Introduction
Gliomas are the most common primary and among the
most fatal types of malignant brain tumors, with an annual
incidence of 5.26 per 100,000 population and an overall 17,000
incident diagnoses per year in the United States; and the number of
patients is expected to increase with increases in population age
(1–3).
Based on the histopathological classification of the 2016 WHO
central nervous system tumors, gliomas are diagnosed from lowest to
highest grades: Grade I angiocentric gliomas; grade II
oligodendrogliomas [isocitrate dehydrogenase (IDH)-mutant and
1p/19q-codeleted] and diffuse astrocytomas (IDH-mutant); grade III
anaplastic oligodendrogliomas (IDH-mutant and 1p/19q-codeleted) and
anaplastic astrocytomas (IDH-mutant); and finally grade IV
glioblastomas (GBM) and diffuse midline gliomas (H3K27m-mutant)
(4). Although histopathological
grades remain useful, the prognoses of patients with glioma are
more closely associated with molecular alterations compared with
grades (5,6). Notably, patients with glioma are
typically associated with heterogeneous tumor morphologies and
variable prognoses. Despite therapeutic advances, gliomas remain
incurable, with the most aggressive forms resulting in mortality
within months (7). Therefore, there
is an urgent requirement to increase the understanding of the
etiology, pathology and taxonomy of glioma, to improve the
selection of treatment options for patients and to develop novel
therapeutic strategies.
High-throughput sequencing and microarray
technologies, for example whole-genome sequencing and RNA
sequencing, provide an opportunity to examine the broad range of
genomic information at an unprecedented high resolution. Previous
technological advances (e.g., RNA-Seq, microarray) and the
exponential decrease in the cost of genome sequencing and
microarray detection have made biological data resources
increasingly accessible (8–10). In previous decades, molecular
biomarker-based diagnostics with glioma have resulted in a change
in the traditional pathological classification methods of glioma,
resulting in more reproducible and precise clinically distinct
subtypes (11–15). At present, IDH mutations and
codeletion of chromosome 1p and 19q arms, as a molecular signature,
are now included in standard glioma diagnosis and classification
methods (4,16–18). In
addition, the roles of several key genes have been described,
including telomerase reverse transcriptase (TERT) (19,20),
phosphatase and tensin homolog (PTEN) (21), alpha thalassemia/mental retardation
syndrome X-linked (ATRX) (22), B-Raf
proto-oncogene, serine/threonine kinase (23) and O-6-methylguanine DNA
methyltransferase (MGMT) (24). In
light of these data, the molecular genetic signatures should be
valuable in the diagnostic evaluation or treatment strategies for
glioma (25).
Large population-based data sources are required to
provide accurate descriptions of the mechanism of glioma. Previous
data collection and integrative data analyses, for example, The
Cancer Genome Atlas (http://cancergenome.nih.gov/) or Chinese Glioma Genome
Atlas (http://cgga.org.cn), have revealed the genomic
landscape of either low-grade or high-grade gliomas. In the present
review, the available data resources and the molecular biomarkers
from precision glioma studies were summarized. The aim of the
present study was to provide a wide overview of existing data and
recent molecular profiling, which may improve the current knowledge
base in this field, and thereby assist in generating novel
pathological classification categories, diagnostic methods, and
novel therapeutic approaches to accelerate studies into the causes
and control of glioma.
Available glioma data resources
Previously described high-throughput technologies
have provided the opportunity for the extensive characterization of
genomic statuses, including, but not limited to, genetic
alterations, methylation modification and gene expression
regulation (26,27). Over the previous decade, several
genome projects were initiated, accelerating the comprehensive
understanding of the genetics of glioma (Table I). Using innovative genome analysis
technologies may assist in generating novel glioma therapies,
diagnostic methods and preventive strategies (12,28,29).
Therefore, the present study reviewed currently available glioma
data resource in this community.
 | Table I.Summary of available omics glioma
data reviewed in the present study. |
Table I.
Summary of available omics glioma
data reviewed in the present study.
