Introduction
A relationship between oral pathogenic bacteria and
systemic and oral diseases has been recognized in elderly and
hospitalized patients (1,2). In particular, elderly patients exhibit
high incidence rates of aspiration pneumonia, which is considered
to be associated with poor oral hygiene and impaired oral function
(1). Additionally, dry mouth is
common among the elderly, and it may lead to oral diseases
including dental caries, oral candidiasis and swallowing disorders
(3). Importantly, oral health care
can aid to remove dental plaques, moisturize the oral cavity,
relieve mucosal inflammation, and promote oral hygiene by reducing
oral bacterial numbers (2,4). Thus, regular oral care serves a vital
role in the maintenance of oral health, which may result in a
decreased risk of systemic disease in elderly subjects.
In a previous study, our group prospectively
investigated the changes in oral bacterial numbers in colorectal
cancer patients during the perioperative period (4). Notably, bacterial numbers on the tongue
dorsum changed less markedly than those of the gingiva of the upper
anterior teeth and palatoglossal arch, even when perioperative oral
health care had been performed (5).
Bacteria of the tongue dorsum may be less susceptible to oral
health care practices such as tongue cleaning with a sponge or
tongue brushes (5,6). This may be because the anatomical
features of the tongue dorsum, namely the papillae, creates
reservoirs for pathogenic bacteria, which may be associated with
oral and systemic disease (7). To
date, the association of between bacteria on the tongue surface
with oral health status and systemic disease remains unclear. Thus,
the correlation between bacteria on the tongue surface and oral and
general health status should be clarified.
In taxonomic studies, sequence analysis of the 16S
ribosomal RNA (rRNA) gene is widely used to identify bacterial
species (8–10). This method enables the measurement of
numerous bacterial strains without specialized culturing
conditions. Bacterial 16S rRNA genes generally contain 9
hypervariable regions that demonstrate considerable sequence
diversity among bacterial species and can be used for species
identification (9,10). Conserved regions of the gene can be
used to design universal primers to amplify various bacteria
(9,10), thus allowing total bacterial numbers
to be estimated using a bacterial DNA sample. Herein, the present
study performed real-time polymerase chain reaction (PCR) using
universal primers for the bacterial 16S rRNA gene to calculate oral
bacterial numbers.
Further clinical investigation is required to
clarify the relationship between oral bacteria and general health
status. Therefore, this study preliminary investigated bacteria on
the tongue surface using PCR amplification of the bacterial 16S
rRNA gene, and examined the correlation between tongue bacteria and
systemic disease in middle-aged and elderly patients.
Materials and methods
Subjects
The current study investigated 70 patients (23
males, 47 females; mean age, 69.5 years; range, 45–92 years) who
visited the Department of Oral Health of Hiroshima University
Hospital, Hiroshima, Japan from March 2018 to April 2018. A 0–3 mm
probing pocket depth was considered to indicate mild or no
periodontitis (11). Moderate
periodontal disease was defined as the presence of ≥4 and <6 mm
probing pocket depth (11) Severe
periodontitis was defined as the presence of ≥6 mm pocket (11). All patients exhibited a mild to
moderate periodontal status. No patients exhibited symptoms of
acute periodontal disease. Regarding toothbrushing frequency, all
participants brushed their teeth more than twice daily. None used a
tongue scraper to clean the surface area of the tongue. The study
design was approved by the Ethics Committee of Hiroshima
University, and all participants signed an informed consent
agreement. Clinical data were obtained regarding age, sex, smoking
status, alcohol consumption and medical history.
Oral sample processing and DNA
extraction
Samples were collected from the tongue dorsum using
a Orcellex® Brush (Rovers Medical Devices, Oss, The
Netherlands) which allows collection of cells or bacteria from oral
epithelial layers. The tongue surface was swabbed prior to tooth
brushing using the Orcellex® 10 times. DNA was extracted
and purified using a PureLink™ Microbiome DNA Purification kit
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). For
PCR analysis, 1 µl was used from a total of 50 µl of oral sample
DNA.
Quantification of bacterial
numbers
The bacterial 16S rRNA gene was used to quantify
bacterial numbers using real-time PCR. Since 16S rRNA gene copy
numbers differ among bacteria, it is difficult to use them in the
accurate quantification of bacteria. Many bacteria have 1 to 10 16S
rRNA gene copies, while some have >10 (12,13). Here,
16S rRNA gene copy numbers were used from Staphylococcus
aureus (5 copies per cell) to quantify approximate oral
bacterial numbers.
