Introduction
Down syndrome (DS) is the most prevalent chromosomal
disorder with an incidence rate between 1 in 1,000 to 1 in 1,100
live births worldwide (1). The
incidence rate is higher in mothers of >35-years-old and
increases with further advances in maternal age (2). DS was clinically described by John
Langdon Down in 1866, prior to the identification of the genetic
basis of the syndrome (3).
Subsequently, an additional chromosome of the later termed
chromosome 21, as the cause of DS, was discovered in 1959 by
Lejeune et al (4). It is
reported that ~95% of patients with DS have this type of trisomy,
and that ~4% of children with DS exhibit trisomy 21 due to
translocation between chromosome 21 and most often an acrocentric
chromosome (5). Additionally, ~1% of
patients are mosaic, and exhibit somatic cells with normal
karyotype alongside others with trisomy (5). These patients typically present with a
less serious phenotype. Rarely, there is partial trisomy 21,
characterized by triplication of only part of chromosome 21
(6). This can aid to determine the
regions of chromosome 21 that are key contributors to the DS
phenotype. Regarding DS phenotype, it has been reported that the
D21S55 region on proximal 21q22.3 contains genes that, when
overexpressed, serve major roles in the pathogenesis of DS
(7). However, genes outside this
region also contribute to DS phenotypes (8). Furthermore, the etiology of the DS
phenotype is complex and includes other mechanisms, including
epigenetic pathways (9). To date,
there has been no pathogenetic model linking specific structural
and functional aspects of chromosome 21 to the DS phenotype.
Nonetheless, in recent years, the involvement of non-coding RNAs,
including microRNAs (miRNA/miRs), and epigenetic mechanisms have
been linked to the DS phenotype, particularly to the intellectual
disability associated with DS.
miRNAs are endogenous RNAs of ~23 nucleotides in
length that pair with the mRNAs of protein-coding genes to direct
post-transcriptional repression, and thus serve important roles in
regulating genes in eukaryotes (10).
Notably, a range of cellular processes, including cell
proliferation, apoptosis and tumorigenesis, organogenesis,
hematopoiesis and developmental timing, are controlled by miRNAs
(11).
In the present review, the possible involvement of
multiple miRNAs in the development of the DS phenotype,
characterized by mental retardation, congenital heart defects,
leukemia, the absence of cardiovascular disease and a low rate of
solid tumor development, is summarized.
Down syndrome
DS is a gene dosage disorder caused by increased
production of the gene products of chromosome 21. For instance, in
DS, the triplicate copy of superoxide dismutase 1 (SOD1) gene,
located on 21q22.11, is responsible for overproduction of SOD1,
which leads to the oxidative stress observed in DS patients
(12). This oxidative stress may
manifest as multiple characteristics of the DS phenotype, including
as cataractogenesis (12) and
premature aging (13). Variable
mental retardation occurs in all patients, and the malformative
features of the phenotype are also variable (3). Generally patients with DS are
underweight and exhibit delayed growth, severe hypotonia and
several dysmorphic features (8). The
latter includes brachycephaly and plagiocephaly, upslanting
palpebral fissures, epicanthus, low-set ears, tongue protrusion,
short hands, single transverse palmar crease and clinodactyly
(14). Individuals with DS develop a
high frequency of infections due to immunological and
non-immunological factors. Among the immunological factors are
suboptimal antibody responses and poor cellular chemotaxis
(15), while the non-immune factors
include airway anomalies, gastro-oesophageal reflux and ear
anomalies (15). Additionally, ~40%
of patients exhibit congenital heart defects (16).
DS patients also have transient leukemoid reactions
(17) and an increased risk of
developing leukemia, most commonly megakaryoblastic (M7) leukemia
(18). This may be due to somatic
mutations of the X-chromosomal gene encoding GATA-binding protein 1
(GATA1), an important transcriptional regulator of normal
megakaryocytic differentiation, which have generally been
identified in DS leukemic cells (19).
Notably, the frequency of solid tumors in DS, namely
neuroblastomas and nephroblastomas in infants and common epithelial
tumors in adults, is reduced compared with normal populations
(20). The involvement of two genes
has been implicated in this lower tumor incidence in individuals
with trisomy 21, namely ETS proto-oncogene 2 (ETS2) (21) and DS candidate region-1 (DSCR1), with
the latter encoding a protein that is able to suppress tumor growth
in mice (22). DSCR1 also regulates
the calcineurin pathway to suppress vascular endothelial growth
factor-mediated angiogenic signalling (22).
Patients with DS develop premature dementia and have
an increased risk of Alzheimer's disease (23), probably due to the roles of amyloid
precursor protein (APP) gene, located on 21q21.3, in DS and AD.