The Cancer Genome Atlas (TCGA)
In 2005, the National Institute of Health and
National Human Genome Research Institute began the TCGA project to
generate comprehensive multi-dimensional maps of the key genomic
alternations that promote malignant transformation. TCGA pilot
project aimed to depict the molecular characteristics of multiple
‘omics’ in human cancer and to generate a data resource for the
scientific community. As the most common and lethal intracranial
tumor, glioma was the first type of cancer studied by TCGA Research
Network. At present, it contains data from GBM and lower grade
gliomas from multiple platforms, including copy number (1,090
samples), DNA methylation (936 samples), RNA-seq profiling (676
samples), mRNA microarray profiling (567 samples), miRNA microarray
profiling (565 samples) data recorded in the Broad FireBrowse
database (http://firebrowse.org/). Integrative
analysis of DNA copy number, gene expression and DNA methylation
data in 206 GBMs revealed that several key genes [including erb-B2
receptor tyrosine kinase 2, neurofibromin 1 (NF1), tumor protein
p53 (TP53), phosphoinositide 3-kinase regulatory subunit 1 and
cyclin dependent kinase inhibitor 2A/B (CDKN2A/B)] and signaling
pathways (including receptor tyrosine kinase/Ras/phosphoinositide
3-kinase, TP53 and CDKN2A) were identified to be frequently mutated
in human GBMs, demonstrating that these data may improve the
understanding of the molecular basis of cancer (30). In addition, a robust gene
expression-based molecular classification of GBMs into the
proneural, neural, classical and mesenchymal subtypes described by
TCGA network provided evidence to support the requirement for
targeted therapeutics (13). This
study unifies genomic and transcriptome dimensions for molecular
GBM stratification by abnormalities in platelet derived growth
factor receptor alpha (PDGFRA), isocitrate dehydrogenase
(NADP(+))1, cytosolic (IDH1), epidermal growth factor receptor
(EGFR) and NF1 genes, which may provide important insight for
future clinical application. Previously, Brennan et al
(28) performed the largest
multi-platform genomic analysis to define the critical genes
associated with gliomas by using 1,122 all-grade gliomas from TCGA.
In this study, they identified six methylation groups and four RNA
expression groups, calculated molecular correlations and provided
understanding of the malignant progression of gliomas. The results
of the whole-genome sequencing revealed that ATRX but not TERT
promoter mutations were associated with increased telomere length,
indicating an alternative mechanism for telomeres lengthening.
Notably, a group of IDH mutant glioma was associated with DNA
demethylation and relatively poor survival; a subtype of IDH-wild
glioma exhibited molecular similarity to pilocytic astrocytoma and
favorable outcomes. This multi-omics glioma analysis provided novel
insights into genomic alternations, emphasized the relevance of DNA
methylation profiles for clinical classification, and associated
somatic alterations involved in telomere maintenance.
At present, several exploratory analysis tools and
databases have also been developed based on TCGA datasets. The cBio
Cancer Genomics Portal (cBioPortal; http://cbioportal.org), an open-access and open-source
resource, was developed as an interactive exploration of cancer
genomics datasets and an intuitive method of presenting data,
including the capacity to quickly view genomic alterations of genes
or pathways of interest across a set of patients, and performing
survival and biological network analysis (31,32). The
portal currently stores DNA copy number variants, methylation, mRNA
and microRNA expression, protein and clinical data. In addition,
these TCGA datasets may be also easily and directly downloaded via
the Broad FireBrowse website, which is updated regularly. These
tools provide rapid, intuitive access to cancer genomics profiling
and matched clinical data, and allows the translation of these
valuable data into biological insights and clinical applications.
To investigate the biological nature of TCGA long non-coding RNAs
(lncRNAs), Cerami et al (31)
and Gao et al (32) developed
the TANRIC and Co-LncRNA databases, respectively, which facilitated
the study of the biological functions of lncRNA and their clinical
applications.
Chinese Glioma Genome Atlas
(CGGA)
The CGGA project (http://cgga.org.cn) is hosted and supported by the
Beijing Neurosurgical Institute and the Chinese Glioma Cooperative
Group Research Network. This project is a comprehensive and
coordinated effort to accelerate the understanding of the molecular
basis of glioma, particularly in secondary GBMs through the
application of high-throughput biotechnologies and bioinformatics.
The project aims to catalogue and identify major genomic
alternations that drive glioma progression, and to provide a
detailed genomic characterization of a large cohort of Chinese
gliomas.
To share this resource, the CGGA portal was
established as an open-access platform for the interactive
exploration of multidimensional glioma genomics datasets. At
present, it provides access to DNA methylation microarray (149
samples), mRNA microarray (305 samples) and sequencing (325
samples), microRNA microarray (198 samples) data and matched
clinical data. Using the gene expression profile from an Agilent
microarray of 225 samples from the CGGA, consensus average linkage
clustering identified three major subgroups (G1, G2 and G3)
(33). The G1 subtype demonstrated
improved clinical outcome, young age and a high frequency of IDH1
mutation. The G3 subtype was characterized by poorer clinical
outcome, older age and low frequency of IDH1 mutation. The
parameters for clinical outcome, age and IDH1 mutation in the G2
subgroup were in between the values for the G1 and G3 subtypes.
