Introduction
Glaucoma causes irreversible blindness and is
characterized by optic nerve degeneration and visual field defects
(1). Primary open-angle glaucoma
(POAG) is the major form of glaucoma. An epidemiological study in
Japan demonstrated that a large number of POAG cases were diagnosed
as normal tension glaucoma (NTG) (2).
Mutations in the optineurin (OPTN) gene that encodes amino
acid substitutions, such as E50K, H486R and R545Q, have been
associated with POAG and NTG (3–6). The
substitution of glutamic acid by lysine at amino acid 50 (E50K) is
exclusively associated with familial and sporadic forms of NTG
(7). Previous studies have shown that
E50K OPTN induces the apoptosis of retinal ganglion cells in
transgenic mice models and progressive retinal degeneration
exclusively in the peripheral region of the retinas, although the
exact mechanism remains unclear (8).
MicroRNAs (miRNAs) bind to target messenger RNAs
(mRNAs) and subsequently suppress protein expression via mRNA
degradation or translational inhibition (9). Several reports have described the
expression of miRNAs in the eye and specific miRNAs have been
indicated in the pathogenesis of glaucoma. In one report, miRNA-24
was shown to regulate transforming growth factor β1 processing
during cyclic mechanical stress in human trabecular network cells
(10). It is unclear whether miRNAs
contribute to pathogenesis in OPTN (E50K) transgenic mice.
To address whether additional miRNAs could play a role in
OPTN (E50K)-induced POAG pathogenesis, miRNA profiling was
performed of the retinal samples from OPTN (E50K) transgenic
mice and the differentially expressed miRNAs were identified.
Differential expression of miRNAs was validated by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). The
identification of the differentially expressed miRNAs due to mutant
OPTN (E50K) expression is likely to provide a foundation for
studying the molecular pathways contributing to POAG.
Materials and methods
Animals and sample collection
All the animal experiments in the study were
performed according to the tenets of the National Institutes of
Health Guidelines and Regulations on the Care and Use of Animals in
Research. Transgenic mice engineered to express the E50K mutant
human OPTN in the retina were as described previously (11). Retinas from 8-month-old transgenic and
wild-type mice were collected for genome-wide miRNA expression
analysis. Retinas from 3 groups of 10 transgenic mice (E1, E2 and
E3) and 3 groups of 10 wild-type mice (W1, W2 and W3) were pooled
for analysis and independently analyzed for miRNA expression.
Retinal samples were frozen at −80°C until analysis.
miRNA microarray study
miRNA microarray experiments were performed at
CapitalBio Corp. (Beijing, China). Total RNA was extracted with
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA)
from retinal samples collected from transgenic and wild-type mice.
Samples were labeled and hybridized to a GeneChip® miRNA Array
(V3.0; Affymetrix, Santa Clara, CA, USA). Raw data were normalized
and analyzed using the miRNA QC Tool software (Affymetrix). The
analysis focused on miRNAs with a ≥1.5-fold expression difference
in retinal samples from OPTN (E50K) transgenic mice as compared to
wild-type control retinal samples.
miRNA target gene prediction and
function analysis
Prediction of miRNA target genes was performed using
a computational approach. The prediction of miRNA target genes was
performed using 3 different miRNA target prediction algorithms:
PicTar, miRanda v5 and TargetScan v6.0. For each algorithm, the
potential binding sites in the mRNA 3′-untranslated region (3′-UTR)
of target genes were identified according to specific base-pairing
rules.
RT-qPCR miRNA analysis
To confirm the results from the miRNA microarray,
RT-qPCR analysis of mmu-miR-141 was performed on the same total RNA
used for microarray analysis. First-strand cDNA synthesis was
performed from equal amounts of total RNA using an All-in-One™
miRNA First-Strand cDNA Synthesis kit (GeneCopoeia, Guangdong,
China) according to the manufacturer's instructions. Primers were
designed and synthesized by GeneCopoeia.
RT-qPCR was performed using the All-in-One™ qPCR Mix
on LightCycler 480 system (Roche, Basel, Switzerland). The levels
of an endogenous control, U6, were used to normalize the expression
levels of each miRNA. All the reactions were performed in
triplicate and included controls. The fold-change in miRNA
expression was calculated using the comparative Ct method. Data are
presented as the fold-change relative to expression in the retinal
samples of wild-type mice.