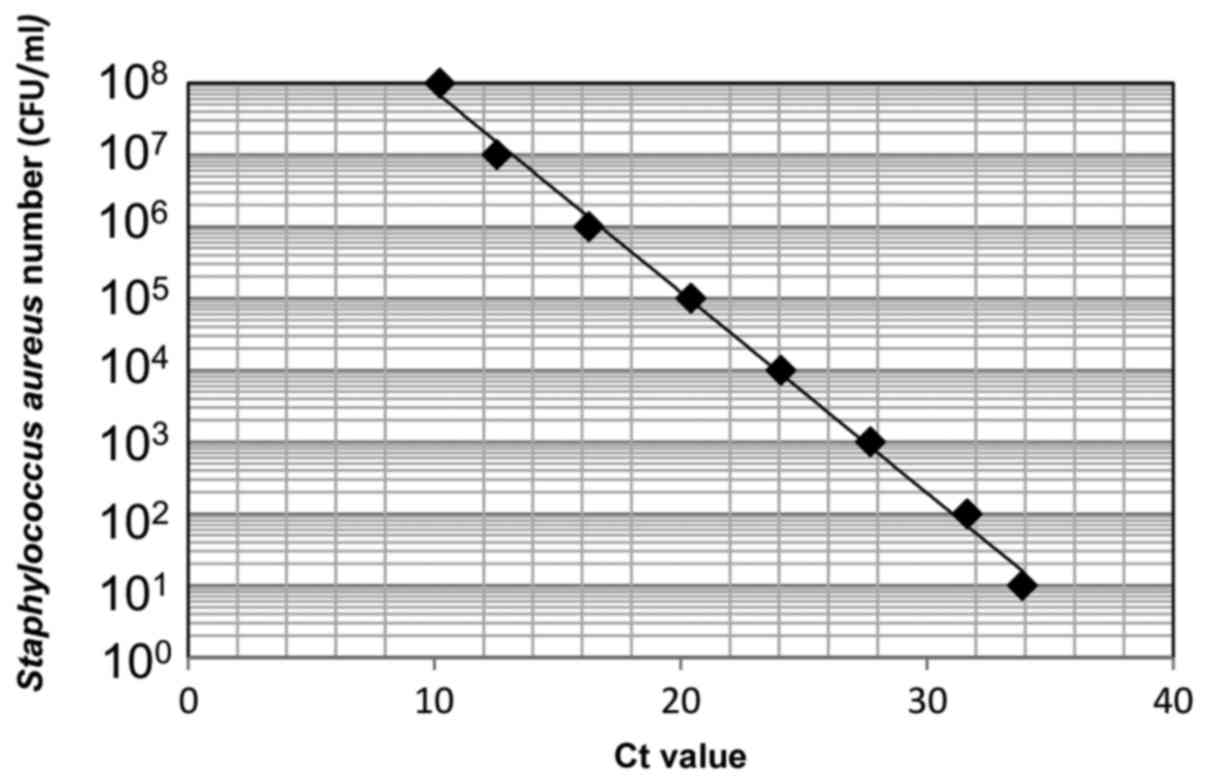
First, DNA was extracted from Staphylococcus
aureus [1.0×108 colony forming units (CFU)/ml] and
used for real-time PCR. Serial 10-fold dilutions of the DNA sample
(ranging from 101 to 108 CFU/ml) were
prepared to make a standard curve (Fig.
1). DNA levels were quantified using a CFX connect real-time
PCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA) and SYBR-Green PCR Master Mix (Toyobo Life Science, Osaka,
Japan), with a reaction mixture containing 1 µl DNA, 9 µl
SYBR-Green Mix, and 10 µmol of each oligonucleotide primer pair.
Amplifications were performed with initial melting at 95°C for 5
min, followed by 40 cycles of denaturing at 95°C for 30 sec,
annealing at 56°C for 30 sec, and extension at 72°C for 1 min. The
16S rRNA primer sequences were forward,
5′-CGTTAGTAATCGTGGATCAGAATG-3′ and reverse,
5′-TGTGACGGGCGGTGTGTA-3′ (14). A
standard curve indicating the cycle threshold value versus the 16S
rRNA gene was obtained to estimate the bacterial number per sample
(15).
Detecting periodontal disease-related
bacteria by PCR
For PCR, 1 µl DNA samples (bacterial numbers:
1.0×103−1.0×106 CFU/ml) were used. Each
mixture was amplified with 10X PCR buffer, dNTPs, Taq DNA
polymerase (both from Toyobo Life Science) and primers. The
following previously reported PCR primer sets were used: For
Porphyromonas gingivalis (P. gingivalis) forward,
5′-AGGCAGCTTGCCATACTGCG-3′ and reverse,
5′-ACTGTTAGCAACTACCGATGT-3′; For Tannerella forsythia (T.
forsythia) forward, 5′-GCGTATGTAACCTGCCCGCA-3′ and reverse,
5′-TGCTTCAGTGTCAGTTATACCT-3′; and for Treponema denticola
(T. denticola) forward, 5′-TAATACCGAATGTGCTCATTTACAT-3′ and
reverse, 5′-TCAAAGAAGCATTCCCTCTTCTTCTTA-3′ (16). The PCR program included initial
melting at 95°C for 5 min, followed by 35 cycles of 95°C for 30
sec, 56°C for 1 min and 72°C for 30 sec. Following the reaction, 10
µl of the PCR product was electrophoresed on 2% agarose gels with
ethidium bromide staining.
Oral examination
Oral examination including periodontal examination
[to assess probing depth (PD) and bleeding on probing (BOP)] was
performed by dentists at Hiroshima University Hospital. Remaining
teeth and denture use were recorded. Oral wetness was evaluated
using an oral moisture-checking device (Moisture Checker
Mucus®; Scalar Corporation, Tokyo, Japan) as previously
described (17). The oral moisture
level was measured at the tongue dorsum 10 mm from the apex linguae
and expressed as a percentage, with mean values calculated from
three independent measurements; a value of >30% was defined as
high moisture level and a value of ≤30% as low moisture level as in
previous study (17). The tongue
coating was classified as none, light-thin (visibly pink
underneath) or heavy-thick (no pink observed). The presence of a
light-thin and/or heavy-thick coating was considered positive
(18).
Statistical analysis
Statistical analysis was performed using SPSS
software, version 24.0 (IBM Corp, Armonk, NY, USA). Determination
of Spearman's rank correlation coefficient was performed to examine
the correlation of bacterial number with age and moisture level.
The Mann-Whitney U test with Bonferroni correction or the
Kruskal-Wallis test was used to evaluate significant differences in
bacterial numbers and clinical factors. All Kruskal-Wallis tests
indicated no significant difference and therefore post-hoc tests
were not applicable. The χ2 test or Fisher's exact test
was used to evaluate significant differences between positive rates
of periodontal bacteria and clinical factors. P<0.05 was
considered to indicate statistical significance.