Combining mutation data of the TERT promoter and IDH from 377 CGGA
grade II/III glioma samples, Brat et al (12) performed a molecular classification of
glioma into IDH-mutation/TERTp-mutation, IDH-mutation only,
TERTp-mutation only and IDH-wild type/TERT promoter-wild type
groups. Patients with only TERTp-mut genotypes exhibited the
poorest prognoses, while patients with an IDH mutation alone
demonstrated more favorable prognoses. This study suggested that
combining mutation data from the TERT promoter and IDH genes
created a novel method of defining glioma subgroups to supplement
the traditional histopathological criteria for disease diagnosis.
To facilitate an increased use of these RNA sequencing data from
the CGGA project, a free, web accessible and use-friendly database
was also constructed (GLIOMASdb; http://cgga.org.cn:9091/gliomasdb/) (34). The GLIOMASdb currently provides data
available to download and analysis of gene patterns in the
malignant progression of gliomas.
Other datasets
The rapid expansion of integrated multi-omics,
bioinformatics analyses and clinical translation has markedly
altered the understanding of glioma. Over the previous decade,
several other glioma genome projects have been undertaken.
The REpository of Molecular BRAin Neoplasia DaTa
(REMBRANDT), a cancer clinical genomics database and an online
mining and analysis platform, aimed to improve understanding of
glioma by effectively combining clinical information and genomic
characteristics (35). To date,
REMBRANDT includes 874 glioma specimens comprising ~566 gene
expression arrays, 834 DNA copy number arrays and 13,472 clinical
data, which may be used as an independent dataset for glioma
research.
To improve glioma stratification standards,
Gravendeel et al (5) performed
gene expression profiling (Affymetrix HU133 Plus 2.0, n=276) from
patients with glioma (GSE16011). In this study, seven distinct
molecular subgroups were identified and correlated with survival.
These contained two subgroups with favorable prognosis (median
survival >4.7 years), two intermediate prognostic subtypes
(median survival of 1–4 years), two with poor prognosis (median
survival of <1 year) and one control group. This result was
validated by 5 other independent datasets, supporting this evidence
that gene expression profiling may be an effective method to
classify gliomas, and that this molecular classification may assist
to diagnose and guide clinical decision making.
In addition, Ballester et al (36) performed a retrospective analysis of
sequencing results of 381 primary gliomas. These cases, including
GBM (n=227), anaplastic astrocytomas (n=46), diffuse astrocytoma
(n=37), anaplastic oligodendrogliomas (n=21) and oligodendrogliomas
(n=11), were used to identify mutations and amplifications in
cancer-associated genes using a validated, commercially-available
panel. The results revealed that the most commonly mutated genes
included TP53 (37.2%), IDH1 (29.4%),
phosphatidylinositol-4,5-biphosphate 3-kinase catalytic subunit
alpha (PIK3CA; 8%), PTEN (8%) and EGFR (7.5%). In addition, 23%
cases (88/381) exhibited genomic amplification in at least 1
cancer-associated gene in the specific panel. The most common genes
that indicated evidence of amplification included EGFR (18.0%),
PDGFRA (2.5%) and KIT proto-oncogene receptor tyrosine kinase
(1.8%). This study demonstrated the utility of next-generation
sequencing for the identification of genetic alterations in brain
tumors in the clinical setting.