Statistical analysis
All the results are expressed as the mean ± standard
deviation. Statistical analysis was performed with the Student's
t-test to identify statistically significant differences using
commercial software (SigmaPlot; Systat Software Inc., San Jose, CA,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Global miRNA profiling in OPTN (E50K)
transgenic and wild-type mice
Retinal miRNA expression patterns were evaluated
using pooled retinal samples for 3 separate groups of 10 transgenic
mice (E1, E2 and E3) and 10 wild-type mice (W1, W2 and W3). For
each pooled sample, >2 µg of retinal RNA was harvested.
Spectrophotometric analysis of RNA purity was performed by
measuring absorbance at 260 and 280 nm. All samples exhibited
260/280 nm absorbance ratios >1.96, indicating isolation of
high-quality retinal RNA.
Global miRNA expression patterns were determined for
each sample by microarray analysis. The analysis focused on
specific miRNAs exhibiting differential expression levels in the
retinas from OPTN (E50K) transgenic mice as compared to
wild-type mice by identifying miRNAs with a ≥1.5-fold-change in
expression level and P≤0.05. Based on this threshold, 48 miRNAs
were differentially expressed in OPTN (E50K) transgenic
retinal samples as compared to wild-type retinal samples,
representing 10% of the 48 miRNAs represented on the array
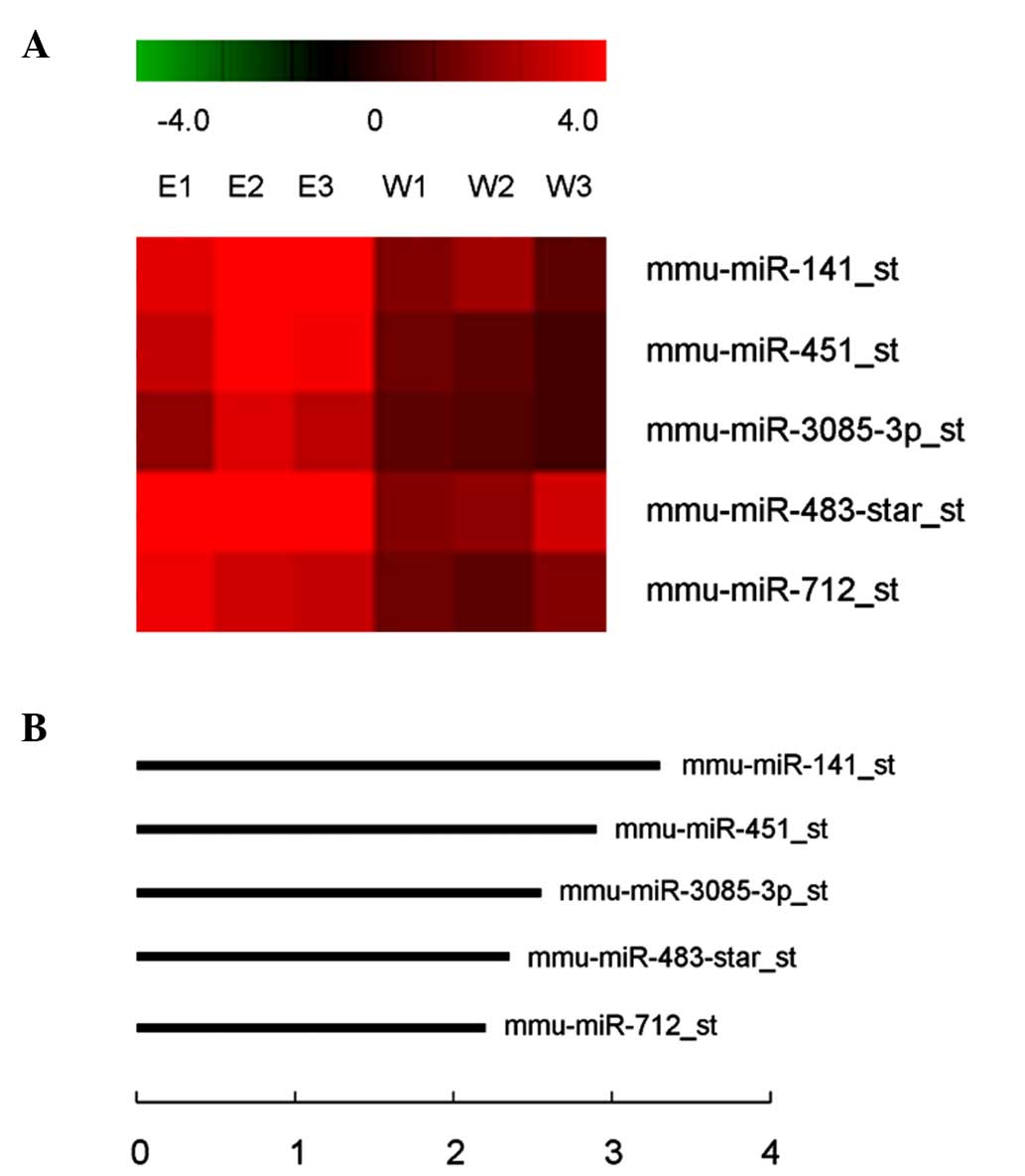
(Table I). Of these, 5 miRNAs passed
the Volcano plot filtering screen to identify miRNAs that were
expressed at significantly different levels in transgenic versus
wild-type retinas (P<0.05; false discovery rate, <0.05;
Fig. 1A). Each of these miRNAs was
upregulated 2.32–3.20-fold in the retinas of OPTN (E50K)
transgenic mice compared to wild-type mice (Fig. 1B). mmu-miR-141 was focused on as it
exhibited the largest increase in expression in retinal samples
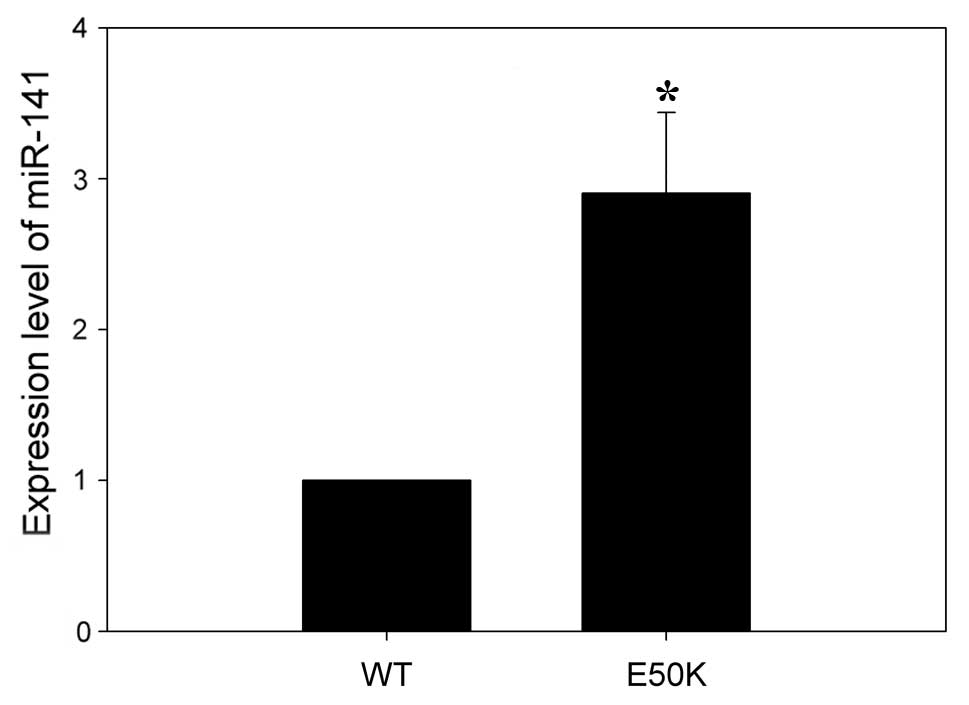
from transgenic mice and RT-qPCR was used to validate the miRNA
microarray results. Consistent with the microarray results, RT-qPCR
analysis demonstrated that mmu-miR-141 was expressed at
significantly higher levels in the retinas of transgenic mice
compared to wild-type mice (Fig.