Results
Correlation between bacterial numbers
on the tongue surface and clinical factors
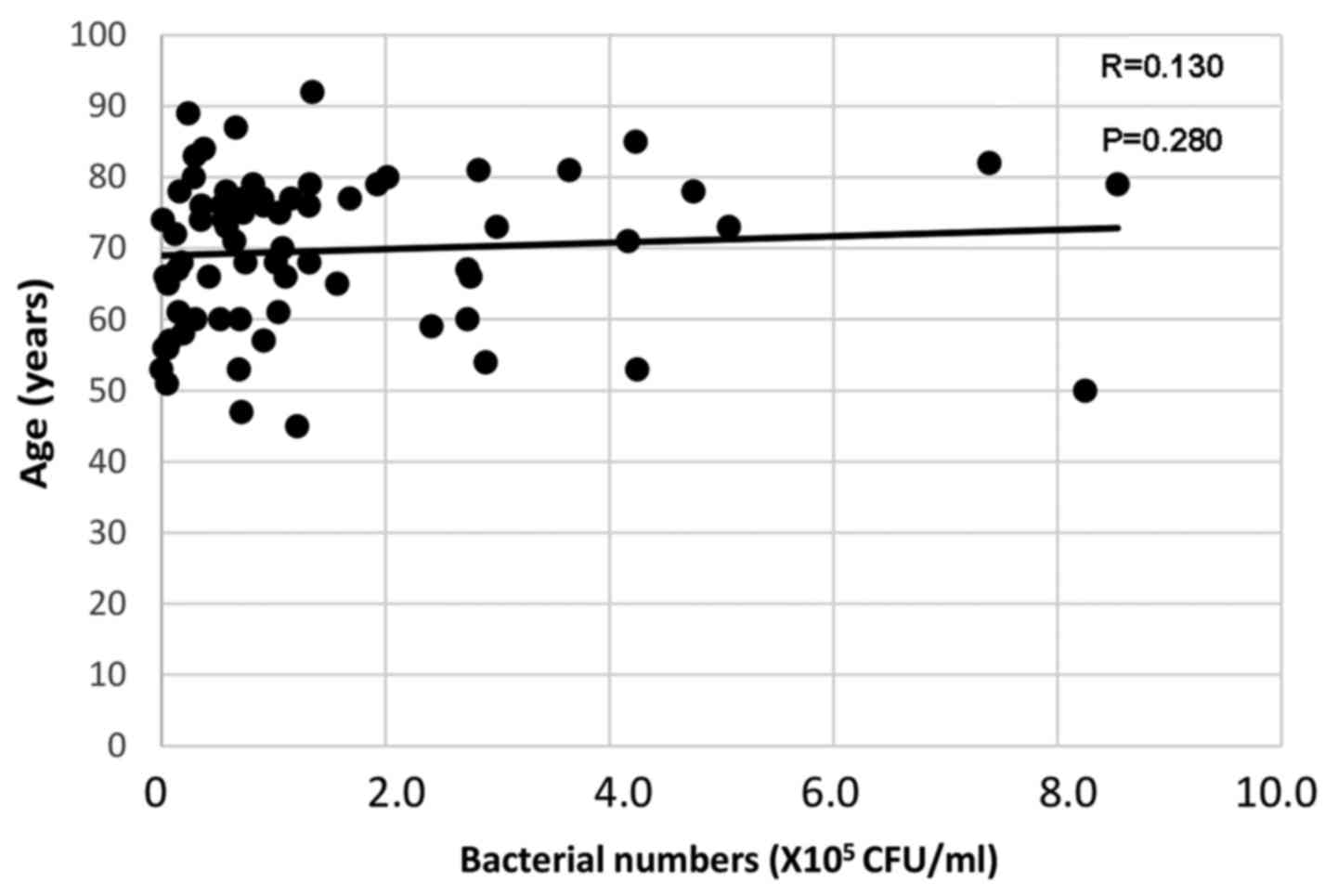
Age was weakly correlated with bacterial numbers on
the tongue surface [Spearman's rank correlation coefficient
(R)=0.130, P=0.280; Fig. 2].
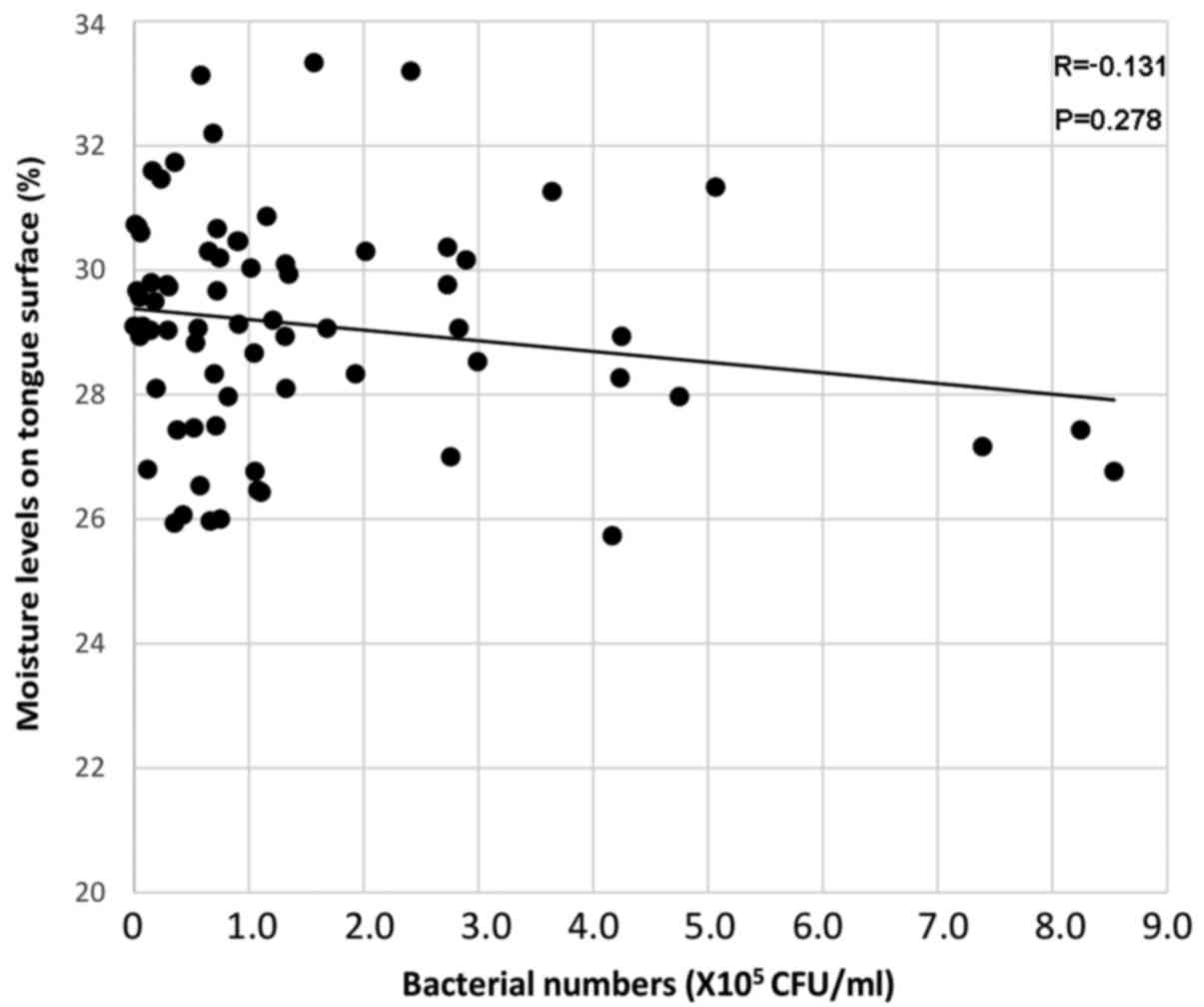
Meanwhile, the oral bacterial numbers were weakly negatively
correlated with moisture levels (R=−0.131, P=0.278; Fig. 3). Table
I summarizes the associations between bacterial counts detected
by real-time PCR and clinical factors. Oral bacterial numbers
appeared to be increased in individuals with fewer than 10
remaining teeth and in denture users; however, bacterial numbers
did not differ significantly according to remaining teeth or
denture use (Table I). Furthermore,
no significant difference was identified between bacterial numbers
on the tongue surface and oral hygiene status [BOP and PD (≥4 mm)].