Molecular profiling as a precise clinical
tool
At present, treatment decisions in patients with
glioma primarily depend on histological classification and clinical
parameters. However, the differences between histological subgroups
and grades are subtle, and are susceptible to high inter-observer
variability. Previously, additional data have suggested that the
molecular profiles of gliomas are an improved predictor of survival
compared with that of histology results (5). At present, most of studies have used
information other than histological or clinical data to establish
the molecular classifiers of glioma. The traditional molecular
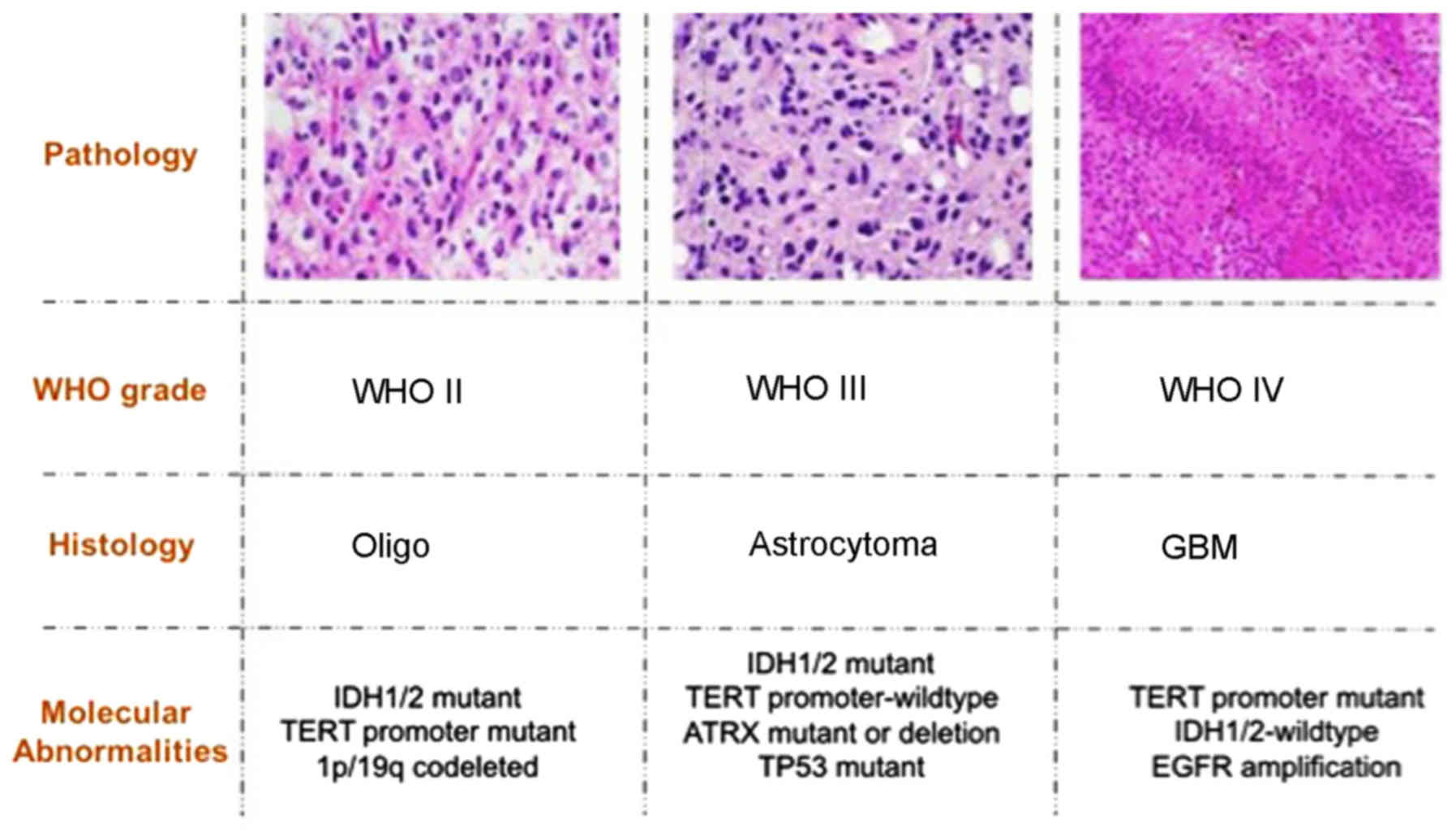
pathology of gliomas are summarized in Fig. 1. The key gene alternations in gliomas
are summarized in Table II.
 | Table II.Summary of key gene alternations in
gliomas. |
Table II.
Summary of key gene alternations in
gliomas.
| Histology | Molecular
abnormalities | WHO grade |
|---|
|
Oligodendroglioma | IDH-mutant &
1p/19q-codeleted | WHO II |
| Astrocytoma | TERT
promoter-wildtype | WHO II |
| (IDH-mutant) | IDH-mutant |
|
| Astrocytoma | IDH-wildtype | WHO II |
| (IDH-wildtype) |
|
|
| Anaplastic
astrocytoma | IDH-mutant | WHO III |
| (IDH-mutant) |
|
|
| Anaplastic
astrocytoma | IDH-wildtype | WHO III |
| (IDH-wildtype) |
|
|
| GBM | TERT
promoter-mutant | WHO IV |
| (IDH-wildtype) | IDH-wildtype |
|
| GBM
(IDH-mutant) | TERT
promoter-mutant | WHO IV |
|
| IDH-mutant |
|
| GBM (F3-T3+) | FGFR3-TACC3 gene
fusion | WHO IV |
| Secondary GBM
(ZM+) | PTPRZ1-MET gene
fusion | WHO IV |
IDH mutation and chromosome 1p/19q codeletion. IDH1
and IDH2 are NADP+-dependent isocitrate dehydrogenases, catalyzing
the oxidative decarboxylation of isocitrate to α-ketoglutarate
(α-KG) and converting NADP to NADPH. Previous data implied that the
mutations in the IDH1 and IDH2 genes were closely associated with
the pathogenesis of malignant gliomas (18,37,38).