Indeed, partial trisomy 21 without triplication of the APP gene
does not lead to AD (24).
Furthermore, compared with healthy individuals, significantly lower
systolic and diastolic blood pressures and absence of
atherosclerosis are reported in DS patients (25,26). This
protection against atherosclerosis may be due to the reduced level
of heart-type fatty acid binding protein (27). A number of DS patients also present
gastrointestinal disorders, namely duodenal stenosis,
gastroesophageal reflux, imperforate anus and Hirschsprung's
disease (28). Hypothyroidism also
frequently develops in DS patients (29). Collectively these reports demonstrate
the complexity and distinct characteristics of the DS
phenotype.
miRNAs
Primary miRNA transcripts (pri-miRNAs) that contain
cap structures and poly(A) tails are generated by RNA polymerase
II, which transcribes miRNA genes (30). The maturation of pri-miRNAs occurs by
two main events: i) Processing of the pri-miRNAs into stem-loop
precursors of ~70 nucleotides (pre-miRNAs) in the nucleus, and ii)
processing of pre-miRNAs into mature miRNAs in the cytoplasm
(31). The initiation step of miRNA
processing in the nucleus is cleavage by the RNase III, human
Drosha (32). This nuclease is a
component of two multi-protein complexes: A larger complex
containing multiple classes of RNA-associated proteins and a
smaller complex composed of Drosha and the
double-stranded-RNA-binding protein DGCR8, the product of the
DiGeorge syndrome critical region gene 8 (33). Exportin-5 mediates the nuclear export
of pre-miRNAs and binds processed pre-miRNAs in a Ran guanosine
triphosphate-dependent manner (34).
In the cytoplasm, the pre-RNA is processed by Dicer, generating an
miRNA of ~22 nucleotides long (35).
These miRNA sequences are incorporated into the RNA-induced silence
complex (RISC) that targets mRNAs for degradation (35). However, only a single strand of the
miRNA duplex remains in the RISC complex to control the expression
of target genes (36).
miRNAs bind to the 3´-untranslated region (3´-UTR)
of target mRNAs to suppress their expression. Interactions among
factors associated with the 3´UTR of the target mRNA, including
translation regulators, RISC and mRNA decay factors, may determine
the trigger event of miRNA-mediated gene silencing (37). Due to the limited complementarity
between miRNAs and their targets, there are hundreds of potential
mRNA targets per miRNA (38). Thus, a