Individuals with a medical history of stroke exhibited increased
bacteria compared with those without, but the difference was not
significant (Table I).
 | Table I.Associations between bacterial
numbers on the tongue surface and clinical factors. |
Table I.
Associations between bacterial
numbers on the tongue surface and clinical factors.
|
| Bacterial
numbers |
|---|
|
|
|
|---|
| Factor (n) | Mean ± SD
(×105) | P-value |
|---|
| Sex |
| Male
(23) | 1.53±1.84 | 0.66 |
| Female
(47) | 1.62±2.17 |
|
| Age (years) |
| ≥65
(50) | 1.58±1.39 | 0.52 |
| <65
(20) | 1.63±2.01 |
|
| Alcohol
consumption |
|
Non-drinker (42) | 1.54±1.93 | 0.06 |
| Social
drinker (15) | 1.00±1.13 |
|
| Every
day (13) | 2.45±2.39 |
|
| Smoking status |
|
Non-smoker (62) | 1.66±2.10 | 0.93 |
| Smoker
(8) | 1.06±0.85 |
|
| Stroke |
| No
(66) | 1.54±1.91 | 0.14 |
| Yes
(4) | 2.54±2.14 |
|
| Heart disease |
| No
(64) | 1.61±1.95 | 0.69 |
| Yes
(6) | 1.54±1.65 |
|
| Hypertension |
| No
(50) | 1.53±1.85 | 0.39 |
| Yes
(20) | 1.74±2.13 |
|
| Diabetes |
| No
(59) | 1.70±2.03 | 0.51 |
| Yes
(11) | 1.00±0.99 |
|
| Hyperlipidemia |
| No
(60) | 1.65±1.99 | 0.77 |
| Yes
(10) | 1.27±1.44 |
|
| Remaining
teeth |
| 0–9
(3) | 2.23±2.46 | 0.73 |
| 10–19
(13) | 1.31±1.39 |
|
| ≥20
(54) | 1.63±2.02 |
|
| Denture use |
|
Non-user (54) | 0.88±1.72 | 0.12 |
| User
(16) | 1.70±1.70 |
|
| Oral wetness |
| Low
(47) | 1.74±2.16 | 0.11 |
| High
(23) | 1.30±1.29 |
|
| Tongue coating |
|
Negative (66) | 1.57±1.92 | 0.53 |
|
Positive (4) | 1.92±2.12 |
|
| Probing depth |
| <4
mm (23) | 1.55±1.83 | 0.93 |
| ≥4 mm
(47) | 1.61±1.98 |
|
| Bleeding on
probing |
| No
(38) | 1.72±2.07 | 0.98 |
| Yes
(32) | 1.44±1.74 |
|
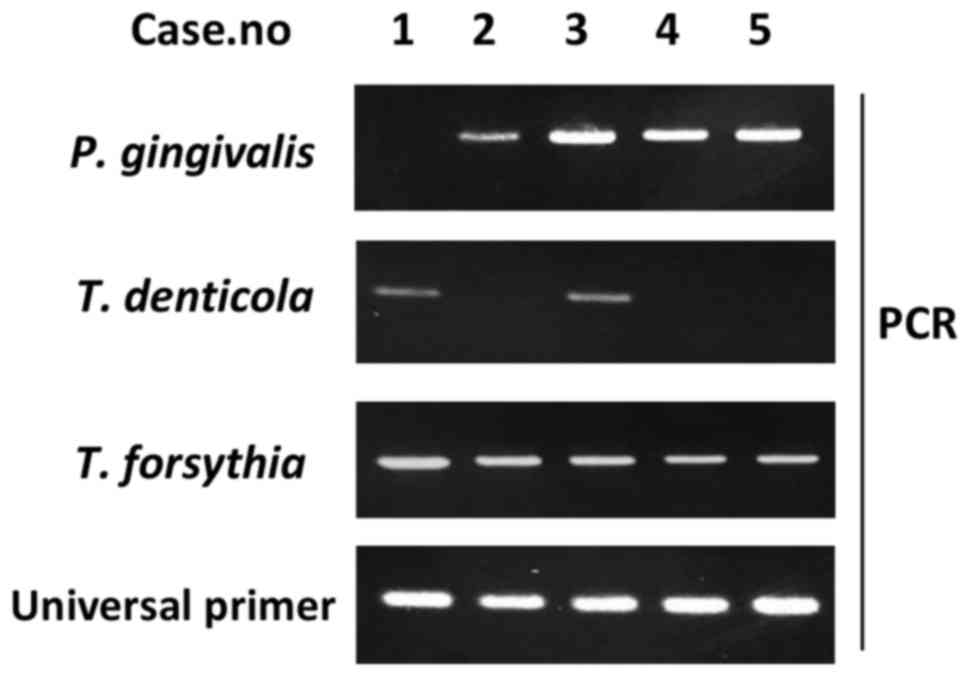
Detecting periodontal disease-related
bacteria by PCR
Red complex bacteria (e.g., T. denticola, T.
forsythia and P. gingivalis) serve major roles in
periodontal disease etiology (19).