Generally, patients with glioma with and without IDH mutations
exhibit significantly different outcomes (12). In particular, IDH mutation, including
R132H in the IDH1 gene, are commonly identified in WHO grade II and
III disease (oligodendrogliomas, astrocytomas and secondary
glioblastomas), suggesting that IDH mutations may be early events
and promote the progression of gliomas (38). In addition, complete deletion of the
short arm of chromosome 1 and the long arm of chromosome 19 (1p/19
co-deletion) is the molecular genetic signature of
oligodendrogliomas, which occurs early in its pathogenesis
(39). The 1p/19q co-deletion occurs
in the majority of WHO grade II tumors, and has been a valuable
diagnostic, prognostic and predictive biomarker for the management
of oligodendroglial tumors (12).
TERT promoter mutations
TERT is a catalytic subunit of the telomerase and
encodes a highly specialized reverse transcriptase. The role of
TERT has been revealed by the frequent mutations of the TERT
promoter (TERTp-mut) in gliomagenesis, but particularly in glioma
(12,18). The mutations of the TERT promoter
frequently occur in 2 loci, C228T and C250T, mapping −124 and
−146bp, respectively, upstream of the TERT ATG site. The presence
of TERTp-mut, creating binding sites for E26
transformation-specific/T-cell factor transcription factors, is
significantly associated with higher mRNA expression. Increasing
evidence has suggested that TERTp-mut also affects cancer
susceptibility and results in poorer prognosis for patients with
glioma (40).
MGMT promoter methylation
DNA methylation of the gene promoter may serve an
important role in carcinogenesis. MGMT, as a coding DNA repair
gene, is crucial for genome stability. During DNA replication and
transcription, the MGMT gene may repair the naturally occurring
mutagenic DNA lesion O-6-methylguanine back to guanine, preventing
mismatch and errors. A number of studies have demonstrated that the
methylation state of the MGMT gene is closely associated with the
response to temozolomide (41).
Specifically, if the promoter is methylated, the treatment is more
effective; otherwise, the patient is not sensitive, suggesting that
MGMT promoter methylation is a favorable predictor for overall
survival and progression-free survival in glioma (42,43). At
present, MGMT has also been revealed to be a useful tool for
increasing gene therapy efficiency, and with applications in
clinical detection (44).
Fusion genes
Gene rearrangements and the consequent fusion
proteins serve an important role in tumorigenesis. It was
previously demonstrated that a recurrent gene fusion event
involving the protein tyrosine phosphatase receptor type Z1
(PTPRZ1) and MET proto-oncogene, receptor tyrosine kinase (MET)
genes, termed the ZM fusion, was identified in 15% of secondary
glioblastomas (45). In this fusion
event, the PTPRZ1 promoter was activated to additionally promote
the expression of full-length MET, leading to MET overexpression.
Patients with ZM fusion exhibited more aggressive phenotypes. The
recurrent nature of the ZM fusion suggests that ZM fusion is
associated with GBM migration and invasion, participates in PIK3CA
signaling, and results in a poorer prognosis, supporting ZM as a
potential GBM therapeutic target. In addition, the fibroblast
growth factor receptor (FGFR)-transforming acidic coiled-coil
containing protein (TACC) was the first well-characterized gene
fusion event in GBMs (46–48). These fusion proteins include the
tyrosine kinase domain of FGFR and the coiled-coil domain of TACC
proteins. There are 2 subtypes of the FGFR-TACC gene fusion
protein, FGFR3-TACC3 (F3-T3) and FGFR3-TACC1, in GBMs. Previously,
Bao et al (45) indicated that
F3-T3 was associated with oxidative phosphorylation and
mitochondrial biogenesis. FGFR-TACC gene fusion may also provide a
promising target for GBM treatment.