single miRNA may regulate multiple biological processes (39), and several miRNAs can regulate an
individual target (38).
Additionally, miRNA expression varies depending on cell type and
cellular conditions (38). The
implications of miRNAs in the pathogenesis of DS are subsequently
addressed.
Down syndrome and chromosome 21 miRNAs
The miRBase (http://www.mirbase.org/search.shtml, accessed on
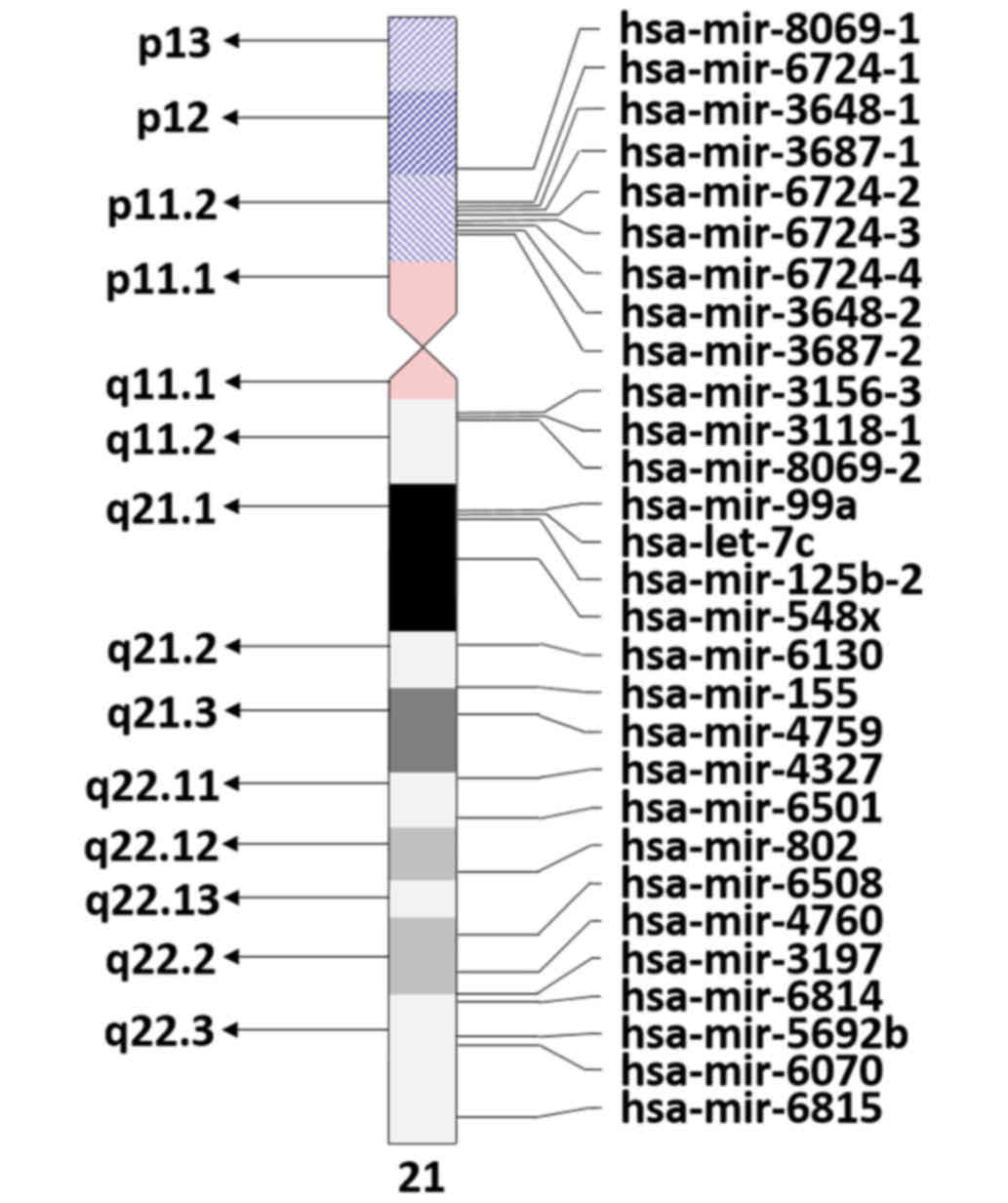
4/9/2017) indicates the presence of 29 Homo sapiens miRNAs on
chromosome 21 (Fig. 1). However, only
some of these miRNAs have been identified to have transcription
levels at the expected 1.5 ratio due to chromosome 21 trisomy in DS
(40). Notably, five of these miRNAs
that meet the overexpression ratio, namely miR-155, miR-802, miR-
125b-2, let-7c and miR-99a, have been implicated to be involved in
DS (41,42). In turn, the specific target genes of
these miRNAs are haploinsufficient in DS (43,44). For
instance, miR-155 targets complement factor H mRNA (CFH), which is
decreased in DS tissues (45). As CFH
protects neurons from axonal injury, complement opsonization, and
leukocyte infiltration in the brain parenchyma (46), overexpression of miR-155 may be
involved in the brain pathology of DS patients. Additionally, CFH
is a repressor of the immune response (45). Thus, among other known factors
described above (15), this
repression may be a cause of susceptibility to infection in DS
patients.
In T21 induced pluripotent stem neuronal progenitor
cells (iPS-NPCs), Lu et al (47) observed the degradation of methyl-CpG
binding protein 2 (MeCP2) following overexpression of miR-155 and
miR-802. Additionally, they observed that T21 iPS-NPCs exhibited
developmental defects and generated fewer neurons than controls
(47). Decreased MeCP2 may also
contribute to the neurochemical abnormalities observed in the
brains of DS individuals (43). In
the study of hippocampal neurons from mice that either lacked
expression or expressed twice the normal levels of MeCP2, Chao
et al (48) identified that
the regulation of glutamatergic synapse number by MeCP2 may be a
mechanism for the altered synaptic strength of neurons in DS.
Keck-Wherley et al (49)
reported that miR-155 and miR-802 were significantly increased in
the DS mouse model Ts65Dn, and that significant overexpression of
these miRNAs may be implicated in hippocampal deficits in DS
phenotypes. Indeed, the hippocampus is an important region involved
in learning and memory and in long-term synaptic plasticity.
