Introduction
Hypertension is the most frequent chronic disease in
the developed world. It is a multisystemic disease that affects the
heart and kidneys among other organs. The overall functional,
structural and biochemical alterations in vasculature have been
extensively studied during hypertension, but the molecular
mechanisms remain unclear. Decreased arterial compliance is one of
the earliest detectable manifestations of adverse structural and
functional changes within the vessel wall (1). It has been shown that the proteomic
approach is a useful technique to analyze a complex mixture of
proteins in various settings, usually by combining two-dimensional
electrophoresis and mass spectrometry (MS) (2). Bian et al (3) recently analyzed the proteome of aorta
from spontaneously hypertensive rats (SHR). They found that SHR
showed a significant alteration in the aortic wall protein profile
compared with normal rats.
Exercise is a key antihypertensive therapy. It is
reported that the physical activity level was negatively associated
with blood pressure. In addition, the blood pressure can be
decreased with long-term physical activity. Blood pressure of SHRs
undergoing the physical activity protocol was lower than that of
the normal SHRs. The functional and structural alterations in
vasculature occurred in hypertensives following exercise training.
Aerobic physical activity may alter the aortic wall remodeling to
adapt the artery to a hyperkinetic blood flow resulting in
alterations of the extracellular matrix modulation and vascular
resistance. Certain data showed that the aorta wall thickness was
smaller in the SHRs undergoing an aerobic physical activity
protocol for 20 weeks (4).
Furthermore, the alteration in the aortic wall protein profile was
shown in SHRs with exercise. Kimura et al (5) demonstrated that the 4-hydroxynonenal and
3-nitrotyrosine levels in the aorta of running-trained SHR were
significantly lower than those in the non-exercised group.
Bobillier et al (6) indicated
that there was an increase in the aortic metallothionein amounts in
swimming-trained SHRs. Swimming-trained SHRs showed an
apelin-immunoreactivity level in the aorta (7), but the overall aortic wall protein
profile remained unclear.
The present study used two-dimensional
polyacrylamide gel electrophoresis (2D-PAGE) for protein separation
and identified some of the proteins by MS. Nine proteins were
identified that had a significant difference between
swimming-exercised SHRs and non-exercised SHRs. The molecular
mechanism of exercise decreasing the blood pressure is also
discussed.
Materials and methods
Animals and research design
Studies were performed with male SHRs and their
normotensive counterpart Wistar-Kyoto (WKY) (180–200 g in weight).
The animals were housed in double cages in a temperature-controlled
room (21–22°C; 50–60% humidity) with a 12-h reversed light cycle
and provided free access to food and tap water. All the experiments
were approved by the Institutional Review Board of the Tianjin
University of Sport Research Animal Resource Center (Tianjin,
China).
Each type of rat was divided into an
exercise-trained and sedentary control group. Thus, the rats were
randomly allocated into four groups (n=8 each): i) Sedentary WKY
(SED-WKY), ii) exercised WKY (EX-WKY), iii) sedentary SHR
(SED-SHRs) and iv) exercised SHR (EX-SHRs).
Exercise training
During week 1, the exercise-trained SHR and WKY were
acclimated to 15-min load-free swimming in a basin (water depth of
50 cm, water temperature of 36°C). The duration was progressively
increased. At the end of week 1 the rats were able to swim for 60
min. This intensity was maintained during the rest of the training
period (5 days/week for 6 weeks). Sedentary rats were kept under
the same living conditions as the exercise-trained rats, except for
the training.
Measurement of blood pressure
Weekly systolic blood pressure (SBP) and diastolic
blood pressure (DBP) were measured with the CODATM2
non-invasive single channel BP measuring instrument (Zenda
Instrument Co., Ltd., Austin, TX, USA).
Sample preparation, two-dimensional
electrophoresis and analysis
Total protein was extracted only from the aorta
(abdominal aorta was not included). Briefly, aorta samples were
pulverized after being frozen by liquid nitrogen. Pulverized tissue
powder was homogenized in lysis buffer (9 M urea, 2 M thiourea, 4%
CHAPS, 2% IPG buffer, 40 mM dithiothreitol and 40 mM Tris-base) and
centrifuged at 20,000 × g for 45 min at 4°C. The supernatant was
collected as a protein sample and the concentration was determined
by the Bradford protein assay (8).
A sample containing 250 µg of protein was applied to
the first-dimensional isoelectric focusing in the ReadyStrip
immobilized pH gradient strips (18 cm; pH 3–10 NL). Separation of
proteins in the second dimension was achieved by SDS-PAGE (10%).