Conclusions
Within the previous decade, the volume of
newly-published biological data has grown rapidly, and has
increased understanding of the key genomic alternations in human
glioma. At present, several glioma-associated genomic studies have
been initiated, including TCGA and the CGGA, aiming to generate and
comprehensively describe the multi-dimensional glioma genome. These
projects provided unprecedented and publicly available glioma
datasets, used widely by the scientific community. To provide
improved understanding of the genetics of glioma, several studies
have classified glioma into subtypes and revealed their
associations with clinical parameters, assisting in the generation
of novel cancer therapies, diagnostic methods and preventive
strategies.
Although malignant glioma remains incurable,
treatment options have been expanding and improving due to improved
understanding of the complex molecular biology of these tumors.
Based on the 2016 WHO organization classification of brain tumors,
diffuse gliomas are defined by histopathology and molecular
pathology, particularly for molecular diagnosis. At present,
several key genes in glioma have been used as biomarkers for
predicting outcome and guiding chemoradiotherapy, including MGMT
promoter methylation. As important biological molecules, non-coding
RNAs are emerging as biomarkers or potential targets for glioma
treatment (49–52). Future studies will also generate
additional biological glioma data that may reveal novel biological
markers as therapeutic targets/candidates in the treatment of
tumors.
Acknowledgements
The authors would like to acknowledge Dr Fanlin Meng
at the School of Medicine, Tsinghua University, Beijing, China for
her assistance in revising the manuscript.
Funding
The present study was supported by The National Key
Research and Development Plan (grant no. 2016YFC0902500).
Availability of data and materials
Not applicable.
Authors' contributions
All authors participated in the design and finial
review of the manuscript. JC and ZZ conceived the review. ZZ and KZ
drafted the manuscript. ZW and KW collected the information on data
resources. XL and FW collected the information on molecular
profiles. All authors reviewed and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Gittleman H, Liao P,
Vecchione-Koval T, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS Statistical Report: Primary brain and other central nervous
system tumors diagnosed in the United States in 2010–2014.
Neuro-oncol. 19 Suppl 5:v1–v88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the Central Nervous System: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gravendeel LA, Kouwenhoven MC, Gevaert O,
de Rooi JJ, Stubbs AP, Duijm JE, Daemen A, Bleeker FE, Bralten LB,
Kloosterhof NK, et al: Intrinsic gene expression profiles of
gliomas are a better predictor of survival than histology. Cancer
Res. 69:9065–9072. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buckner J, Giannini C, Eckel-Passow J,
Lachance D, Parney I, Laack N and Jenkins R: Management of diffuse
low-grade gliomas in adults - use of molecular diagnostics. Nat Rev
Neurol. 13:340–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao S, Cai J, Li J, Bao G, Li D, Li Y,
Zhai X, Jiang C and Fan L: Bioinformatic Profiling Identifies a
Glucose-Related Risk Signature for the Malignancy of Glioma and the
Survival of Patients. Mol Neurobiol. 54:8203–8210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19(1A): A68–A77. 2015.PubMed/NCBI
|
|
9
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The Cancer Cell Line Encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Consortium EP: ENCODE Project Consortium:
An integrated encyclopedia of DNA elements in the human genome.
Nature. 489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arita H, Yamasaki K, Matsushita Y,
Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M,
Shimizu S, et al: A combination of TERT promoter mutation and MGMT
methylation status predicts clinically relevant subgroups of newly
diagnosed glioblastomas. Acta Neuropathol Commun. 4:792016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brat DJ, Verhaak RG, Aldape KD, Yung WK,
Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O,
et al: Cancer Genome Atlas Research Network: Comprehensive,
Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl
J Med. 372:2481–2498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Verhaak RG, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Cancer Genome Atlas Research Network: Integrated genomic analysis
identifies clinically relevant subtypes of glioblastoma
characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1.
Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Diamandis P and Aldape KD: Insights From
Molecular Profiling of Adult Glioma. J Clin Oncol. 35:2386–2393.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang P, Cai J, Yan W, Zhang W, Wang Y,
Chen B, Li G, Li S, Wu C, Yao K, et al: CGGA project:
Classification based on mutations of TERT promoter and IDH
characterizes subtypes in grade II/III gliomas. Neuro-oncol.
