify the amount of BA, c-Met siRNA and
streptavidin used for conjugation. When the final ratio for BA,
streptavidin and siRNA was 2:1:2, a single and distinct band was
obversed (Fig. 1A), suggesting that
it was the optimal ratio for BA, streptavidin and siRNA
conjugation. Western blotting results showed that 72 h after
transfection, with a c-Met siRNA final concentration of 100 nM, the
c-Met protein expression in the liposome- and BA-mediated groups
was significantly reduced compared with cells transfected with
control siRNA (0.418±0.18, P=0.026; 0.57±0.09, P=0.039) (Fig. 1B–C). The presence of streptavidin or
BA alone had no effect on the protein levels of c-Met. The
liposome-mediated siRNA delivery downregulated the target gene
expression by >50%. Similarly, BA-mediated siRNA delivery
reduced the target gene expression by ~50%. The results showed that
BA mediated the delivery of siRNA into U87-EGFRvIII cells, leading
to target gene silencing.
c-Met gene silencing as a result of
BA-mediated c-Met siRNA delivery increases apoptosis of glioma
cells
The percentage of apoptotic glioma cells was
determined using flow cytometry. The results showed that in the
U87-EGFRvIII cells, the liposome- and BA-mediated groups had an
apoptotic rate of 26.47±7.49 and 23.23±10.14%, respectively,
revealing a significant increase over their respective negative
control group (P=0.011 and P=0.025). There was no difference in the
apoptotic rate between the liposome- and BA-mediated groups
(P=0.573). However, in the U87MG cells, the apoptotic rate of the
liposome-mediated group (19.1±5.15%) increased significantly
compared with that of its control group (P=0.004) and differed
significantly from the BA-mediated group (8.33±3.3%) (P=0.006). The
apoptotic rate of the BA-mediated group exhibited no difference
from its control group (P=0.991) (Fig.
2). These results indicated that BA specifically mediated the
delivery of c-Met siRNA into U87-EGFRvIII cells but not U87MG
cells, and therefore, reduced c-Met gene expression and induced
U87-EGFRvIII apoptosis.
c-Met gene silencing as a result of
BA-mediated c-Met siRNA delivery decreases the proliferation of
glioma cells
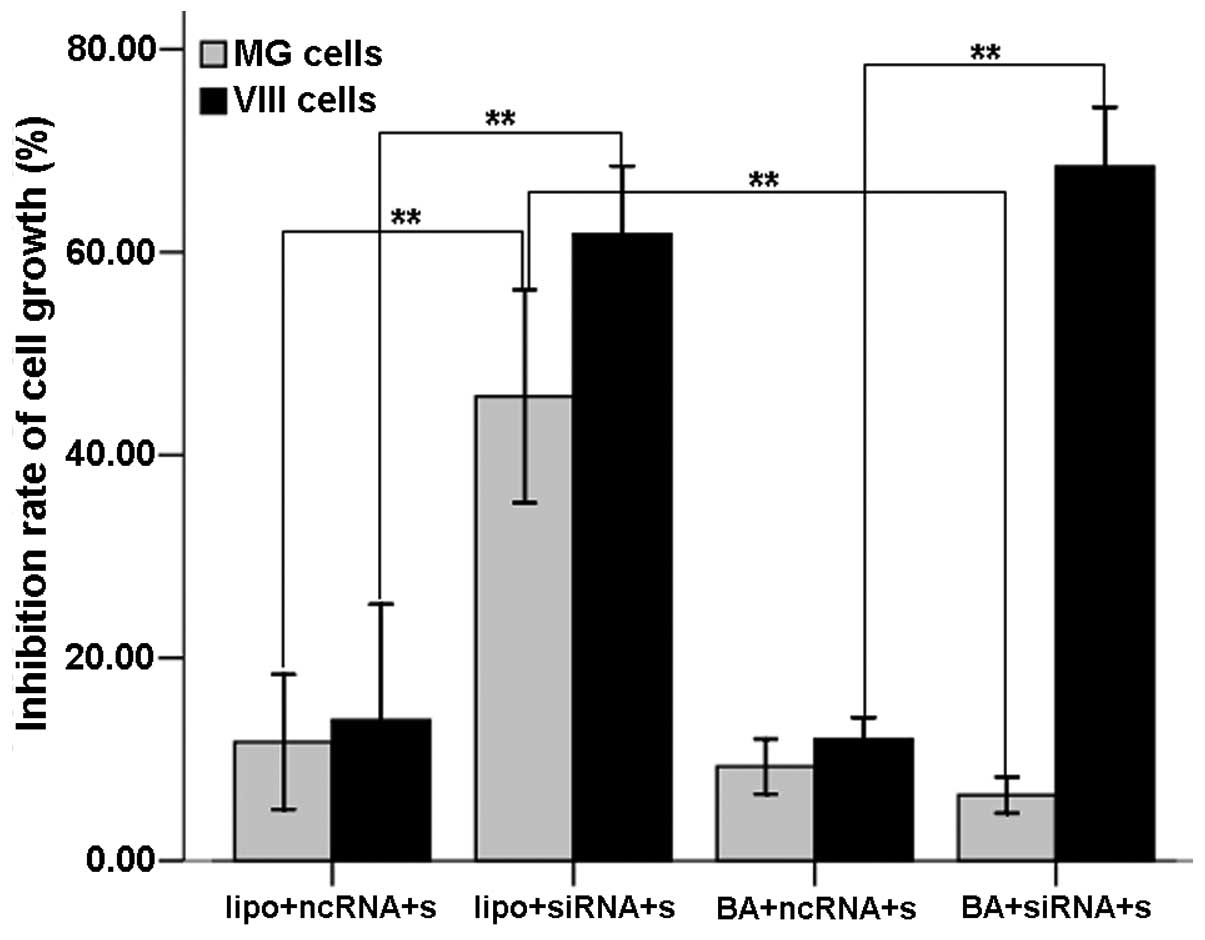
The OD values of each group were determined with the
MTT assays. Similar to the flow cytometry results, in the
U87-EGFRvIII cells the inhibition rate of the liposome- and
BA-mediated groups was 61.77±6.73 and 68.43±5.87%, respectively,