Two-dimensional gels were stained by coomassie brilliant blue and
scanned by Imagescanner (GE-Amersham Biosciences Corp., Piscataway,
NJ, USA). Digitized images were recorded and imported to
ImageMaster 2D Platinum 6.0 software (GE Healthcare Life Sciences,
Fairfield, CT, USA). Analysis was performed matching the spots from
different gels of different animals for each group.
Trypsin digestion and protein
identification by MS
Protein spots of interest were excised manually from
the gels and digested with trypsin as described previously
(9). The identification of protein was
performed by peptide mass fingerprinting using LCQ Deca XP (Thermo
Electron Corp, San Jose, CA, USA) mass spectrometers.
Statistical analyses
Results of blood pressure are expressed as means ±
standard error of the mean. Comparisons between multiple groups
were performed by or two-way analysis of variance. Tests were
performed using the SPSS 12.0 software package (SPSS, Inc.,
Chicago, IL, USA). Proteomic statistical analysis was performed
using TurboSEQUEST 3.3 software (Bioworks, Thermo Electron,
Marietta, OH, USA), which includes a statistical package. Spots
were tested with the t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Blood pressure decrease in SHR induced
by 6-week load-free swimming
The blood pressure of 12-week-old SED-SHR was
significantly higher than that of the age-matched SED-WKY [SBP,
195.78±11.46 vs. 126.63±11.70 mmHg; DBP, 144.05±21.47 vs.
85.43±9.73 mmHg; mean arterial pressure (MAP), 166.32±11.93 vs.
104.59±16.23 mmHg; P<0.01]. Contrasted to that of SED-SHR, the
blood pressure of the EX-SHR that underwent 6-week load-free
swimming was reduced (SBP, 163.44±12.90 vs. 195.78±11.46 mmHg; DBP,
121.19±12.61 vs. 144.05±21.47 mmHg; MAP, 134.13±18.31 vs.
166.32±11.93 mmHg; P<0.01). Six-week load-free swimming did not
reduce the blood pressure in exercised WKY rats (SBP, 123.92±12.55
vs. 126.63±11.70 mmHg; DBP, 85.20±12.54 vs. 85.43±9.73 mmHg; MAP,
98.28 ±9.19 vs. 104.59±16.23 mmHg). The results are shown in
Table I.
 | Table I.Effects of 6-week load-free swimming
on the blood pressure in SHR and WKY rats. |
Table I.
Effects of 6-week load-free swimming
on the blood pressure in SHR and WKY rats.
|
| SED-WKY (n=8) | EX-WKY (n=8) | SED-SHR (n=8) | EX-SHR (n=8) |
|---|
|
|
|
|
|
|
|---|
| Pressure | 0 weeks | 6 weeks | 0 weeks | 6 weeks | 0 weeks | 6 weeks | 0 weeks | 6 weeks |
|---|
| SBP | 119.62±8.03 | 126.63±11.70 | 117.14±12.15 | 123.92±12.55 |
153.17±9.91a |
195.78±11.46a,b | 153.17±17.08 |
163.44±12.90c |
| DBP |
93.79±6.90 | 85.43±9.73 | 87.58±7.40 |
85.20±12.54 |
110.69±11.92a |
144.05±21.47a,b | 112.74±7.65 |
121.19±12.61c |
| MAP |
81.25±7.31 | 104.59±16.23 | 75.17±6.79 | 98.28±9.19 |
97.67±11.40a |
166.32±11.93a,b | 98.70±6.30 |
134.13±18.31c |
Protein expression profile
Comparison among SED-WKY, EX-WKY, SED-SHR and EX-SHR
was performed by the replicate group option and the statistical
software package of ImageMaster 2D Platium 6.0. Spot quantification
was normalized on the basis of the total staining density of the
image to compensate for any variation in protein loading and
development level of coomassie brilliant blue staining. The
differential expression was calculated for every spot that could be
matched in all the samples of ≥1 group.