18:1099–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang P, Cai J, Yan W, Zhang W, Wang Y,
Chen B, Li G, Li S, Wu, Yao K, et al: CGGA project: Classification
based on mutations of TERT promoter and IDH characterizes subtypes
in grade II/III gliomas. Neuro-oncol. 18:1099–1108. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang L, Jiang T, Yuan F, Li GL, Cui Y,
Liu EZ and Wang ZC: Correlation of chromosomes 1p and 19q status
and expressions of O6-methylguanine DNA methyltransferase (MGMT),
p53 and Ki-67 in diffuse gliomas of World Health Organization (WHO)
grades II and III: A clinicopathological study. Neuropathol Appl
Neurobiol. 35:367–379. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eckel-Passow JE, Lachance DH, Molinaro AM,
Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML,
Smirnov IV, et al: Glioma Groups Based on 1p/19q, IDH, and TERT
Promoter Mutations in Tumors. N Engl J Med. 372:2499–2508. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Killela PJ, Reitman ZJ, Jiao Y, Bettegowda
C, Agrawal N, Diaz LA Jr, Friedman AH, Friedman H, Gallia GL,
Giovanella BC, et al: TERT promoter mutations occur frequently in
gliomas and a subset of tumors derived from cells with low rates of
self-renewal. Proc Natl Acad Sci USA. 110:6021–6026. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pekmezci M, Rice T, Molinaro AM, Walsh KM,
Decker PA, Hansen H, Sicotte H, Kollmeyer TM, McCoy LS, Sarkar G,
et al: Adult infiltrating gliomas with WHO 2016 integrated
diagnosis: Additional prognostic roles of ATRX and TERT. Acta
Neuropathol. 133:1001–1016. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang SI, Puc J, Li J, Bruce JN, Cairns P,
Sidransky D and Parsons R: Somatic mutations of PTEN in
glioblastoma multiforme. Cancer Res. 57:4183–4186. 1997.PubMed/NCBI
|
|
22
|
Koschmann C, Calinescu AA, Nunez FJ,
Mackay A, Fazal-Salom J, Thomas D, Mendez F, Kamran N, Dzaman M,
Mulpuri L, et al: ATRX loss promotes tumor growth and impairs
nonhomologous end joining DNA repair in glioma. Sci Transl Med.
8:328ra282016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dahiya S, Emnett RJ, Haydon DH, Leonard
JR, Phillips JJ, Perry A and Gutmann DH: BRAF-V600E mutation in
pediatric and adult glioblastoma. Neuro-oncol. 16:318–319. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wick W, Weller M, van den Bent M, Sanson
M, Weiler M, von Deimling A, Plass C, Hegi M, Platten M and
Reifenberger G: MGMT testing - the challenges for biomarker-based
glioma treatment. Nat Rev Neurol. 10:372–385. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Frattini V, Trifonov V, Chan JM, Castano
A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, et al: The
integrated landscape of driver genomic alterations in glioblastoma.
Nat Genet. 45:1141–1149. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
No authors listed. The future of cancer
genomics. Nat Med. 21:992015. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stratton MR, Campbell PJ and Futreal PA:
The cancer genome. Nature. 458:719–724. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brennan CW, Verhaak RG, McKenna A, Campos
B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ,
Berman SH, et al: TCGA Research Network: The somatic genomic
landscape of glioblastoma. Cell. 155:462–477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee JK, Wang J, Sa JK, Ladewig E, Lee HO,
Lee IH, Kang HJ, Rosenbloom DS, Camara PG, Liu Z, et al:
Spatiotemporal genomic architecture informs precision oncology in
glioblastoma. Nat Genet. 49:594–599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cancer Genome Atlas Research Network:
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan W, Zhang W, You G, Zhang J, Han L, Bao
Z, Wang Y, Liu Y, Jiang C, Kang C, et al: Molecular classification
of gliomas based on whole genome gene expression: A systematic
report of 225 samples from the Chinese Glioma Cooperative Group.
Neuro-oncol. 14:1432–1440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao Z, Meng F, Wang W, Wang Z, Zhang C
and Jiang T: Comprehensive RNA-seq transcriptomic profiling in the
malignant progression of gliomas. Sci Data. 4:1700242017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Madhavan S, Zenklusen JC, Kotliarov Y,
Sahni H, Fine HA and Buetow K: Rembrandt: Helping personalized
medicine become a reality through integrative translational
research. Mol Cancer Res. 7:157–167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ballester LY, Fuller GN, Powell SZ, Sulman
EP, Patel KP, Luthra R and Routbort MJ: Retrospective Analysis of
Molecular and Immunohistochemical Characterization of 381 Primary
Brain Tumors. J Neuropathol Exp Neurol. 76:179–188. 2017.PubMed/NCBI
|
|
37
|
Ducray F, Marie Y and Sanson M: IDH1 and
IDH2 mutations in gliomas. N Engl J Med. 360:2248–2249; author
reply 2249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vijayakumar V, Liebisch G, Buer B, Xue L,
Gerlach N, Blau S, Schmitz J and Bucher M: Integrated multi-omics
analysis supports role of lysophosphatidylcholine and related
glycerophospholipids in the Lotus japonicus-Glomus intraradices
mycorrhizal symbiosis. Plant Cell Environ. 39:393–415. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Diplas BH, He X, Brosnan-Cashman JA, Liu
H, Chen LH, Wang Z, Moure CJ, Killela PJ, Loriaux DB, Lipp ES, et
al: The genomic landscape of TERT promoter wildtype-IDH wildtype
glioblastoma. Nat Commun. 9:20872018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Perry JR, Laperriere N, O'Callaghan CJ,
Brandes AA, Menten J, Phillips C, Fay M, Nishikawa R, Cairncross
JG, Roa W, et al: Trial Investigators: Short-Course Radiation plus
Temozolomide in Elderly Patients with Glioblastoma. N Engl J Med.