Therefore, the present study performed PCR to detect DNA from these
bacteria (Fig. 4), to examine the
relationship between periodontal bacteria of the tongue dorsum and
clinical factors. A total of 31 out of the 70 participants (57.1%)
were P. gingivalis-positive (Table II), with the rate appearing higher in
people aged 65 years and over (50.0%) compared with in those aged
under 65 (30.0%). Additionally, smokers exhibited higher P.
gingivalis-positive rates than nonsmokers. Subjects with
medical histories of hypertension, diabetes, stroke and heart
disease exhibited higher P. gingivalis-positive rates than
those without these disorders. Subjects with BOP and ≥4 mm PD
exhibited higher P. gingivalis-positive rates (50.0 and
51.1%, respectively) than those with no BOP and <4 mm PD (39.5
and 30.4%, respectively), but these differences were not
significant.
 | Table II.Associations between positive rate of
P. gingivalis and clinical factors. |
Table II.
Associations between positive rate of
P. gingivalis and clinical factors.
|
| P. gingivalis |
|
|---|
|
|
|
|
|---|
| Factor (n) | (−) | (+) | P-value |
|---|
| Sex |
| Male
(23) | 11 (47.8%) | 12 (52.2%) | 0.44 |
| Female
(47) | 28 (59.6%) | 19 (40.4%) |
|
| Age (years) |
| ≥65
(50) | 25 (50.0%) | 25 (50.0%) | 0.18 |
| <65
(20) | 14 (70.0%) | 6 (30.0%) |
|
| Alcohol
consumption |
|
Non-drinker (42) | 26 (61.9%) | 16 (38.1%) | 0.14 |
| Social
drinker (15) | 9 (60.0%) | 6 (40.0%) |
|
| Every
day (13) | 4 (30.8%) | 9 (69.2%) |
|
| Smoking status |
|
Non-Smoker (62) | 37 (59.7%) | 25 (40.3%) | 0.13 |
| Smoker
(8) | 2 (25.0%) | 6 (75.0%) |
|
| Stroke |
| No
(66) | 38 (57.6%) | 28 (42.4%) | 0.32 |
| Yes
(4) | 1 (25.0%) | 3 (75.0%) |
|
| Heart disease |
| No
(64) | 37 (57.8%) | 27 (42.2%) | 0.40 |
| Yes
(6) | 2 (33.3%) | 4 (66.7%) |
|
| Hypertension |
| No
(50) | 31 (62.0%) | 19 (38.0%) | 0.12 |
| Yes
(20) | 8 (40.0%) | 12 (60.0%) |
|
| Diabetes |
| No
(59) | 36 (61.0%) | 23 (39.0%) | 0.05 |
| Yes
(11) | 3 (27.3%) | 8 (72.7%) |
|
| Hyperlipidemia |
| No
(60) | 35 (58.3%) | 25 (41.7%) | 0.30 |
| Yes
(10) | 4 (40.0%) | 6 (60.0%) |
|
| Remaining
teeth |
| 0–9
(3) | 1 (33.3%) | 2 (66.7%) | 0.26 |
| 10–19
(13) | 5 (38.5%) | 8 (61.5%) |
|
| ≥20
(54) | 33 (61.1%) | 21 (38.9%) |
|
| Denture user |
|
Non-user (54) | 32 (59.3%) | 22 (40.7%) | 0.41 |
| User
(16) | 7 (43.8%) | 9 (56.2%) |
|
| Oral wetness |
| Low
(47) | 25 (53.2%) | 22 (46.8%) | 0.73 |
| High
(23) | 14 (60.9%) | 9 (39.1%) |
|
| Tongue coating |
|
Negative (66) | 37 (56.0%) | 29 (44.0%) | 1.00 |
|
Positive (4) | 2 (50.0%) | 2 (50.0%) |
|
| Probing depth |
| <4
mm (23) | 16 (69.6%) | 7 (30.4%) | 0.17 |
| ≥4 mm
(47) | 23 (48.9%) | 24 (51.1%)) |
|
| Bleeding on
probing |
| No
(38) | 23 (60.5%) | 15 (39.5%) | 0.47 |
| Yes
(32) | 16 (50.0%) | 16 (50.0%) |
|
In contrast to the results for P. gingivalis, T.
forsythia exhibited no apparent differences in positive rates
between those with and without factors associated with systemic and
oral disease (Table III). All
smokers were T. forsythia-positive, but no significant
difference was identified in positive rate between smokers and
never-smokers.
 | Table III.Associations between positive rate of
T. forsythia and clinical factors. |
Table III.
Associations between positive rate of
T. forsythia and clinical factors.
|
| T.