From all the spots resolved in the pH 4 to 7 range
by two-dimensional electrophoresis of aortic wall tissue from WKY
and SHR, the same 453 well-resolved spots were focused on in a 12-
to 90-kDa molecular weight, as previously analyzed (6). Significant differences were shown in 11
analyzed spots in EX-SHR versus SED-SHR (P<0.05); 5 increased
and 6 decreased. Ten protein spots were found to change
significantly in EX-SHR versus EX-WKY (P<0.05); 5 increased and
5 decreased. There were 8 protein spots that showed significant
differences in EX-WKY versus SED-WKY (P<0.05); 1 increased and 7
decreased. Thirteen protein spots were found to change
significantly in EX-WKY versus SED-WKY (P<0.05); 11 increased
and 2 decreased. Accurate protein identification was achieved for 9
spots and failed in 2 in EX-SHR versus SED-SHR. The spots are all
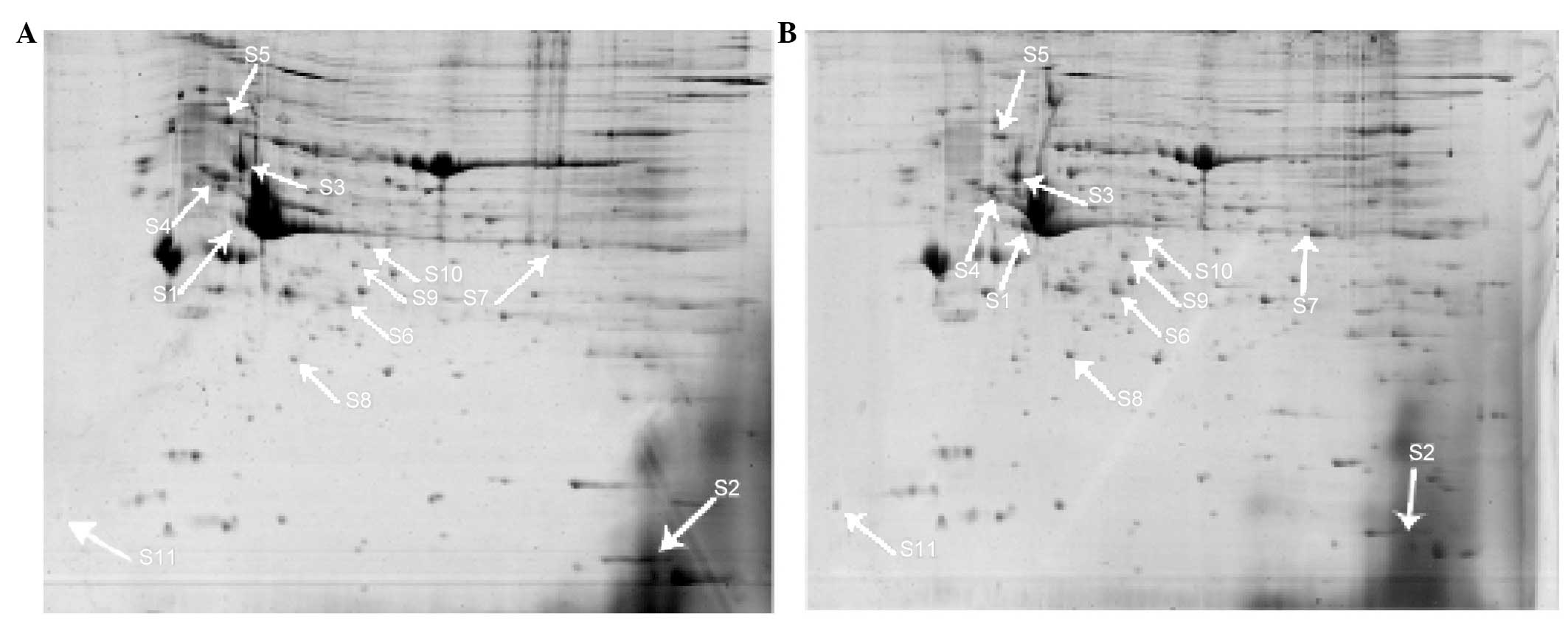
indicated with arrows in Fig. 1.
Protein identification
All the 11 spots were excised from the rehydrated
gels and subjected to in-gel trypsin digestion for subsequent
identification by MS. Nine of them contained peptides and 2 failed
identification. Five upregulated spots and 4 downregulated spots
were identified finally. The results are shown in Table II.
 | Table II.Identification of the differentially
expressed proteins in EX-SHR versus SED-SHR. |
Table II.