Underexpression of angiotensin II type 1 receptor, a
target of miR-155, may explain the absence of cardiovascular
disease in DS individuals (43), as
this receptor has been implicated in cardiovascular pathologies
(50). Additionally, Coppola et
al (51) observed overexpression
of the miR-99a/let-7c cluster and subsequent decrease of their
targets in fetal DS heart tissue, suggesting that the cluster may
contribute to congenital heart defects in DS.
miR-125b-2 may serve a role in the regulation of
megakaryopoiesis and may be an oncogenic miRNA involved in the
pathogenesis of megakaryoblastic leukemia observed in DS (52). Zhang et al (53) demonstrated that the most expressed
miRNAs in pediatric acute myeloid leukemia (AML) were miR-100,
miR-125b, miR-335, miR-146, and miR-99a. MiR-155 was also
identified to be elevated in the bone marrow of some patients with
AML (54). Furthermore, leukemic
cells from DS patients with acute megakaryoblastic leukemia have
been demonstrated to contain acquired mutations in GATA1, which
serves as an important hematopoietic transcription factor (55). Shaham et al (56) also documented a cooperation between
GATA1 and miR-486-5p and observed that miR-486-5p enhanced the
survival of leukemic cells from DS patients.
Overexpression of miR-125a or miR-125b in an Erb-B2
receptor tyrosine kinase 2-dependent human breast cancer cell line
impaired its growth potential and reduced its motility and invasive
capabilities (57). Overexpression of
the miRNA let-7 has also been identified in breast cancer, and may
regulate the tumorigenicity of breast cancer cells (58). In particular, let-7c inhibited the
tumor formation capacity of breast cancer stem cells (59). These results may explain the low rate
of breast cancer among women with DS.
Furthermore, Johnson et al (60) demonstrated that let-7 was highly
expressed in lung tissue, repressed cell proliferation in lung
cells and affected cell cycle progression in a liver cancer cell
line. They also identified that let-7 regulated cell cycle-related
genes involved in the repression of cell proliferation pathways
(60). In particular, let-7c may
inhibit lung adenocarcinoma proliferation (61).
Prostate cancer is also less common in patients with
DS compared with healthy individuals (62). The miR-99 family of miRNAs have been
reported to inhibit the proliferation of prostate cancer cells and
decrease the expression of prostate-specific antigen, a biomarker
for prostate cancer diagnosis (63).
Collectively these reports may explain the low risk
of solid tumor development in patients with DS (64). However, other factors have been
suggested to explain this low tumor risk, including high expression
of the calcineurin inhibitor DSCR1 on the basis of its inhibition
of vascular endothelial growth factor-mediated angiogenic
signalling (22).
A previous study identified two novel miRNAs,
miR-nov1 and miR-nov2, on chromosome 21, located up- and downstream
of the annotated miR-802 loci (40).
miR-nov2 is located in the ‘DS critical region’ (chr21q22.2) and
its overexpression has been identified in DS lymphocytes (40). Xu et al (40) predicted that the 97 mRNA targets of
miR-nov2 were associated with cell growth, cell death, cellular
localization and protein transport. In a subsequent study, they
also confirmed the identification of miR-nov1 and miRnov2 in cord
blood mononuclear cells of DS fetuses (65). Notably, it was observed that miR-99a,
let-7c, miR-125b-2 and miR-155 were downregulated in DS cells
(65). Thus, the role of these miRNAs
in the development of the DS phenotype should be investigated.
miRNAs derived from other chromosomes
associated with DS phenotype
Using microarray technology to identify miRNAs that
were aberrantly expressed, Lim et al (66) compared genome-wide miRNA expression in
the placentas of normal and DS fetuses. They observed that no
chromosome 21-derived miRNAs were differentially expressed.
However, of the 584 genes on chromosome 21, 76 were differentially
expressed and possible targets of miRNAs. These target genes on
chromosome 21 were significantly associated with DS phenotypes,
including mental retardation and congenital abnormalities (66). Nevertheless, the absence of
differentially expressed miRNAs on chromosome 21 between DS and
normal placentas disagrees with other studies conducted in fetal
cord blood cells (65). Lim et
al (66) proposed that this
variance may be due to differences in the characteristics of
tissues reported in previous studies. Indeed, Liang et al
(67) identified that numerous miRNAs
had distinct expression in the placenta compared with other
tissues.
However, the results of Lim et al are also
contrasting to the results of Svobodová et al (68), who observed that three miRNAs located
on chromosome 21 (miR-99a, miR-125b and let-7c) and four miRNAs
located on other chromosomes (miR-542-5p, miR-10b, miR-615 and
miR-654) were upregulated in DS placentas. Additionally, Lim et
al (69) reported that miRNA
expression was significantly different between blood and placenta
samples, and that mir-1973 and mir-3196 were overexpressed in the
trisomy 21 placenta. These two miRNAs may regulate target genes
involved in development of the nervous system (69).