forsythia |
|
|---|
|
|
|
|
|---|
| Factor (n) | (−) | (+) | P-value |
|---|
| Sex |
| Male
(23) | 4 (17.4%) | 19 (82.6%) | 0.17 |
| Female
(47) | 16 (34.0%) | 31 (66.0%) |
|
| Age (years) |
| ≥65
(50) | 12 (24.0%) | 38 (76.0%) | 0.24 |
| <65
(20) | 8 (40.0%) | 12 (60.0%) |
|
| Alcohol
consumption |
|
Non-drinker (42) | 15 (26.3%) | 42 (73.7%) | 0.57 |
| Social
drinker (15) | 4 (40.0%) | 6 (60.0%) |
|
| Every
day (13) | 1 (33.3%) | 2 (66.7%) |
|
| Smoking status |
|
Non-smoker (62) | 20 (32.3%) | 42 (67.7%) | 0.05 |
| Smoker
(8) | 0 (0.0%) | 8 (100.0%) |
|
| Stroke |
| No
(66) | 19 (28.8%) | 47 (71.2%) | 1.00 |
| Yes
(4) | 1 (25.0%) | 3 (75.0%) |
|
| Heart disease |
| No
(64) | 37 (57.8%) | 27 (42.2%) | 0.40 |
| Yes
(6) | 2 (33.3%) | 4 (66.7%) |
|
| Hypertension |
| No
(50) | 16 (32.0%) | 34 (68.0%) | 0.39 |
| Yes
(20) | 4 (20.0%) | 16 (80%) |
|
| Diabetes |
| No
(59) | 17 (32.2%) | 42 (67.8%) | 1.00 |
| Yes
(11) | 3 (27.3%) | 8 (72.7%) |
|
| Hyperlipidemia |
| No
(60) | 17 (28.3%) | 43 (71.7%) | 1.00 |
| Yes
(10) | 3 (30.0%) | 7 (70.0%) |
|
| Remaining
teeth |
| 0–9
(3) | 0 (0.0%) | 3 (100.0%) | 0.68 |
| 10–19
(13) | 3 (23.1%) | 10 (76.9%) |
|
| ≥20
(54) | 17 (31.5%) | 37 (68.5%) |
|
| Denture use |
|
Non-user (54) | 17 (31.5%) | 37 (68.5%) | 0.36 |
| User
(16) | 3 (18.8%) | 14 (81.2%) |
|
| Oral wetness |
| Low
(47) | 13 (27.7%) | 34 (72.3%) | 1.00 |
| High
(23) | 7 (30.4%) | 16 (69.6%) |
|
| Tongue coating |
|
Negative (66) | 20 (30.3%) | 46 (69.7%) | 0.32 |
|
Positive (4) | 0 (0.0%) | 4 (100.0%) |
|
| Probing depth |
| <4
mm (23) | 9 (39.1%) | 14 (60.9%) | 0.26 |
| ≥4 mm
(47) | 23 (48.9%) | 24 (51.1%) |
|
| Bleeding on
probing |
| No
(38) | 13 (34.2%) | 25 (65.8%) | 0.30 |
| Yes
(32) | 7 (21.9%) | 25 (78.1%) |
|
For T. denticola, men were indicated to
exhibit significantly higher positive rates than women (Table IV). T. denticola positivity
did not differ between those with and without a medical history of
hypertension, diabetes, stroke or heart disease. Interestingly,
there appeared to be a significant association between the T.
denticola-positive rate and the number of remaining teeth. All
individuals with fewer than nine teeth exhibited T.
denticola-positivity, while the positive rates in those with 10
or more teeth were <40%. These results indicate that T.
denticola on the tongue surface may be associated with tooth
loss.
 | Table IV.Associations between positive rate of
T. denticola and clinical factors. |
Table IV.
Associations between positive rate of
T. denticola and clinical factors.
|
| T.
denticola |
|
|---|
|
|
|
|
|---|
| Factor (n) | (−) | (+) | P-value |
|---|
| Sex |
| Male
(23) | 11 (47.8%) | 12 (52.2%) | 0.03 |
| Female
(47) | 36 (76.6%) | 11 (23.4%) |
|
| Age (years) |
| ≥65
(50) | 32 (64.0%) | 18 (36.0%) | 0.42 |
| <65
(20) | 15 (75.0%) | 5 (25.0%) |
|
| Alcohol
consumption |
|
Non-drinker (42) | 32 (56.1%) | 10 (43.9%) | 0.05 |
| Social
drinker (15) | 10 (66.7%) | 5 (33.3%) |
|
| Every
day (13) | 5 (38.5%) | 8 (61.5%) |
|
| Smoking status |
|
Non-smoker (62) | 43 (69.4%) | 19 (30.6%) | 0.43 |
| Smoker
(8) | 4 (50.0%) | 4 (50.0%) |
|
| Stroke |
| No
(66) | 44 (66.7%) | 22 (33.3%) | 1.00 |
| Yes
(4) | 3 (75.0%) | 1 (25.0%) |
|
| Heart disease |
| No
(64) | 42 (65.6%) | 22 (34.4%) | 0.66 |
| Yes
(6) | 5 (83.3%) | 1 (16.7%) |
|
| Hypertension |
| No
(50) | 35 (70.0%) | 15 (30.0%) | 0.58 |
| Yes
(20) | 12 (60.0%) | 8 (40.0%) |
|
| Diabetes |
| No
(59) | 41 (69.5%) | 18 (30.5%) | 0.49 |
| Yes
(11) | 6 (54.5%) | 5 (45.5%) |
|
| Hyperlipidemia |
| No
(60) | 41 (68.3%) | 19 (31.7%) | 0.72 |
| Yes
(10) | 6 (60.0%) | 4 (40.0%) |
|
| Remaining
teeth |
| 0–9
(3) | 0 (0.0%) | 3 (100.0%) | 0.04 |
| 10–19
(13) | 8 (61.6%) | 5 (38.4%) |
|
| ≥20
(54) | 39 (72.2%) | 15 (27.8%) |
|
| Denture use |
|
Non-user (54) | 37 (68.5%) | 17 (31.5%) | 0.88 |
| User
(16) | 10 (62.5%) | 6 (37.5%) |
|
| Oral wetness |
| Low
(47) | 30 (63.8%) | 17 (36.2%) | 0.43 |
| High
(23) | 17 (73.9%) | 6 (26.1%) |
|
| Tongue coating |
|
Negative (66) | 45 (68.2%) | 21 (31.8%) | 0.59 |
|
Positive (4) | 2 (50.0%) | 2 (50.0%) |
|
| Probing depth |
| <4
mm (23) | 17 (73.9%) | 6 (26.1%) | 0.43 |
| ≥4 mm
(47) | 30 (63.8%) | 17 (36.2%) |
|
| Bleeding on
probing |
| No
(38) | 25 (65.8%) | 13 (34.2%) | 1.00 |
| Yes
(32) | 22 (68.8%) | 10 (31.2%) |
|
Discussion
Real-time PCR has advantages in calculating total
bacterial numbers in terms of sensitivity and rapidity compared
with other methods (e.g., DNA hybridization and flow cytometry)
(20,21). Therefore, in the present study
real-time PCR methods were employed using universal primers for the
16S rRNA gene to quantitate approximate oral bacterial numbers.