Identification of the differentially
expressed proteins in EX-SHR versus SED-SHR.
| Protein number | Ms
identification | NCBInr index
score | Top score | pI | Mw (kDa) |
|---|
| S2 | Adipocyte-type
fatty acid-binding protein | gi|2494405 | 155 | 7.71 | 14.699 |
| S4 | Tubulin β-2C
chain | gi|5174735 | 276 | 4.79 | 49.799 |
| S5 | 78 kDa
glucose-regulated protein precursor | gi|25742763 | 261 | 5.07 | 72.302 |
| S6 | Mimecan | gi|157824206 | 133 | 5.85 | 34.048 |
| S7 | Long-chain specific
acyl-CoA dehydrogenase | gi|6978431 | 228 | 7.63 | 47.842 |
| S8 | Heat shock protein
β-1 | gi|94400790 | 147 | 6.12 | 22.808 |
| S9 | NAD subunit α | gi|16758446 | 130 | 6.47 | 39.588 |
| S10 | Actin, α cardiac
muscle 1 preprotein | gi|4885049 | 116 | 5.23 | 41.992 |
| S11 | Calmodulin isoform
2 | gi|71664 | 62 | 4.09 | 16.696 |
Discussion
The aim of the present study was to screen and
identify proteins secreted by the aortic wall as potential
biomarkers of susceptibility to exercise-induced blood pressure
decreasing. SHR and WKY rats were chosen for the contrasting
susceptibilities of different blood pressure level. The results
also showed that the blood pressure is normal in WKY rats while
hypertension occurs in SHR. After 6-week load-free swimming, the
blood pressure decreased significantly in SHR, which was consistent
with other studies (10,11).
Recent research found that aerobic physical activity
may alter the aortic wall remodeling to adapt the artery to a
hyperkinetic blood flow resulting in alterations of the
extracellular matrix modulation and vascular resistance (4). Certain studies identified that the
expression of several proteins changed in the aortic wall induced
by exercise in SHRs (5,6). However, the overall aortic wall protein
profile remains unclear.
In the present study, the proteomics of the aortic
wall in SED-SHRs and EX-SHRs were presented to explore the
molecular mechanism for exercise decreasing the blood pressure.
Differentially expressed proteins were detected in the aortic wall
from SED-SHRs compared with EX-SHRs. Five upregulated proteins and
4 downregulated proteins were obtained in the aortic wall of
EX-SHRs. An association between these proteins and decreasing blood
pressure was suggested.
Spot S2 was identified as the adipocyte-type fatty
acid-binding protein (A-FABP). A-FABP was downregulated 12.8-fold
in the EX-SHR group compared with the SED-SHR group. FABPs are
abundantly expressed 14–15 kDa proteins that reversibly bind
hydrophobic ligands (12). A-FABP is
highly expressed in adipocytes and macrophages (13). It is involved in insulin resistance,
lipid metabolism and atherosclerosis. Expression of A-FABP is
highly regulated during differentiation of adipocytes and its mRNA
is transcriptionally controlled by fatty acids, PPAR-γ agonists and
insulin (14,15).
Spot S4 was identified as tubulin β-2C chain.
Tubulin β-2C chain was downregulated 2.3-fold in the EX-SHR group
compared with the SED-SHR group. Exercise training decreased
β-tubulin protein expression in the kidney of chronic heart failure
(CHF) rats, which suggests that exercise training can significantly
improve the renal dysfunction in CHF rats (16).
Spot S5 was identified as the 78 kDa
glucose-regulated protein precursor. In the present study,
glucose-regulated protein 78 precursor was downregulated 2.2-fold
in the EX-SHR group compared with the SED-SHR group as determined
by 2D-PAGE. Glucose-regulated protein 78 (GRP78) is a
well-characterized molecular chaperone that is ubiquitously
expressed in mammalian cells. GRP78 is best known for binding to
hydrophobic patches on nascent polypeptides within the endoplasmic
reticulum and for its role in signaling the unfolded protein
response. Studies have shown that GRP78 is expressed on the cell
surface in numerous tissue types in vitro and in
vivo. GRP78 disregulation is also indicated in atherosclerotic,
thrombotic and auto-immune disease (17). Previous studies showed that the levels
of GRP78 were upregulated in the soleus muscle of Wistar rats and
the brain of Alzheimer's disease mice adhering to treadmill running
for 3 to 4 months duration (18,19).
However, investigators found that treadmill running for 5 days did
not increase GRP78 expression in cardiac muscle (20).