Shi et al (70)
studied the microRNA expression profile of hippocampal tissues from
DS fetuses using miRNA microarray, and reported that the function
of miR-138-5p and the downregulation of its target, enhancer of
zeste homolog 2, in the hippocampus may be involved in the
intellectual disability of DS patients. Furthermore, Wang et
al (71) reported that
interleukin (IL)-1β, IL-12 receptor subunit β2, autism
susceptibility candidate 2 (AUTS2) and KIAA2022 may be involved in
atrioventricular septal defect in DS patients, and that AUTS2 and
KIAA2022 may be targeted by miR-518a, miR-518e, miR-518f, miR-528a
and miR-96.
Lin et al (72)
studied the expression profiles of miRNA and protein in cord blood
samples from DS and normal fetuses, and reported that three miRNAs
(miR-329, miR-27b and miR-27a) and seven proteins (growth factor
receptor-bound protein 2, thymosin β10, RuvB-like 2,
mitogen-activated protein kinase 1, tyrosine-protein phosphatase
non-receptor type 11, α-actin-2 and protein tyrosine kinase 2)
exhibited high levels of differential expression in DS fetuses.
This differential expression may serve a role in the pathogenesis
of DS. More recently, Arena et al (73) reported a higher level of miR-146a
expression in astroglial cells within the hippocampal white matter
of DS fetuses compared with normal fetuses, and identified
persistence of this elevated expression postnatally. This may be a
key finding, as the expression level of miR-146a has been suggested
as an important determinant for neuronal development (74).
Thus, the study of these miRNAs in DS cells may
contribute towards greater comprehension of the DS phenotype.
Conclusion
The overexpression of several miRNAs, including
miR-155, miR-802, miR-99 and let-7c, and the consequent
haploinsufficiency of their specific target proteins are
potentially involved in the DS phenotype. In particular, miR-155
and miR-802 may be involved in neuropathology, the cluster
miR-99/let-7c in congenital heart defects and miR-155 in the
absence of cardiovascular disease observed in DS. Additionally,
miR-125b-2, miR-155 and miR-99a possibly serve roles in the
pathogenesis of megakaryoblastic leukemia in DS patients. A number
of miRNAs expressed in DS patients may also be implicated in the
low rate of solid tumor development in DS patients, including
miR-125b and let-7c in breast cancer, miR-99 in prostate cancer and
let-7c in lung cancer. Nevertheless, the role of miRNAs located on
other chromosomes, and with target genes are on or off chromosome
21, should not be excluded from the DS phenotype.
Acknowledgements
The present review was financed by National Funds
through the FCT Foundation for Science and Technology (project no.
UID/BIM/00009/2016).
References
|
1
|
World Health Organization, . Genomic
Resource Centre: Genes and human disease. http://www.who.int/genomics/public/geneticdiseases/en/index1.htmlSeptember
4–2017
|
|
2
|
Hultén MA, Patel S, Jonasson J and
Iwarsson E: On the origin of the maternal age effect in trisomy 21
Down syndrome: The Oocyte Mosaicism Selection model. Reproduction.
139:1–9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Speicher MR: ChromosomesVogel and
Motulsky's Human Genetics Problems and Approaches. Speicher MR,
Antonarakis SE and Motulsky AG: Springer-Verlag; Berlin,
Heidelberg: pp. 55–138. 2010, View Article : Google Scholar
|
|
4
|
Lejeune J, Gauthier M and Turpin R: Human
chromosomes in tissue cultures. C R Hebd Seances Acad Sci.
248:602–603. 1959.(In French). PubMed/NCBI
|
|
5
|
Mutton D, Alberman E and Hook EB: National
Down Syndrome Cytogenetic Register and the Association of Clinical
Cytogeneticists: Cytogenetic and epidemiological findings in Down
syndrome, England and Wales 1989 to 1993. J Med Genet. 33:387–394.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raoul O, Carpentier S, Dutrillaux B,
Mallet R and Lejeune J: Partial trisomy of chromosome 21 by
maternal translocation t(15;21) (q26.2; q21). Ann Genet.
19:187–190. 1976.(In French). PubMed/NCBI
|
|
7
|
Rahmani Z, Blouin JL, Créau-Goldberg N,
Watkins PC, Mattei JF, Poissonnier M, Prieur M, Chettouh Z, Nicole
A, Aurias A, et al: Down syndrome critical region around D21S55 on
proximal 21q22.3. Am J Med Genet Suppl. 7:98–103. 1990.PubMed/NCBI
|
|
8
|
Korenberg JR, Chen XN, Schipper R, Sun Z,
Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P and Disteche
C: Down syndrome phenotypes: The consequences of chromosomal
imbalance. Proc Natl Acad Sci USA. 91:pp. 4997–5001. 1994;
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Do C, Xing Z, Yu YE and Tycko B:
Trans-acting epigenetic effects of chromosomal aneuploidies:
Lessons from Down syndrome and mouse models. Epigenomics.