showing a significant increase compared with that of the respective
control groups (P<0.01 and P<0.01). The inhibition rate did
not differ between the two groups (P=0.762). In the U87MG cells,
the inhibition rate of the liposome-mediated group (45.78±10.53%)
increased significantly compared with its control group and the
BA-mediated group (6.46±1.78%) (P<0.01 and P<0.01,
respectively). The inhibition rate of the BA-mediated group showed
no difference from the control group (P=0.971) (Fig. 3). These findings demonstrated that
the BA-mediated c-Met siRNA delivery into U87-EGFRvIII cells
suppressed U87-EGFRvIII cell proliferation while producing no
impact on that of U87MG cells.
Discussion
GBM patients have extremely poor prognosis and short
median survival rates (16).
EGFR-targeting protein tyrosine kinase inhibitors are highly
immunogenic and are associated with various degrees of resistance,
leading to the failure of cancer chemotherapy. The type and level
of biomarkers varies in different types of tumors with different
degrees of differentiation and location. Aptamers selected against
living cells by cell-SELEX can recognize specific cell surface
receptors and can therefore be used to identify tumor cell surface
biomarkers (4,17). Following binding to surface proteins
on target cells, certain aptamers are internalized within cells
(18–20). The advantages of aptamers over
conventional antibodies include low molecular weight, rapid
synthesis in vitro, low toxicity, low immunogenicity, good
tissue permeability and ease of chemical modification, which
minimizes degradation and enhances pharmacokinetic parameters in
vivo. There are two key issues in the application of RNAi in
the clinical trials of human diseases. The first issue is an
effective means for siRNA delivery, and the second issue is the
specificity of siRNA-targeted delivery and the safety of the
delivery vehicle in humans. The aforementioned characteristics of
aptamers make them promising siRNA delivery vehicles (21). An aptamer for human epidermal growth
factor receptor 2 (HER2) can carry siRNA into HER2-expressing
breast cancer cells to mediate target gene silencing (13). Aptamers targeting human
immunodeficiency virus (HIV) glycoprotein 120 and cluster of
differentiation 4 can also mediate the delivery of their
corresponding siRNAs into target cells in the form of aptamer-siRNA
chimeras, thereby inhibiting HIV-1 viral replication (10,11).