Bacterial counters use a dielectrophoretic impedance measurement
method via a microelectrode chip on which bacteria from liquids are
captured by dielectrophoresis (22).
Initially, the current study compared oral bacteria numbers
obtained from real-time PCR using 16S rRNA gene copy numbers with
those obtained from a bacterial counter (Panasonic Healthcare Co.,
Ltd., Tokyo, Japan) and identified a significant positive
association between these methods (data not shown). This confirmed
that real-time PCR targeting the 16S rRNA gene can be used to
estimate approximate oral bacterial numbers.
Bacterial numbers were weakly negatively correlated
with moisture levels on the tongue surface. This indicated that low
moisture levels of the tongue may be associated with bacterial
growth on the tongue surface. Kobayashi et al (6) previously demonstrated that moisturizing
the tongue is important for reducing bacterial numbers on the
tongue surface. They determined that combining tongue cleaning with
a mouthwash and moisturizing gel was most effective for reducing
bacterial numbers on the tongue (6).
Thus, moisturizing the tongue surface may serve an important role
in inhibiting oral bacterial growth.
Regarding associations between systemic disease and
oral health, all diabetic subjects exhibited ≥4-mm periodontal
pockets (not shown). Although no significant association was
observed between diabetes and oral health status, periodontitis
likely serves a role in developing diabetes (23). Periodontal disease-associated
cytokines may induce systemic inflammation leading to insulin
resistance in type 2 diabetic patients (24). Indeed, the host inflammatory response
may be involved in the mechanisms of diabetes (23,24).
In the present study, P. gingivalis infection
was more common among elderly (≥65 years old) subjects. Previously,
the P. gingivalis detection rate was observed to
significantly increase with age, while that of T. denticola
and T. forsythia was comparably high across all age groups
(25). In the current study,
participants with ≥4-mm pockets exhibited higher positive rates of
the red complex bacteria P. gingivalis and T.
denticola compared with those with <4-mm pockets. Faveri
et al (26) reported that the
tongue surface is an important reservoir for periodontal pathogens
including P. gingivalis. These results indicate that
periodontal disease-associated bacteria (i.e., P. gingivalis
and T. denticola) on the tongue surface may be associated
with deep periodontal pockets and periodontal tissue
inflammation.
Regarding the association between systemic disease
and red complex bacteria, subjects with medical histories of
hypertension, diabetes, stroke and heart disease exhibited higher
P. gingivalis-positive rates than those without such
diseases. P. gingivalis may be associated with heart disease
by inducing endothelial cell dysfunction and atherosclerosis via
increased cytokine production, direct bacterial infection and
induction of an immune response to bacterial heat-shock proteins
(27,28). Additionally, P. gingivalis is
likely involved in both diabetes and oral health conditions
(29). Among participants with
systemic disease, diabetic patients exhibited increased P.
gingivalis-positive rates (72.7%) than those without (39.0%),
indicating that there is an important association between P.
gingivalis and diabetes. These findings and other previous
results (29) suggest that P.
gingivalis on the tongue surface serves a crucial role in the
development of diabetes. However, it remains unclear whether there
is a significant association between other red complex bacteria
including T. forsythia and T. denticola and systemic
disease.
Interestingly, males exhibited a greater T.
denticola-positive rate than females, although the reason for
this is unknown. Additionally, people with fewer than 10 teeth
exhibited a higher T. denticola-positive rate than those
with 10 teeth or more. T. denticola has been detected more
frequently and in higher counts in people with recurring
periodontal disease (30). Thus,
T. denticola may serve an important role in deteriorating
periodontal tissue, which may be associated with greater tooth
loss.
In conclusion, 16S rRNA gene-targeted PCR using swab
samples from the tongue dorsum may be useful for counting
approximate oral bacterial numbers and detecting periodontal
disease-associated bacteria to predict the condition of periodontal
tissue. The number of participants in the current study was
relatively small; therefore, further investigation in a large-scale
study is necessary to clarify the correlations between oral
bacterial numbers and systemic disease and factors associated with
oral health, particularly in elderly subjects.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
All data generated or analyzed in this study are
included in this article.
Authors' contributions
CYS designed the study and performed experiments,
analyzed and interpreted the data and was involved in manuscript
writing. HS performed experiments, analyzed and interpreted the
data and aided to write the paper. RN performed experiments. KO
analyzed and interpreted the data. MS designed the study, analyzed
and interpreted the data and aided to write the paper.
Ethics approval and consent to
participate
The study design was approved by the Ethics
Committee of Hiroshima University and all participants signed an
informed consent agreement prior to participation.
Patient consent for publication
All participants consented to the publication of
relevant data following anonymization of personal information.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
16S rRNA
|
16S ribosomal RNA
|
|
BOP
|
bleeding on probing
|
|
PD
|
probing depth
|
|
CFU
|
colony forming units
|
|
P. gingivalis
|
Porphyromonas gingivalis
|
|
T. forsythia
|
Tannerella forsythia
|
|
T. denticola
|
Treponema denticola
|
|
PCR
|
polymerase chain reaction
|
References
|
1
|
Terpenning M: Geriatric oral health and
pneumonia risk. Clin Infect Dis. 40:1807–1810. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shigeishi H, Ohta K, Fujimoto S, Nakagawa
T, Mizuta K, Ono S, Shimasue H, Ninomiya Y, Higashikawa K, Tada M,
et al: Preoperative oral health care reduces postoperative
inflammation and complications in oral cancer patients. Exp Ther
Med. 12:1922–1928. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Turner MD and Ship JA: Dry mouth and its
effects on the oral health of elderly people. J Am Dent Assoc.
138:15S–20S. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kawano T, Shigeishi H, Fukada E,
Yanagisawa T, Kuroda N, Takemoto T and Sugiyama M: Changes in
bacterial number at different sites of oral cavity during
perioperative oral care management in gastrointestinal cancer
patients: Preliminary study. J Appl Oral Sci. 26:e201705162018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayashida S, Funahara M, Sekino M,
Yamaguchi N, Kosai K, Yanamoto S, Yanagihara K and Umeda M: The
effect of tooth brushing, irrigation, and topical tetracycline
administration on the reduction of oral bacteria in mechanically
ventilated patients: A preliminary study. BMC Oral Health.