Spot S6 was identified as mimecan. Mimecan was
downregulated 2.1-fold in the EX-SHR group compared with the
SED-SHR group as determined by 2D-PAGE. Mimecan (also known as
osteoglycin) belongs to the family of small leucine-rich
proteoglycans, a group of 11 proteins, characterized by
leucine-rich repeats in their central domain. These proteins are
located in the extracellular matrix and are important for the
regulation of the matrix structure, but also in the regulation of
cell cycle and growth factor actions (21). Mimecan in the aorta was mainly produced
by VSMCs and perivascular fibroblasts. A downregulation was
confirmed following the onset of arteriogenesis (22). Its expression was also regulated during
atherosclerosis in patients and animals. Differential expression of
mimecan was identified in VSMCs during neointima formation and in
atherosclerosis plaques (23,24). The downregulation of osteoglycin
expression in the collateral arteries of rabbits subjected to
femoral artery occlusion indicated a function of osteoglycin as a
negative regulator of the mitotic activity in the wall of
collateral arteries (22).
Spot S7 was identified as the long-chain specific
acyl-CoA dehydrogenase (LCAD). In the present study, LCAD was
upregulated 2.0-fold in the EX-SHR group compared with the SED-SHR
group as determined by 2D-PAGE. LCAD catalyzes the
α,β-dehydrogenation of acyl-CoAs, the initial step of mitochondrial
β-oxidation. LCAD has been shown to be involved in the degradation
of branched-chain fatty acids originating from peroxisomal
catabolism of phytanic acid, but LCAD is also able to handle
straight-chain and certain monounsaturated acyl-CoAs (25). The phenotype of the LCAD−/−
mouse is characterized by unprovoked sudden death, fasting and cold
intolerance, hypoketotic hypoglycaemia and marked fatty changes in
liver and heart (26). A previous
study found that the 3-hydroxyacylcarnitines were absent in
LCAD−/− tissues and a profound deficiency of
acetylcarnitine was observed in LCAD−/− hearts,
indicating that the cardiomyopathy in the LCAD−/− mouse
is caused primarily by a severe energy deficiency in the heart,
stressing the important role of LCAD in cardiac fatty acid
metabolism in the mouse (27). LCAD
was elevated in SHR cardiac mitochondria (121%) (28).
Spot S8 was identified as heat-shock protein β-1
(HSPB1). HSPB1 was upregulated 1.9-fold in the EX-SHR group
compared with the SED-SHR group as determined by 2D-PAGE. HSPB1,
also known as HSP27, is a member of the mammalian heat-shock
protein family (29). In smooth muscle
cells, HSP27 appears to be the link between the signal transduction
cascade and the contractile machinery (30). Park et al (31) reported lower HSP27 levels in plaques,
lower levels of phosphorylation of HSP27 and higher plasma levels
of secreted HSP27 in humans with acute coronary syndrome raised, in
regards to the contribution of HSP27 in atherogenesis. Expression
can increase in response to physical and chemical stressors
including heat, mechanical strain, oxidative stress and
proinflammatory mediators. Expression of small HSPs in striated and
smooth muscle is frequently constitutive, but can also be modified
by chemical or physical stressors (32).
Spot S9 was identified as isocitrate dehydrogenase
(NAD) subunit α. NAD subunit α was upregulated 1.9-fold in the
EX-SHR group compared with the SED-SHR group, as determined by
2D-PAGE. NAD-dependent isocitrate dehydrogenase, Idh, one of the
eight enzymes of the Krebs cycle, is an octamer composed of Idh1p
and Idh2p (encoded by IDH1 and IDH2, respectively) (33). In a previous study, the yeast enzyme
was shown to be composed of two non-identical subunits, IDH1 and
IDH2, with both being equally represented in the holoenzyme
(34). These two subunits were shown
to be essential for holoenzyme activity, since disruption of either
or both genes encoding the subunits results in yeast strains that
exhibit no detectable NAD1-specific isocitrate dehydrogenase
activity and that are unable to grow with acetate as a carbon
source (33). Research demonstrated
that the yeast Krebs cycle enzyme IDH binds specifically and with
high affinity to the 5-untranslated leader sequences of
mitochondrial mRNAs in vitro and have proposed a role for
the enzyme in the regulation of mitochondrial translation. Cells
disrupted for the IDH genes exhibit a strong increase in
mitochondrial translation activity and the newly synthesized
products are also more rapidly degraded, which suggested that
binding of Idh to mitochondrial mRNAs may suppress inappropriate
translation of mitochondrial mRNAs (35). Research found significantly lower
protein levels of IDH2 (93%) and depressed total IDH activities
(68%) in SHR heart mitochondria. IDH1 appeared to have a higher
level in SD rats undergoing 8 weeks of swimming training (36). IDH activity increased 32% in the 6
weeks of treadmill running in trained intermyofibrillar
mitochondria (37).