376:1027–1037. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Weller M, Stupp R, Reifenberger G, Brandes
AA, van den Bent MJ, Wick W and Hegi ME: MGMT promoter methylation
in malignant gliomas: Ready for personalized medicine? Nat Rev
Neurol. 6:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Petrova RS, Webb KF, Vaghefi E, Walker K,
Schey KL and Donaldson PJ: Dynamic functional contribution of the
water channel AQP5 to the water permeability of peripheral lens
fiber cells. Am J Physiol Cell Physiol. 314:C191–C201. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Binabaj MM, Bahrami A, ShahidSales S,
Joodi M, Mashhad Joudi M, Hassanian SM, Anvari K and Avan A: The
prognostic value of MGMT promoter methylation in glioblastoma: A
meta-analysis of clinical trials. J Cell Physiol. 233:378–386.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K,
Ye WL, Hu BQ, Yan W, Zhang W, Akers J, et al: RNA-seq of 272
gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in
secondary glioblastomas. Genome Res. 24:1765–1773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Singh D, Chan JM, Zoppoli P, Niola F,
Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S,
et al: Transforming fusions of FGFR and TACC genes in human
glioblastoma. Science. 337:1231–1235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Di Stefano AL, Fucci A, Frattini V,
Labussiere M, Mokhtari K, Zoppoli P, Marie Y, Bruno A, Boisselier
B, Giry M, et al: Detection, Characterization, and Inhibition of
FGFR-TACC Fusions in IDH Wild-type Glioma. Clin Cancer Res.
21:3307–3317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lasorella A, Sanson M and Iavarone A:
FGFR-TACC gene fusions in human glioma. Neuro Oncol. 19:475–483.
2017.PubMed/NCBI
|
|
49
|
Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z,
Shao T, Jiang T, Ren H, Kang C, et al: Comprehensive analysis of
the functional microRNA-mRNA regulatory network identifies miRNA
signatures associated with glioma malignant progression. Nucleic
Acids Res. 41:e2032013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Luo H, Chen Z, Wang S, Zhang R, Qiu W,
Zhao L, Peng C, Xu R, Chen W, Wang HW, et al: c-Myc-miR-29c-REV3L
signalling pathway drives the acquisition of temozolomide
resistance in glioblastoma. Brain. 138:3654–3672. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Z, Zhang C, Liu X, Wang Z, Sun L, Li
G, Liang J, Hu H, Liu Y, Zhang W, et al: Molecular and clinical
characterization of PD-L1 expression at transcriptional level via
976 samples of brain glioma. OncoImmunology. 5:e11963102016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang CB, Zhu P, Yang P, Cai JQ, Wang ZL,
Li QB, Bao ZS, Zhang W and Jiang T: Identification of high risk
anaplastic gliomas by a diagnostic and prognostic signature derived
from mRNA expression profiling. Oncotarget. 6:36643–36651.
2015.PubMed/NCBI
|
|
53
|
Lee Y, Scheck AC, Cloughesy TF, Lai A,
Dong J, Farooqi HK, Liau LM, Horvath S, Mischel PS and Nelson SF:
Gene expression analysis of glioblastomas identifies the major
molecular basis for the prognostic benefit of younger age. BMC Med
Genomics. 1:522008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Phillips HS, Kharbanda S, Chen R, Forrest
WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et
al: Molecular subclasses of high-grade glioma predict prognosis,
delineate a pattern of disease progression, and resemble stages in
neurogenesis. Cancer Cell. 9:157–173. 2006. View Article : Google Scholar : PubMed/NCBI
|