16:672016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kobayashi K, Ryu M, Izumi S, Ueda T and
Sakurai K: Effect of oral cleaning using mouthwash and a mouth
moisturizing gel on bacterial number and moisture level of the
tongue surface of older adults requiring nursing care. Geriatr
Gerontol Int. 17:116–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Danser MM, Gómez SM and Van der Weijden
GA: Tongue coating and tongue brushing: A literature review. Int J
Dent Hyg. 1:151–158. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Segata N, Haake SK, Mannon P, Lemon KP,
Waldron L, Gevers D, Huttenhower C and Izard J: Composition of the
adult digestive tract bacterial microbiome based on seven mouth
surfaces, tonsils, throat and stool samples. Genome Biol.
13:R422012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Horz HP, Vianna ME, Gomes BP and Conrads
G: Evaluation of universal probes and primer sets for assessing
total bacterial load in clinical samples: General implications and
practical use in endodontic antimicrobial therapy. J Clin
Microbiol. 43:5332–5337. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suzuki N, Yoshida A and Nakano Y:
Quantitative analysis of multi-species oral biofilms by TaqMan
Real-Time PCR. Clin Med Res. 3:176–185. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cutress TW, Ainamo J and Sardo-Infirri J:
The community periodontal index of treatment needs (CPITN)
procedure for population groups and individuals. Int Dent J.
37:222–233. 1987.PubMed/NCBI
|
|
12
|
Kembel SW, Wu M, Eisen JA and Green JL:
Incorporating 16S gene copy number information improves estimates
of microbial diversity and abundance. PLOS Comput Biol.
8:e10027432012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Perisin M, Vetter M, Gilbert JA and
Bergelson J: 16Stimator: Statistical estimation of ribosomal gene
copy numbers from draft genome assemblies. ISME J. 10:1020–1024.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoshida A, Suzuki N, Nakano Y, Oho T,
Kawada M and Koga T: Development of a 5 fluorogenic nuclease-based
real-time PCR assay for quantitative detection of Actinobacillus
actinomycetemcomitans Porphyromonas gingivalis. J Clin
Microbiol. 41:863–866. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yasukawa T, Ohmori M and Sato S: The
relationship between physiologic halitosis and periodontopathic
bacteria of the tongue and gingival sulcus. Odontology. 98:44–51.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takahashi F, Koji T and Morita O: Oral
dryness examinations: Use of an oral moisture checking device and
the usefulness of an oral moisture checking device and a modified
cotton method. Prosthodont Res Pract. 5:26–30. 2006. View Article : Google Scholar
|
|
18
|
Winkel EG, Roldán S, Van Winkelhoff AJ,
Herrera D and Sanz M: Clinical effects of a new mouthrinse
containing chlorhexidine, cetylpyridinium chloride and zinc-lactate
on oral halitosis. A dual-center, double-blind placebo-controlled
study. J Clin Periodontol. 30:300–306. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Socransky SS, Haffajee AD, Cugini MA,
Smith C and Kent RL Jr: Microbial complexes in subgingival plaque.
J Clin Periodontol. 25:134–144. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Veal DA, Deere D, Ferrari B, Piper J and
Attfield PV: Fluorescence staining and flow cytometry for
monitoring microbial cells. J Immunol Methods. 243:191–210. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Socransky SS, Smith C, Martin L, Paster
BJ, Dewhirst FE and Levin AE: ‘Checkerboard’ DNA-DNA hybridization.
Biotechniques. 17:788–792. 1994.PubMed/NCBI
|
|
22
|
Hamada R, Suehiro J, Nakano M, Kikutani T
and Konishi K: Development of rapid oral bacteria detection
apparatus based on dielectrophoretic impedance measurement method.
IET Nanobiotechnol. 5:25–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Genco RJ and Borgnakke WS: Risk factors
for periodontal disease. Periodontol 2000. 62:59–94. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nishimura F, Iwamoto Y, Mineshiba J,
Shimizu A, Soga Y and Murayama Y: Periodontal disease and diabetes
mellitus: The role of tumor necrosis factor-alpha in a 2-way
relationship. J Periodontol. 74:97–102. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chigasaki O, Takeuchi Y, Aoki A, Sasaki Y,
Mizutani K, Aoyama N, Ikeda Y, Gokyu M, Umeda M, Ishikawa I, et al:
A cross-sectional study on the periodontal status and prevalence of
red complex periodontal pathogens in a Japanese population. J Oral
Sci. 60:293–303. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Faveri M, Feres M, Shibli JA, Hayacibara
RF, Hayacibara MM and de Figueiredo LC: Microbiota of the dorsum of
the tongue after plaque accumulation: An experimental study in
humans. J Periodontol. 77:1539–1546. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ford PJ, Gemmell E, Hamlet SM, Hasan A,
Walker PJ, West MJ, Cullinan MP and Seymour GJ: Cross-reactivity of
GroEL antibodies with human heat shock protein 60 and
quantification of pathogens in atherosclerosis. Oral Microbiol
Immunol. 20:296–302. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seymour GJ, Ford PJ, Cullinan MP, Leishman
S and Yamazaki K: Relationship between periodontal infections and
systemic disease. Clin Microbiol Infect. 13 Suppl 4:3–10. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Yang H, Ding Y, Aprecio R, Zhang W,
Wang Q and Li Y: Experimental periodontitis induced by
Porphyromonas gingivalis does not alter the onset or
severity of diabetes in mice. J Periodontal Res. 48:582–590. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meyer-Bäumer A, Eick S, Mertens C, Uhlmann
L, Hagenfeld D, Eickholz P, Kim TS and Cosgarea R: Periodontal
pathogens and associated factors in aggressive periodontitis:
Results 5-17 years after active periodontal therapy. J Clin
Periodontol. 41:662–672. 2014. View Article : Google Scholar : PubMed/NCBI
|