Spot S10 was identified as actin, α cardiac muscle 1
preprotein. This preprotein was upregulated 1.9-fold in the EX-SHR
group compared with the SED-SHR group as determined by 2D-PAGE. The
highly conserved actins are the major constituents of the thin
filaments in the muscle sarcomere. They are involved in force
generation within the sarcomere and force transmission from the
sarcomere to the surrounding syncytium (38). The α-cardiac actin gene (ACTC) 1 is the
major component of the sarcomeric thin filaments and is essential
for cardiac muscle contraction. Knockdown of ACTC1 in chicks shows
less developed atrial septa supporting a dose-dependent effect of
ACTC1 on cardiac development. It has been suggested that the lack
of ACTC1 may induce apoptosis leading to disrupted cardiac
differentiation (39). ACTC was the
first gene identified to harbor HCM and DCM mutations, with 6
mutations leading to HCM and 2 mutations leading to DCM (40).
Spot S11 was identified as calmodulin (CaM) isoform
2. CaM isoform 2 was downregulated by 1.8-fold in the EX-SHR group
compared with the SED-SHR group as determined by 2D-PAGE. CaM is a
calcium-binding protein which, when complexed with calcium, may
mediate numerous calcium-dependent cellular activities (41). CaM is associated with a variety of cell
functions including inflammation, apoptosis and muscular
contraction. A previous study has shown that in the aorta from SHR,
expression levels of several CaM-related proteins, including
eukaryotic elongation factor kinase and death-associated protein
kinase protein, increased, while Ca(2+)/CaM-dependent protein
kinase IIδ and histone deacetylases 4 protein decreased.
CaM-related proteins may at least be in part associated with the
pathogenesis of hypertensive vascular diseases (42).
In conclusion, proteomic analysis of proteins in the
aortic wall from SHRs performing 6-week load-free swimming provided
an effective approach for elucidating the molecular mechanisms of
the exercise-induced blood pressure decrease. Eleven protein spots
with different abundance were found, of which 9 differentially
expressed proteins were identified by MALDI-TOF MS. The roles of
these identified proteins in the exercise-induced blood pressure
decrease were discussed. These findings provide information for
understanding the mechanism of exercise decreasing blood pressure
in the aortic wall.
Acknowledgements
The present study is supported in part by a grant
from the National Natural Science Foundation of China (nos.
81171870 and 31470061), the Key Projects of Applied Basic and
Frontier Technology Research of Tianjin (grant no. 14JCZDJC32700)
and the Innovation Platform Special Program of Tianjin Science and
Technology Innovation System.
References
|
1
|
Cavalcante JL, Lima JA, Redheuil A and
Al-Mallah MH: Aortic stiffness: Current understanding and future
directions. J Am Coll Cardiol. 57:1511–1522. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aebersold R and Mann M: Mass
spectrometry-based proteomics. Nature. 422:198–207. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bian YL, Qi YX, Yan ZQ, Long DK, Shen BR
and Jiang ZL: A proteomic analysis of aorta from spontaneously
hypertensive rat: RhoGDI alpha upregulation by angiotensin II via
AT(1) receptor. Eur J Cell Biol. 87:101–110. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Horta PP, de Carvalho JJ and
Mandarim-de-Lacerda CA: Exercise training attenuates blood pressure
elevation and adverse remodeling in the aorta of spontaneously
hypertensive rats. Life Sci. 77:3336–3343. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kimura H, Kon N, Furukawa S, Mukaida M,
Yamakura F, Matsumoto K, Sone H and Murakami-Murofushi K: Effect of
endurance exercise training on oxidative stress in spontaneously
hypertensive rats (SHR) after emergence of hypertension. Clin Exp
Hypertens. 32:407–415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bobillier Chaumont S, Maupoil V, Jacques
Lahet J and Berthelot A: Effect of exercise training on
metallothionein levels of hypertensive rats. Med Sci Sports Exerc.
