Introduction
Colorectal cancer (CRC) is the third most common
cancer in Brazil and globally (1,2). The most
prevalent molecular pathways for colorectal cancer development are
APC, TP53, and KRAS mutations. The KRAS
gene is 47,305 bp long and contains 6 exons. It plays the role of a
GTPase in the transduction of signals (3). The activation of KRAS forms a GTP
complex, which can then be inactivated by hydrolysis to GDP. The
mutated form of KRAS renders the complex less susceptible to
hydrolysis, remaining in an activated form which induces the cell
to proliferate via several signal pathways, including MAPK
(3,4).
The RAS family includes three subunits: Kirsten-RAS
(KRAS), Neuroblastome-RAS (NRAS), and Harvey-RAS (HRAS). Mutations
of KRAS are found in 34.7%, of NRAS in 7%, and of HRAS in 0.5% of
CRC. Mutations in KRAS and NRAS confer a poor
prognosis, even at the metastatic stage or early stage colorectal
cancers. Additionally, they can lead to the development of
resistance against anti-EGFR molecules (5-9).
By contrast, EGFR inhibitors confer a positive predictive value
response and increased overall survival in KRAS and NRAS wild-type
tumors (10). Venook et al
suggested that sidedness of the primary tumor greatly affects the
clinical outcomes in an advanced or metastatic setting (11). The median survival for primary tumors
located on the left side was significantly longer than that for
tumors of the right side (overall survival: 33.3 vs. 19.4 months;
P<0.0001). Patients treated with cetuximab with wild-type KRAS
and left-sided primary tumors had an OS of 37.5 months versus those
with right-sided primary tumors who had an OS of 16.4 months (HR =
1.97; 95% CI = 1.56-2.48). This suggests that patients with primary
tumors on the right side of the colon should not be treated with
anti-EGFRs (11).
DNA sequencing is the gold standard for detecting
mutations. Originally, Sanger sequencing constituted standard
usage; however, next-generation sequencing (NGS) has allowed for
faster and high-throughput screening for mutations in several types
of cancer (12). High-resolution
melting (HRM) has recently been used as an alternative strategy to
DNA sequencing (13). Through
differences in DNA melting temperatures and curve profiles, it is
possible to distinguish mutant samples from controls. HRM analyzers
allow for a more accurate detection of differences in melting
temperatures between two samples (13). In the present study, it was
hypothesized that HRM could be used as an effective alternative to
next-generation sequencing for the detection of KRAS mutations in
colorectal cancer.
Materials and methods
General
The present study was performed in accordance with
the Declaration of Helsinki and approved by the Universidade
Federal de Sao Paulo Ethics Committee Plataforma Brasil CAAE:
55446116000005505, Biobank BR080. All the patients signed an
informed consent allowing tumor samples to be used in our study.
The study included 47 patients, with a mean age of 62 years. Of the
47 patients, 24 were male.
A 25 mg sample from each confirmed colorectal
adenocarcinoma was collected for DNA sequencing using the Illumina
HiSeq 2500 Platform and for DNA analyses for HRM via the StepOne
Plus® Real-Time PCR Systems.
In addition, using a second-generation Illumina DNA
sequencing platform as a reference, we compared the HRM capacity to
identify mutations in exons 2, 3, and 4 of KRAS. A total of 47
fresh-frozen tissue samples were obtained.
DNA extraction and quantification
DNA was extracted from 25 mg of fresh-frozen
colorectal adenocarcinoma tissue using the QIAmp® DNA
Mini Kit (Qiagen®) according to the manufacturer's
instructions. Samples were cut into small fragments, incubated at
56˚C for 6 h, and vortexed every 30 min. DNA was eluted in
nuclease-free water and quantified by spectro-photometry
(NanoDrop®; Thermo Fisher Scientific) and fluorimetry
(Qubit®; Thermo Fisher Scientific) and stored in
cryotubes at -20˚C. Every 25 mg of the tumor sample yielded
approximately 30 µg of DNA.