9:189–207. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brás A, Monteiro C and Rueff J: Oxidative
stress in trisomy 21. A possible role in cataractogenesis.
Ophthalmic Paediatr Genet. 10:271–277. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Campos C, Guzmán R, López-Fernández E and
Casado A: Urinary uric acid and antioxidant capacity in children
and adults with Down syndrome. Clin Biochem. 43:228–233. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clinical Cytogenetics: Disorders of the
autosomes and the sex chromosomesThompson & Thompson Genetics
in Medicine. Nussbaum RL, McInnes RR and Willard HF: Saunders
Elsevier; pp. 89–113. 2007, View Article : Google Scholar
|
|
15
|
Ram G and Chinen J: Infections and
immunodeficiency in Down syndrome. Clin Exp Immunol. 164:9–16.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Freeman SB, Bean LH, Allen EG, Tinker SW,
Locke AE, Druschel C, Hobbs CA, Romitti PA, Royle MH, Torfs CP, et
al: Ethnicity, sex, and the incidence of congenital heart defects:
A report from the National Down Syndrome Project. Genet Med.
10:173–180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fong CT and Brodeur GM: Down's syndrome
and leukemia: Epidemiology, genetics, cytogenetics and mechanisms
of leukemogenesis. Cancer Genet Cytogenet. 28:55–76. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Creutzig U, Ritter J, Vormoor J, Ludwig
WD, Niemeyer C, Reinisch I, Stollmann-Gibbels B, Zimmermann M and
Harbott J: Myelodysplasia and acute myelogenous leukemia in Down's
syndrome. A report of 40 children of the AML-BFM study group.
Leukemia. 10:1677–1686. 1996.PubMed/NCBI
|
|
19
|
Hitzler JK and Zipursky A: Origins of
leukaemia in children with Down syndrome. Nat Rev Cancer. 5:11–20.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Satgé D, Sommelet D, Geneix A, Nishi M,
Malet P and Vekemans M: A tumor profile in Down syndrome. Am J Med
Genet. 78:207–216. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sussan TE, Yang A, Li F, Ostrowski MC and
Reeves RH: Trisomy represses Apc(Min)-mediated tumours in mouse
models of Down's syndrome. Nature. 451:73–75. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baek KH, Zaslavsky A, Lynch RC, Britt C,
Okada Y, Siarey RJ, Lensch MW, Park IH, Yoon SS, Minami T, et al:
Down's syndrome suppression of tumour growth and the role of the
calcineurin inhibitor DSCR1. Nature. 459:1126–1130. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hartley D, Blumenthal T, Carrillo M,
DiPaolo G, Esralew L, Gardiner K, Granholm AC, Iqbal K, Krams M,
Lemere C, et al: Down syndrome and Alzheimer's disease: Common
pathways, common goals. Alzheimers Dement. 11:700–709. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Prasher VP, Farrer MJ, Kessling AM, Fisher
EM, West RJ, Barber PC and Butler AC: Molecular mapping of
Alzheimer-type dementia in Down's syndrome. Ann Neurol. 43:380–383.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Murdoch JC, Rodger JC, Rao SS, Fletcher CD
and Dunnigan MG: Down's syndrome: An atheroma-free model? BMJ.
2:226–228. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ylä-Herttuala S, Luoma J, Nikkari T and
Kivimäki T: Down's syndrome and atherosclerosis. Atherosclerosis.
76:269–272. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vianello E, Dogliotti G, Dozio E and Corsi
Romanelli MM: Low heart-type fatty acid binding protein level
during aging may protect down syndrome people against
atherosclerosis. Immun Ageing. 10:22013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Buchin PJ, Levy JS and Schullinger JN:
Down's syndrome and the gastrointestinal tract. J Clin
Gastroenterol. 8:111–114. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Purdy IB, Singh N, Brown WL, Vangala S and
Devaskar UP: Revisiting early hypothyroidism screening in infants
with Down syndrome. J Perinatol. 34:936–940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee Y, Jeon K, Lee JT, Kim S and Kim VN:
MicroRNA maturation: Stepwise processing and subcellular
localization. EMBO J. 21:4663–4670. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gregory RI, Yan KP, Amuthan G, Chendrimada
T, Doratotaj B, Cooch N and Shiekhattar R: The Microprocessor
complex mediates the genesis of microRNAs. Nature. 432:235–240.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lund E, Güttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bernstein E, Caudy AA, Hammond SM and
Hannon GJ: Role for a bidentate ribonuclease in the initiation step
of RNA interference. Nature. 409:363–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schwarz DS, Hutvágner G, Du T, Xu Z,
Aronin N and Zamore PD: Asymmetry in the assembly of the RNAi
enzyme complex. Cell. 115:199–208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu W and Coller J: What comes first:
Translational repression or mRNA degradation? The deepening mystery
of microRNA function. Cell Res. 22:1322–1324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saito T and Saetrom P: MicroRNAs-targeting
and target prediction. N Biotechnol. 27:243–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Min A, Zhu C, Peng S, Rajthala S, Costea
DE and Sapkota D: MicroRNAs as important players and biomarkers in
oral carcinogenesis. BioMed Res Int. 2015:1869042015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu Y, Li W, Liu X, Chen H, Tan K, Chen Y,
Tu Z and Dai Y: Identification of dysregulated microRNAs in
lymphocytes from children with Down syndrome. Gene. 530:278–286.