2).
 | Table I.MicroRNA (miRNA) profiles were
performed on the retinas of optineurin (E50K) transgenic and
wild-type mice. miRNAs are ranked in order of fold-change, from the
largest to the smallest. |
Table I.
MicroRNA (miRNA) profiles were
performed on the retinas of optineurin (E50K) transgenic and
wild-type mice. miRNAs are ranked in order of fold-change, from the
largest to the smallest.
| miRNA ID | Fold-change | miRNA sequence |
|---|
| mmu-miR-141 | 3.2023 |
UAACACUGUCUGGUAAAGAUGG |
| mmu-miR-451 | 2.8473 |
AAACCGUUACCAUUACUGAGUU |
| mmu-miR-200a | 2.6426 |
UAACACUGUCUGGUAACGAUGU |
| mmu-miR-483* | 2.5317 |
UCACUCCUCCCCUCCCGUCUU |
| mmu-miR-3057-3p | 2.4620 |
UCCCACAGGCCCAGCUCAUAGC |
| mmu-miR-877* | 2.4260 |
UGUCCUCUUCUCCCUCCUCCCA |
|
mmu-miR-3102-3p.2 | 2.4235 |
CUCUACUCCCUGCCCCAGCCA |
| mmu-miR-429 | 2.3851 |
UAAUACUGUCUGGUAAUGCCGU |
| mmu-miR-3085-3p | 2.3604 |
UCUGGCUGCUAUGGCCCCCUC |
| mmu-miR-712 | 2.3241 |
CUCCUUCACCCGGGCGGUACC |
| mmu-miR-223 | 2.2929 |
UGUCAGUUUGUCAAAUACCCCA |
| mmu-miR-211 | 2.2731 |
UUCCCUUUGUCAUCCUUUGCCU |
| mmu-miR-31* | 2.2721 |
UGCUAUGCCAACAUAUUGCCAUC |
| mmu-miR-27a | 2.0945 |
UUCACAGUGGCUAAGUUCCGC |
| mmu-miR-3963 | 2.0212 |
UGUAUCCCACUUCUGACAC |
| mmu-miR-1a | 2.0141 |
UGGAAUGUAAAGAAGUAUGUAU |
| mmu-miR-3968 | 1.9957 |
CGAAUCCCACUCCAGACACCA |
| mmu-miR-200b | 1.9956 |
UAAUACUGCCUGGUAAUGAUGA |
| mmu-miR-200a* | 1.9578 |
CAUCUUACCGGACAGUGCUGGA |
| mmu-let-7b* | 1.9529 |
CUAUACAACCUACUGCCUUCCC |
| mmu-miR-3092 | 1.8662 |
GAAUGGGGCUGUUUCCCCUCC |
| mmu-miR-200b* | 1.8444 |
CAUCUUACUGGGCAGCAUUGGA |
| mmu-miR-21 | 1.8270 |
UAGCUUAUCAGACUGAUGUUGA |
| mmu-miR-205 | 1.7966 |
UCCUUCAUUCCACCGGAGUCUG |
| mmu-miR-203 | 1.7911 |
GUGAAAUGUUUAGGACCACUAG |
| mmu-miR-3072 | 1.7820 |
UGCCCCCUCCAGGAAGCCUUCU |
| mmu-miR-27a* | 1.7546 |
AGGGCUUAGCUGCUUGUGAGCA |
| mmu-miR-30e | 1.7120 |
UGUAAACAUCCUUGACUGGAAG |
| mmu-miR-28c | 1.6958 |
AGGAGCUCACAGUCUAUUGA |
| mmu-miR-1843b-5p | 1.6886 |
AUGGAGGUCUCUGUCUGACUU |
| mmu-miR-122 | 1.6574 |
UGGAGUGUGACAAUGGUGUUUG |
| mmu-miR-467d* | 1.6427 |
AUAUACAUACACACACCUACAC |
| mmu-miR-34c* | 1.6416 |
AAUCACUAACCACACAGCCAGG |
| mmu-miR-148a | 1.6200 |
UCAGUGCACUACAGAACUUUGU |
| mmu-miR-1298* | 1.5997 |
CAUCUGGGCAACUGAUUGAACU |
| mmu-miR-1949 | 1.5995 |
CUAUACCAGGAUGUCAGCAUAGUU |
| mmu-miR-15a | 1.5988 |
UAGCAGCACAUAAUGGUUUGUG |
| mmu-miR-497 | 1.5903 |
CAGCAGCACACUGUGGUUUGUA |
| mmu-miR-5127 | 1.5743 |
UCUCCCAACCCUUUUCCCA |
| mmu-miR-200c | 1.5620 |
UAAUACUGCCGGGUAAUGAUGGA |
| mmu-miR-467b* | 1.5491 |
AUAUACAUACACACACCAACAC |
| mmu-miR-205* | 1.5424 |
GAUUUCAGUGGAGUGAAGCUCA |
| mmu-miR-744* | 1.5254 |
CUGUUGCCACUAACCUCAACCU |
| mmu-miR-140 | 1.5219 |
CAGUGGUUUUACCCUAUGGUAG |
| mmu-miR-143 | 1.5215 |
UGAGAUGAAGCACUGUAGCUC |