33:724–728. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Ren CX, Qi YF, Lou LX, Chen L,
Zhang LK, Wang X and Tang C: Exercise training promotes expression
of apelin and APJ of cardiovascular tissues in spontaneously
hypertensive rats. Life Sci. 79:1153–1159. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jenö P, Mini T, Moes S, Hintermann E and
Horst M: Internal sequences from proteins digested in
polyacrylamide gels. Anal Biochem. 224:75–82. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song YJ, Sawamura M, Ikeda K, et al:
Training in swimming reduces blood pressure and increase muscle
transportactivity as well as GLUT 4 contents in stroke-prone
spontaneously hypertensive rats. Appl Human Sci. 17:275–280. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vogt M, Ott B, Rupp H and Jacob R:
Significance of physical exercise in hypertension. Influence of
water temperature and beta-blockade on blood pressure, degree of
cardiac hypertrophy and cardiac function in swimming training of
spontaneously hypertensive rats. Basic Res Cardiol. 81 (Suppl
1):157–169. 1986.PubMed/NCBI
|
|
12
|
Zimmerman AW and Veerkamp JH: New insights
into the structure and function of fatty acid-binding proteins.
Cell Mol Life Sci. 59:1096–1116. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu Y, Luo N and Lopes-Virella MF: Oxidized
LDL induces the expression of ALBP/aP2 mRNA and protein in human
THP-1 macrophages. J Lipid Res. 41:2017–2023. 2000.PubMed/NCBI
|
|
14
|
Haunerland NH and Spener F: Fatty
acid-binding proteins-insights from genetic manipulations. Prog
Lipid Res. 43:328–349. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Makowski L and Hotamisligil GS: The role
of fatty acid binding proteins in metabolic syndrome and
atherosclerosis. Curr Opin Lipidol. 16:543–548. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin QQ, Lin R, Ji QL, Zhang JY, Wang WR,
Yang LN and Zhang KF: Effect of exercise training on renal function
and renal aquaporin-2 expression in rats with chronic heart
failure. Clin Exp Pharmacol Physiol. 38:179–185. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Quinones QJ, de Ridder GG and Pizzo SV:
GRP78: A chaperone with diverse roles beyond the endoplasmic
reticulum. Histol Histopathol. 23:1409–1416. 2008.PubMed/NCBI
|
|
18
|
González B, Hernando R and Manso R: Stress
proteins of 70 kDa in chronically exercised skeletal muscle.
Pflugers Arch. 440:42–49. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Um HS, Kang EB, Leem YH, Cho IH, Yang CH,
Chae KR, Hwang DY and Cho JY: Exercise training acts as a
therapeutic strategy for reduction of the pathogenic phenotypes for
Alzheimer's disease in an NSE/APPsw-transgenic model. Int J Mol
Med. 22:529–539. 2008.PubMed/NCBI
|
|
20
|
Murlasits Z, Lee Y and Powers SK:
Short-term exercise does not increase ER stress protein expression
in cardiac muscle. Med Sci Sports Exerc. 39:1522–1528. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iozzo RV: The family of the small
leucine-rich proteoglycans: Key regulators of matrix assembly and
cellular growth. Crit Rev Biochem Mol Biol. 32:141–174. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kampmann A, Fernández B, Deindl E, Kubin
T, Pipp F, Eitenmüller I, Hoefer IE, Schaper W and Zimmermann R:
The proteoglycan osteoglycin/mimecan is correlated with
arteriogenesis. Mol Cell Biochem. 322:15–23. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shanahan CM, Cary NR, Osbourn JK and
Weissberg PL: Identification of osteoglycin as a component of the
vascular matrix. Differential expression by vascular smooth muscle
cells during neointima formation and in atherosclerotic plaques.
Arterioscler Thromb Vasc Biol. 17:2437–2447. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fernández B, Kampmann A, Pipp F,
Zimmermann R and Schaper W: Osteoglycin expression and localization
in rabbit tissues and atherosclerotic plaques. Mol Cell Biochem.
246:3–11. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lea W, Abbas AS, Sprecher H, Vockley J and
Schulz H: Long-chain acyl-CoA dehydrogenase is a key enzyme in the
mitochondrial beta-oxidation of unsaturated fatty acids. Biochim
Biophys Acta. 1485:121–128. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cox KB, Hamm DA, Millington DS, Matern D,
Vockley J, Rinaldo P, Pinkert CA, Rhead WJ, Lindsey JR and Wood PA:
Gestational, pathologic and biochemical differences between very
long-chain acyl-CoA dehydrogenase deficiency and long-chain
acyl-CoA dehydrogenase deficiency in the mouse. Hum Mol Genet.