In the present study, an aptamer obtained through a cell-SELEX
procedure mediated the delivery of siRNA into glioma cells with
EGFRvIII overexpression, specifically silenced target genes and
modulated the apoptosis and proliferation of U87-EGFRvIII cells.
This aptamer did not affect the survival rate of U87MG cells as it
did not bind and enter the U87MG cells. In a previous report, we
proved that EGFRvIII is the target receptor of BA (14), and we therefore hypothesized that
receptor-mediated endocytosis is a possible mechanism for siRNA
delivery. The present study provides evidence for the feasibility
of aptamers as siRNA delivery vehicles.
Abnormal HGF/c-Met signals can lead to the
stimulation of tumor cell functions, including cell proliferation,
survival, cell dispersion and movement, epithelial to mesenchymal
transition, angiogenesis, invasion and metastasis (22,23).
HGF/c-Met overexpression and EGFRvIII high expression in gliomas
are closely associated with the development and progression of
gliomas, complicating the diagnosis and treatment of gliomas. The
synergy among these molecules attenuates the response of gliomas to
any single chemotherapeutic agent and tends to induce drug
resistance as a result. It has been reported that the
overexpression of c-Met can activate HER3 and cause resistance to
the chemotherapeutic agent, fibrate gemfibrozil, which is an EGFR
inhibitor used in the treatment of patients with lung cancer
(24). The combination of c-Met and
EGFR kinase inhibitors can enhance the therapeutic effect on
EGFRvIII-positive GBM and overcome its chemoresistance (25). In glioma cells overexpressing
EGFRvIII, c-Met activation is not dependent on its ligand HGF, but
is dependent on the activation of the EGFRvIII-mediated tyrosine
kinase pathway. This indicates that the c-Met receptor may be
another significant target for the treatment of EGFRvIII-positive
GBM. In the case of a high EGFRvIII expression, c-Met is also
highly activated in tumors and is detrimental to chemotherapy in
cancer patients (26). In the
present study, c-Met gene silencing inhibited EGFRvIII
overexpressed glioma cell proliferation and induced apoptosis,
indicating that the co-suppression of HGF/c-Met and EGFRvIII
signaling pathways can be more effective in glioma treatment.
Notably, aptamer-mediated c-Met siRNA delivery into U87-EGFRvIII
cells offers a combined treatment regimen for patients with a high
expression of c-Met and EGFRvIII.
Chu et al (7)
created a biotin-labeled anti-prostate-specific membrane antigen
(PSMA) aptamer and a biotin-labeled siRNA, which were incubated
with streptavidin containing four binding sites. The siRNA,
streptavidin and aptamer formed stable conjugates via the
association between streptavidin and biotin, resulting in target
gene silencing in PSMA-positive cells. Biotinylated aptamers link
to biotinylated siRNA via a streptavidin bridge, offering a simple
yet effective approach for siRNA delivery. In the present study,
c-Met expression did not differ in the presence or absence of
streptavidin during liposomal transfection of c-Met siRNA, and the
addition of streptavidin alone had no effect on the expression of
c-Met, indicating that streptavidin has no cytotoxicity. Future
studies should examine the role of aptamer-mediated c-Met siRNA
delivery in tumor suppression in an animal model of glioma.
Acknowledgements
The present study was supported by grants from the
National Science Foundation of China (grant nos. 30973481 and
81272509) and the Guangdong Natural Science Foundation (grant nos.
9151051501000053 and grant no. 07005145).
References
|
1
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Burnett JC and Rossi JJ: RNA-based
therapeutics: current progress and future prospects. Chem Biol.
19:60–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tuerk C and Gold L: Systematic evolution
of ligands by exponential enrichment: RNA ligands to bacteriophage
T4 DNA polymerase. Science. 249:505–510. 1990. View Article : Google Scholar
|
|
4
|
Sefah K, Shangguan D, Xiong X, O’Donoghue
MB and Tan W: Development of DNA aptamers using Cell-SELEX. Nature
Protoc. 5:1169–1185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gan HK, Kaye AH and Luwor RB: The EGFRvIII
variant in glioblastoma multiforme. J Clin Neurosci. 16:748–754.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu W, Fu Y, Xu S, et al: c-Met expression
is associated with time to recurrence in patients with glioblastoma
multiforme. J Clin Neurosci. 18:119–121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chu TC, Twu KY, Ellington AD and Levy M:
Aptamer mediated siRNA delivery. Nucleic Acids Res. 34:e732006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McNamara JO II, Andrechek ER, Wang Y, et
al: Cell type-specific delivery of siRNAs with aptamer-siRNA
chimeras. Nat Biotechnol. 24:1005–1015. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dassie JP, Liu XY, Thomas GS, et al:
Systemic administration of optimized aptamer-siRNA chimeras
promotes regression of PSMA-expressing tumors. Nat Biotechnol.