| mmu-miR-19b | 1.5059 |
UGUGCAAAUCCAUGCAAAACUGA |
| mmu-miR-106b | 1.5015 |
UAAAGUGCUGACAGUGCAGAU |
| mmu-miR-335-5p | 1.5000 |
UCAAGAGCAAUAACGAAAAAUGU |
Computational prediction of potential
target genes and network analysis
Candidate target genes for mmu-miR-141 were
identified using three commonly used prediction algorithms to
reduce the unpredictable number of false positives: PicTar, miRanda
and TargetScan. The results of the miRNA-mRNA regulatory network
analysis indicated that mmu-miR-141 belonged to the miR-8 family.
The miR-8 family also includes mmu-miR-200a, mmu-miR-200b,
mmu-miR-200c and mmu-miR-429, all of which were upregulated in the
retinas of OPTN (E50K) transgenic mice by microarray but did not
achieve statistical significance.
Discussion
The OPTN E50K mutation is the only mutation
currently confirmed to play a causative role in NTG pathogenesis
(2). Transgenic mice that overexpress
OPTN E50K provide a model system for elucidating the
molecular mechanisms leading to POAG (8). While the majority of previous studies
have focused on direct protein-protein interactions, differences in
the expression levels of core proteins have also been noted and may
contribute to POAG pathogenesis. The mechanisms accounting for
differential expression of proteins indicated in POAG are not
clear.
miRNAs can regulate gene expression through sequence
complementarity to the 3′-UTRs of target mRNAs. miRNA binding to
target mRNAs can result in translational repression through mRNA
degradation or translational inhibition (12). Accumulating evidence suggests that
miRNAs act as novel cellular senescence regulators. However, there
is little information regarding the potential involvement of miRNAs
in the mediating effects originating from the expression of mutant
proteins associated with disease processes, such as OPTN
(E50K). The present results suggest that the miRNAs miR-141,
miR-200a, miR-200b, miR-200c and miR-429, which all belong to the
miR-8 family, may be critical regulators of POAG induced by the
OPTN (E50K) mutation.
The miR-8 family has been demonstrated to regulate
numerous biological processes, including the response to osmotic
stress in zebrafish embryos (13) and
the response to steroid signaling in Drosophila (14). miR-200b, miR-200c and miR-429 are
TBK1 gene-related miRNAs according to the miRbase database.
The E50K mutation in OPTN may enhance its interaction with
TBK1 and affect OPTN function by rendering OPTN insoluble
(15). However, no experiments have
been performed to fully elucidate the mechanisms by which the
OPTN (E50K) mutation induces POAG to date. Therefore,
further research to understand the interaction of the OPTN
(E50K) mutation with TBK1 could improve the management of POAG.