2013a. View Article : Google Scholar
|
|
41
|
Siew WH, Tan KL, Babaei MA, Cheah PS and
Ling KH: MicroRNAs and intellectual disability (ID) in Down
syndrome, X-linked ID, and Fragile X syndrome. Front Cell Neurosci.
7:412013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alexandrov PN, Percy ME and Lukiw WJ:
Chromosome 21-Encoded microRNAs (mRNAs): Impact on Down's syndrome
and trisomy-21 linked disease. Cell Mol Neurobiol. July
7–2017.(Epub ahead of print). doi.org/10.1007/s10571-017-0514-0.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Elton TS, Sansom SE and Martin MM:
Trisomy-21 gene dosage over-expression of miRNAs results in the
haploinsufficiency of specific target proteins. RNA Biol.
7:540–547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Elton TS, Selemon H, Elton SM and
Parinandi NL: Regulation of the MIR155 host gene in physiological
and pathological processes. Gene. 532:1–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li YY, Alexandrov PN, Pogue AI, Zhao Y,
Bhattacharjee S and Lukiw WJ: miRNA-155 upregulation and complement
factor H deficits in Down's syndrome. Neuroreport. 23:168–173.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Griffiths MR, Neal JW, Fontaine M, Das T
and Gasque P: Complement factor H, a marker of self protects
against experimental autoimmune encephalomyelitis. J Immunol.
182:4368–4377. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lu HE, Yang YC, Chen SM, Su HL, Huang PC,
Tsai MS, Wang TH, Tseng CP and Hwang SM: Modeling neurogenesis
impairment in Down syndrome with induced pluripotent stem cells
from Trisomy 21 amniotic fluid cells. Exp Cell Res. 319:498–505.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chao HT, Zoghbi HY and Rosenmund C: MeCP2
controls excitatory synaptic strength by regulating glutamatergic
synapse number. Neuron. 56:58–65. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Keck-Wherley J, Grover D, Bhattacharyya S,
Xu X, Holman D, Lombardini ED, Verma R, Biswas R and Galdzicki Z:
Abnormal microRNA expression in Ts65Dn hippocampus and whole blood:
Contributions to Down syndrome phenotypes. Dev Neurosci.
33:451–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Billet S, Aguilar F, Baudry C and Clauser
E: Role of angiotensin II AT1 receptor activation in cardiovascular
diseases. Kidney Int. 74:1379–1384. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Coppola A, Romito A, Borel C, Gehrig C,
Gagnebin M, Falconnet E, Izzo A, Altucci L, Banfi S, Antonarakis
SE, et al: Cardiomyogenesis is controlled by the miR-99a/let-7c
cluster and epigenetic modifications. Stem Cell Res (Amst).
12:323–337. 2014. View Article : Google Scholar
|
|
52
|
Klusmann JH, Li Z, Böhmer K, Maroz A, Koch
ML, Emmrich S, Godinho FJ, Orkin SH and Reinhardt D: miR-125b-2 is
a potential oncomiR on human chromosome 21 in megakaryoblastic
leukemia. Genes Dev. 24:478–490. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang H, Luo XQ, Zhang P, Huang LB, Zheng
YS, Wu J, Zhou H, Qu LH, Xu L and Chen YQ: MicroRNA patterns
associated with clinical prognostic parameters and CNS relapse
prediction in pediatric acute leukemia. PLoS One. 4:e78262009.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
O'Connell RM, Rao DS, Chaudhuri AA, Boldin
MP, Taganov KD, Nicoll J, Paquette RL and Baltimore D: Sustained
expression of microRNA-155 in hematopoietic stem cells causes a
myeloproliferative disorder. J Exp Med. 205:585–594. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wechsler J, Greene M, McDevitt MA,
Anastasi J, Karp JE, Le Beau MM and Crispino JD: Acquired mutations
in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat
Genet. 32:148–152. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shaham L, Vendramini E, Ge Y, Goren Y,
Birger Y, Tijssen MR, McNulty M, Geron I, Schwartzman O, Goldberg
L, et al: MicroRNA-486-5p is an erythroid oncomiR of the myeloid
leukemias of Down syndrome. Blood. 125:1292–1301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong
C, Huang Y, Hu X, Su F, Lieberman J, et al: let-7 regulates self
renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sun X, Xu C, Tang SC, Wang J, Wang H, Wang
P, Du N, Qin S, Li G, Xu S, et al: Let-7c blocks estrogen-activated
Wnt signaling in induction of self-renewal of breast cancer stem
cells. Cancer Gene Ther. 23:83–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang PY, Sun YX, Zhang S, Pang M, Zhang
HH, Gao SY, Zhang C, Lv CJ and Xie SY: Let-7c inhibits A549 cell
proliferation through oncogenic TRIB2 related factors. FEBS Lett.