27:839–849. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou J, Li H, Zhang J, Piotr S and Rossi
J: Development of cell-type specific anti-HIV gp120 aptamers for
siRNA delivery. J Vis Exp. 52:29542011.PubMed/NCBI
|
|
11
|
Wheeler LA, Trifonova R, Vrbanac V, et al:
Inhibition of HIV transmission in human cervicovaginal explants and
humanized mice using CD4 aptamer-siRNA chimeras. J Clin Invest.
121:2401–2412. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou J, Swiderski P, Li H, et al:
Selection, characterization and application of new RNA HIV gp 120
aptamers for facile delivery of Dicer substrate siRNAs into HIV
infected cells. Nucleic Acids Res. 37:3094–3109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thiel KW, Hernandez LI, Dassie JP, et al:
Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells
using RNA aptamers. Nucleic Acids Res. 40:6319–6337.
2012.PubMed/NCBI
|
|
14
|
Tan Y, Shi YS, Wu XD, et al: DNA aptamers
that target human glioblastoma multiforme cells overexpressing
epidermal growth factor receptor variant III in vitro. Acta
Pharmacol Sin. 34:1491–1498. 2013. View Article : Google Scholar
|
|
15
|
Zhan Y and O’Rourke DM: SHP-2-dependent
mitogen-activated protein kinase activation regulates EGFRvIII but
not wild-type epidermal growth factor receptor phosphorylation and
glioblastoma cell survival. Cancer Res. 64:8292–8298. 2004.
View Article : Google Scholar
|
|
16
|
Nicholas MK, Lukas RV, Chmura S, Yamini B,
Lesniak M and Pytel P: Molecular heterogeneity in glioblastoma:
therapeutic opportunities and challenges. Semin Oncol. 38:243–253.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shangguan D, Cao ZC, Li Y and Tan W:
Aptamers evolved from cultured cancer cells reveal molecular
differences of cancer cells in patient samples. Clin Chem.
53:1153–1155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang YF, Shangguan D, Liu H, et al:
Molecular assembly of an aptamer-drug conjugate for targeted drug
delivery to tumor cells. Chembiochem. 10:862–868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Y, Sefah K, Liu H, Wang R and Tan W:
DNA aptamer-micelle as an efficient detection/delivery vehicle
toward cancer cells. Proc Natl Acad Sci USA. 107:5–10. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang K, Sefah K, Tang L, et al: A novel
aptamer developed for breast cancer cell internalization.
ChemMedChem. 7:79–84. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thiel KW and Giangrande PH: Intracellular
delivery of RNA-based therapeutics using aptamers. Ther Deliv.
1:849–861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McCarty JH: Glioblastoma resistance to
anti-VEGF therapy: has the challenge been MET? Clin Cancer Res.
19:1631–1633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peruzzi B and Bottaro DP: Targeting the
c-Met signaling pathway in cancer. Clin Cancer Res. 12:3657–3660.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Engelman JA, Zejnullahu K, Mitsudomi T, et
al: MET amplification leads to gefitinib resistance in lung cancer
by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pillay V, Allaf L, Wilding AL, et al: The
plasticity of oncogene addiction: implications for targeted
therapies directed to receptor tyrosine kinases. Neoplasia.
11:448–458, 2 p following 458. 2009.PubMed/NCBI
|
|
26
|
Huang PH, Mukasa A, Bonavia R, et al:
Quantitative analysis of EGFRvIII cellular signaling networks
reveals a combinatorial therapeutic strategy for glioblastoma. Proc
Natl Acad Sci USA. 104:12867–12872. 2007. View Article : Google Scholar : PubMed/NCBI
|