Quantitative PCR and analysis of DNA
melting temperatures
Quantitative PCR with the StepOne Plus®
Real-Time PCR Systems (Applied Biosystems), using Meltdoctor™ HRM
Master Mix (Applied Biosystems) and forward (f) and reverse (r)
primers of exon 2 of KRAS, was carried for DNA amplifications and
post-PCR melt analyses. Exon 2f: TTA TAA GGC CTG CTG AAA ATG ACT
GAA; exon 2r: TGA ATT AGC TGT ATC GTC AAG GCA CT. The PCR assays
were carried out in a final reaction volume of 25 µl containing
12.5 µl of Meltdoctor™, 1 µl each of forward and reverse primers at
10 pmol/ml, respectively, and 1 µl of DNA with a concentration
range of 20-100 ng. The PCR conditions were: denaturation at 95˚C
for 10 min, followed by 50 cycles at 95˚C for 15 sec, annealing at
62˚C for 10 sec, and an extension at 72˚C for 10 sec. For the HRM
assays, we performed denaturation at 95˚C for 15 sec, and obtained
high-resolution melting profiles from 60 to 95˚C at intervals of
0.5˚C. All the HRM reactions were performed in triplicate. After
the PCR runs, the melting curve profiles were analyzed by HRM
Software v3.01 (Thermo Fisher Scientific).
DNA next-generation sequencing
NGS for exon 2 was conducted using the TruSight™
Tumor Sample Preparation Kit (Illumina®; San Diego) and
the TruSight™ Tumor 26 (Illumina®). The steps were
performed according to the manufacturer's recommendations. These
included: hybridization of the oligo pool for 15 min and incubation
for 10 h, removal of unbound oligos for 20 min, extension-ligation
of bound oligos for 5 min and incubation for 45 min, PCR
amplification, PCR cleanup, library quantification and
normalization, and library denaturing and pooling. To identify the
somatic mutations, we used the Catalogue of Somatic Mutations in
Cancer (COSMIC, www.cancer.sanger.ac.uk). Patients were divided into
wild-type and mutant groups as per the results of NGS.
Statistical analysis
Statistical analyses were performed using SPSS
software v.21 (SPSS, Inc.; Chicago, IL, USA). The κ coefficient
with a 95% CI was used to assess the agreement between the two
methods. A t-test for independent samples was carried out to
compare the mean melting temperatures between wild-type and mutant
groups, and the McNemar test was used to describe the sensitivity
and specificity of HRM at exon 2 for the KRAS mutations.
Results
Patient characteristics
Forty-seven patients were included, the mean age was
62 years and 24 were males. The pathological features included
mucinous characteristics (19%); well-differentiated (25.5%), and
poorly differentiated (6.3%). Most of the patients had stage II or
III tumors. Table I summarizes the
baseline characteristics of the patients.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Characteristic | N / mean (range) | (%) |
|---|
| Sex |
|
Female | 23 | 48.9 |
|
Male | 24 | 51.1 |
| Age | 62.39 (35-86) | - |
| Pathological
features |
|
Mucinous | 9 | 19.1 |
|
Well-differentiated | 12 | 25.5 |
|
Poorly
differentiated | 3 | 6.3 |
|
Moderately
differentiated | 18 | 38.3 |
|
Perineural
invasion | 15 | 31.9 |
|
Vascular
invasion | 18 | 38.3 |
|
Inflammatory
process | 10 | 21.2 |
| Staging |
|
I | 1 | 2 |
|
II | 24 | 50 |
|
III | 14 | 29.2 |
|
IV | 5 | 10.4 |
The tumors were sequenced on the HiSeq Sequencing
Platform and then subjected to PCR amplification and DNA
melting.
Next-generation sequencing
In total, 16 tumors (34%) had mutations in
KRAS; 78% of these mutations were in exon 2, 11% in exon 3
and 11% in exon 4. The mutation in exon 2 of KRAS included
c.35C > T (G12D); c.34C > A(G12C), and c.38C > T(G13D)
(Fig. 1).