587:2675–2681. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Patja K, Pukkala E, Sund R, Iivanainen M
and Kaski M: Cancer incidence of persons with Down syndrome in
Finland: A population-based study. Int J Cancer. 118:1769–1772.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sun D, Lee YS, Malhotra A, Kim HK, Matecic
M, Evans C, Jensen RV, Moskaluk CA and Dutta A: miR-99 family of
MicroRNAs suppresses the expression of prostate-specific antigen
and prostate cancer cell proliferation. Cancer Res. 71:1313–1324.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hasle H: Pattern of malignant disorders in
individuals with Down's syndrome. Lancet Oncol. 2:429–436. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xu Y, Li W, Liu X, Ma H, Tu Z and Dai Y:
Analysis of microRNA expression profile by small RNA sequencing in
Down syndrome fetuses. Int J Mol Med. 32:1115–1125. 2013b.
View Article : Google Scholar
|
|
66
|
Lim JH, Kim DJ, Lee DE, Han JY, Chung JH,
Ahn HK, Lee SW, Lim DH, Lee YS, Park SY, et al: Genome-wide
microRNA expression profiling in placentas of fetuses with Down
syndrome. Placenta. 36:322–328. 2015a. View Article : Google Scholar
|
|
67
|
Liang Y, Ridzon D, Wong L and Chen C:
Characterization of microRNA expression profiles in normal human
tissues. BMC Genomics. 8:1662007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Svobodová I, Korabečná M, Calda P, Břešťák
M, Pazourková E, Pospíšilová Š, Krkavcová M, Novotná M and Hořínek
A: Differentially expressed miRNAs in trisomy 21 placentas. Prenat
Diagn. 36:775–784. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lim JH, Lee DE, Kim SY, Kim HJ, Kim KS,
Han YJ, Kim MH, Choi JS, Kim MY, Ryu HM, et al: MicroRNAs as
potential biomarkers for noninvasive detection of fetal trisomy 21.
J Assist Reprod Genet. 32:827–837. 2015b. View Article : Google Scholar
|
|
70
|
Shi WL, Liu ZZ, Wang HD, Wu D, Zhang H,
Xiao H, Chu Y, Hou QF and Liao SX: Integrated miRNA and mRNA
expression profiling in fetal hippocampus with Down syndrome. J
Biomed Sci. 23:482016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang L, Li Z, Song X, Liu L, Su G and Cui
Y: Bioinformatic analysis of genes and microRNAs associated with
atrioventricular septal defect in Down syndrome patients. Int Heart
J. 57:490–495. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lin H, Sui W, Li W, Tan Q, Chen J, Lin X,
Guo H, Ou M, Xue W, Zhang R, et al: Integrated microRNA and protein
expression analysis reveals novel microRNA regulation of targets in
fetal down syndrome. Mol Med Rep. 14:4109–4118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Arena A, Iyer AM, Milenkovic I, Kovács GG,
Ferrer I, Perluigi M and Aronica E: Developmental expression and
dysregulation of miR-146a and miR-155 in Down's syndrome and mouse
models of Down's syndrome and Alzheimer's disease. Curr Alzheimer
Res. 14:July 6–2017.(Epub ahead of print).
doi.org/10.2174/1567205014666170706112701. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Nguyen LS, Lepleux M, Makhlouf M, Martin
C, Fregeac J, Siquier-Pernet K, Philippe A, Feron F, Gepner B,
Rougeulle C, et al: Profiling olfactory stem cells from living
patients identifies miRNAs relevant for autism pathophysiology. Mol
Autism. 7:12016. View Article : Google Scholar : PubMed/NCBI
|