In conclusion, the present study identified a number
of differentially expressed miRNAs in the retinas of OPTN
(E50K) mutant transgenic mice. The results suggest several
noteworthy directions for future research aimed at elucidating the
role of miRNAs in glaucoma pathogenesis. Detailed analyses of miRNA
expression patterns and the impact on retinal cell function may
contribute to our understanding of the pathophysiological processes
leading to POAG.
Acknowledgements
The present study was funded by the National Natural
Science Foundation of China (grant no. 81271000) and the Major
State Basic Research Development Program of China (973 Program,
grant no. 2011CB707502). The authors thank Dr Peter Wilker for
editing the manuscript.
References
|
1
|
Quigley HA and Broman AT: The number of
people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol.
90:262–267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iwase A, Suzuki Y, Araie M, Yamamoto T,
Abe H, Shirato S, Kuwayama Y, Mishima HK, Shimizu H, Tomita G, et
al: Tajimi Study Group, Japan Glaucoma Society: The prevalence of
primary open-angle glaucoma in Japanese: The Tajimi Study.
Ophthalmology. 111:1641–1648. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rezaie T, Child A, Hitchings R, Brice G,
Miller L, Coca-Prados M, Héon E, Krupin T, Ritch R, Kreutzer D, et
al: Adult-onset primary open-angle glaucoma caused by mutations in
optineurin. Science. 295:1077–1079. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aung T, Rezaie T, Okada K, Viswanathan AC,
Child AH, Brice G, Bhattacharya SS, Lehmann OJ, Sarfarazi M and
Hitchings RA: Clinical features and course of patients with
glaucoma with the E50K mutation in the optineurin gene. Invest
Ophthalmol Vis Sci. 46:2816–2822. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Leung YF, Fan BJ, Lam DS, Lee WS, Tam PO,
Chua JK, Tham CC, Lai JS, Fan DS and Pang CP: Different optineurin
mutation pattern in primary open-angle glaucoma. Invest Ophthalmol
Vis Sci. 44:3880–3884. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fuse N, Takahashi K, Akiyama H, Nakazawa
T, Seimiya M, Kuwahara S and Tamai M: Molecular genetic analysis of
optineurin gene for primary open-angle and normal tension glaucoma
in the Japanese population. J Glaucoma. 13:299–303. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alward WL, Kwon YH, Kawase K, Craig JE,
Hayreh SS, Johnson AT, Khanna CL, Yamamoto T, Mackey DA, Roos BR,
et al: Evaluation of optineurin sequence variations in 1,048
patients with open-angle glaucoma. Am J Ophthalmol. 136:904–910.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chi ZL, Akahori M, Obazawa M, Minami M,
Noda T, Nakaya N, Tomarev S, Kawase K, Yamamoto T, Noda S, et al:
Overexpression of optineurin E50K disrupts Rab8 interaction and
leads to a progressive retinal degeneration in mice. Hum Mol Genet.
19:2606–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:(Database). D105–D110. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luna C, Li G, Qiu J, Epstein DL and
Gonzalez P: MicroRNA-24 regulates the processing of latent TGFβ1
during cyclic mechanical stress in human trabecular meshwork cells
through direct targeting of FURIN. J Cell Physiol. 226:1407–1414.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meng Q, Xiao Z, Yuan H, Xue F, Zhu Y, Zhou
X, Yang B, Sun J, Meng B, Sun X, et al: Transgenic mice with
overexpression of mutated human optineurin(E50K) in the retina. Mol
Biol Rep. 39:1119–1124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: MicroRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Flynt AS, Thatcher EJ, Burkewitz K, Li N,
Liu Y and Patton JG: miR-8 microRNAs regulate the response to
osmotic stress in zebrafish embryos. J Cell Biol. 185:115–127.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin H, Kim VN and Hyun S: Conserved
microRNA miR-8 controls body size in response to steroid signaling
in Drosophila. Genes Dev. 26:1427–1432. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Minegishi Y, Iejima D, Kobayashi H, Chi
Z-L, Kawase K, Yamamoto T, Seki T, Yuasa S, Fukuda K and Iwata T:
Enhanced optineurin E50K-TBK1 interaction evokes protein
insolubility and initiates familial primary open-angle glaucoma.
Hum Mol Genet. 22:3559–3567. 2013. View Article : Google Scholar : PubMed/NCBI
|