10:2069–2077. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van Vlies N, Tian L, Overmars H, Bootsma
AH, Kulik W, Wanders RJ, Wood PA and Vaz FM: Characterization of
carnitine and fatty acid metabolism in the long-chain acyl-CoA
dehydrogenase-deficient mouse. Biochem J. 387:185–193. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jüllig M, Hickey AJ, Chai CC, Skea GL,
Middleditch MJ, Costa S, Choong SY, Philips AR and Cooper GJ: Is
the failing heart out of fuel or a worn engine running rich? A
study of mitochondria in old spontaneously hypertensive rats.
Proteomics. 8:2556–2572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ciocca DR, Oesterreich S, Chamness GC,
McGuire WL and Fuqua SA: Biological and clinical implications of
heat shock protein 27,000 (Hsp27): A review. J Natl Cancer Inst.
85:1558–1570. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ibitayo AI, Sladick J, Tuteja S,
Louis-Jacques O, Yamada H, Groblewski G, Welsh M and Bitar KN:
HSP27 in signal transduction and association with contractile
proteins in smooth muscle cells. Am J Physiol. 277:G445–G454.
1999.PubMed/NCBI
|
|
31
|
Park HK, Park EC, Bae SW, Park MY, Kim SW,
Yoo HS, Tudev M, Ko YH, Choi YH, Kim S, et al: Expression of heat
shock protein 27 in human atherosclerotic plaques and increased
plasma level of heat shock protein 27 in patients with acute
coronary syndrome. Circulation. 114:886–893. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Salinthone S, Tyagi M and Gerthoffer WT:
Small heat shock proteins in smooth muscle. Pharmacol Ther.
119:44–54. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cupp JR and McAlister-Henn L: Cloning and
characterization of the gene encoding the IDH1 subunit of
NAD(+)-dependent isocitrate dehydrogenase from Saccharomyces
cerevisiae. J Biol Chem. 267:16417–16423. 1992.PubMed/NCBI
|
|
34
|
Keys DA and McAlister-Henn L: Subunit
structure, expression, and function of NAD(H)-specific isocitrate
dehydrogenase in Saccharomyces cerevisiae. J Bacteriol.
172:4280–4287. 1990.PubMed/NCBI
|
|
35
|
de Jong L, Elzinga SD, McCammon MT,
Grivell LA and van der Spek H: Increased synthesis and decreased
stability of mitochondrial translation products in yeast as a
result of loss of mitochondrial (NAD(+))-dependent isocitrate
dehydrogenase. FEBS Lett. 483:62–66. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun B, Wang JH, Lv YY, Zhu SS, Yang J and
Ma JZ: Proteomic adaptation to chronic high intensity swimming
training in the rat heart. Comp Biochem Physiol Part D Genomics
Proteomics. 3:108–117. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bizeau ME, Willis WT and Hazel JR:
Differential responses to endurance training in subsarcolemmal and
intermyofibrillar mitochondria. J Appl Physiol (1985).
85:1279–1284. 1998.PubMed/NCBI
|
|
38
|
Mogensen J, Klausen IC, Pedersen AK,
Egeblad H, Bross P, Kruse TA, Gregersen N, Hansen PS, Baandrup U
and Borglum AD: Alpha-cardiac actin is a novel disease gene in
familial hypertrophic cardiomyopathy. J Clin Invest. 103:R39–R43.
1999. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Matsson H, Eason J, Bookwalter CS, Klar J,
Gustavsson P, Sunnegårdh J, Enell H, Jonzon A, Vikkula M, Gutierrez
I, et al: Alpha-cardiac actin mutations produce atrial septal
defects. Hum Mol Genet. 17:256–265. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mogensen J, Perrot A, Andersen PS,
Havndrup O, Klausen IC, Christiansen M, Bross P, Egeblad H,
Bundgaard H, Osterziel KJ, et al: Clinical and genetic
characteristics of alpha cardiac actin gene mutations in
hypertrophic cardiomyopathy. J Med Genet. 41:e102004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Van Breemen C, Aaronson P and Loutzenhiser
R: Sodium-calcium interactions in mammalian smooth muscle.
Pharmacol Rev. 30:167–208. 1978.PubMed/NCBI
|
|
42
|
Usui T, Okada M, Hara Y and Yamawaki H:
Exploring calmodulin-related proteins, which mediate development of
hypertension, in vascular tissues of spontaneous hypertensive rats.
Biochem Biophys Res Commun. 405:47–51. 2011. View Article : Google Scholar : PubMed/NCBI
|