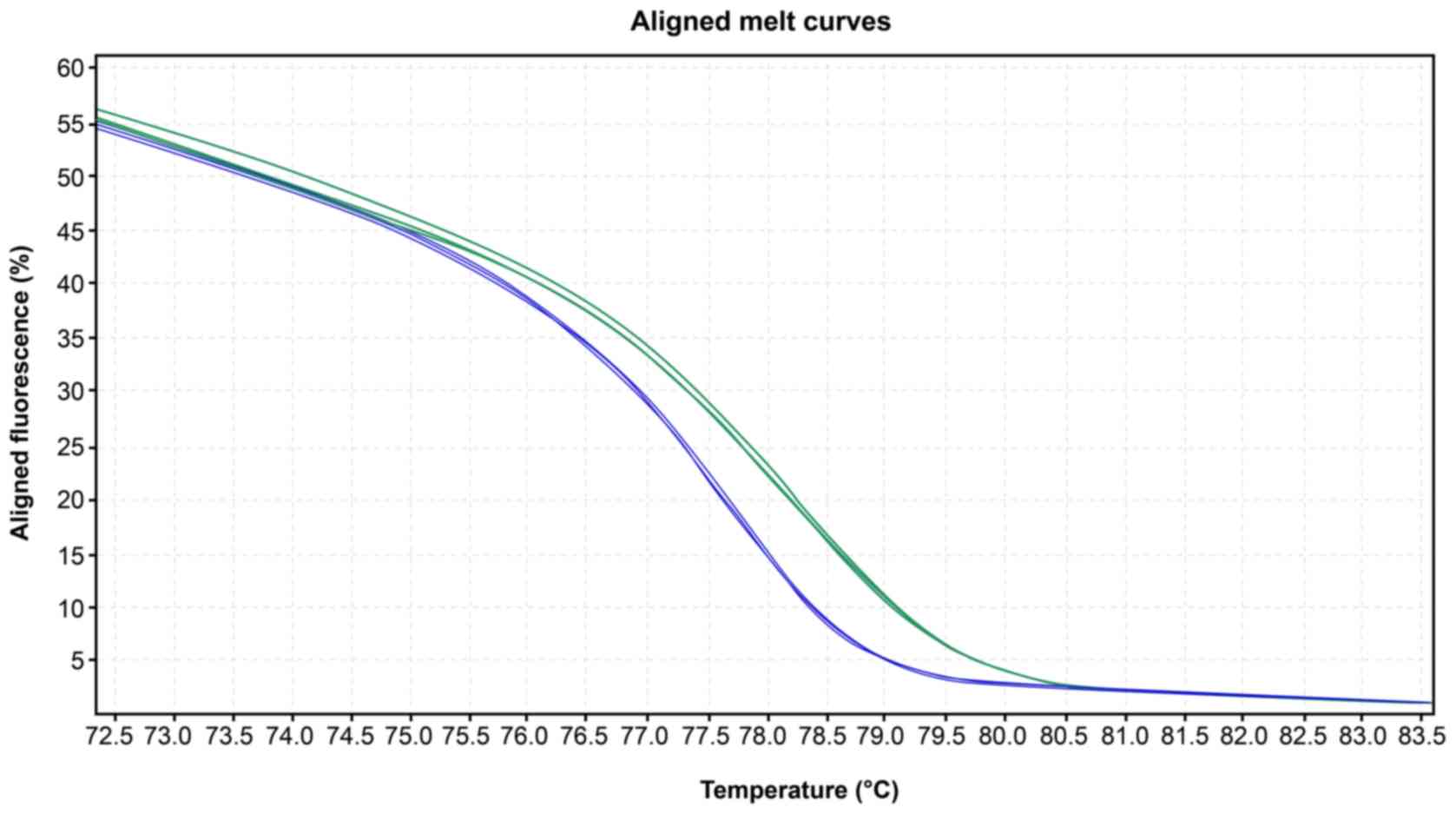
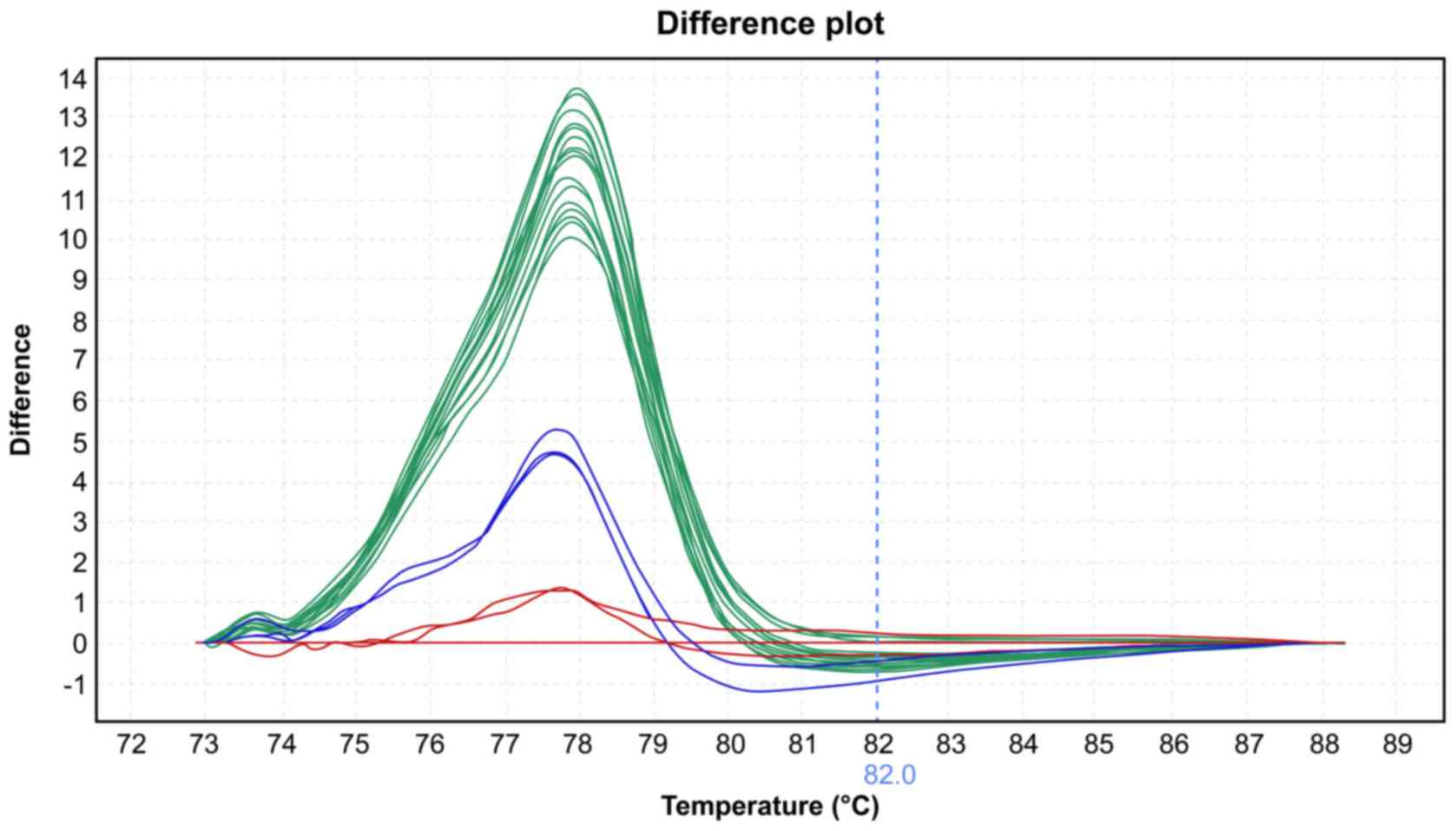
High-resolution melting
The mean melting temperatures for the wild-type and
mutated group at exon 2 were 78.13˚C and 77.87˚C, respectively
(P=0.001, Table II). The melting
curve profiles are shown in Figs. 1
and 2.
 | Table IIMean temperature (˚C) of melting for
the wild-type and mutated groups according to next-generation
sequencing. |
Table II
Mean temperature (˚C) of melting for
the wild-type and mutated groups according to next-generation
sequencing.
| Item | Wild-type | Mutated | 95% CI | P-value |
|---|
| Mean ± SD | 78.1352±15 | 77.87±0.28 | 0.11-0.41 | 0.001 |
Sensibility and specificity for
HRM
The sensitivity and specificity of high-resolution
melting were 83.3 and 96.6%, respectively, with a high concordance
between the methods (κ =0.816; P=0.625) (Table III). Of the 16 samples diagnosed as
mutant by HRM, one was not considered mutated by NGS. Furthermore,
we identified 29 samples as wild-type via HRM and one of those
samples was considered mutated via sequencing.
 | Table IIIMutation on KRAS exon 2 by HRM and
NGS. |
Table III
Mutation on KRAS exon 2 by HRM and
NGS.
| | HRM | |
|---|
| NGS | Wild-type | Mutated | P-valuea |
|---|
| Wild-type [N
(%)] | 26 (96.6) | 1 (3.4) | 0.625 |
| Mutated [N
(%)] | 3 (16.7) | 15 (83.3) | |
Discussion
Personalized oncology claims that biomarkers predict
a likely response to chemotherapy. In CRCs, somatic mutations in
exon 2 of KRAS are predictive for resistance to anti-EGFR
antibodies. Several studies have reported mutations in exon 2,
ranging from 36 to 45% (14).
Next-generation sequencing, which demonstrates a high concordance
with Sanger sequencing methods, has been used for faster detection
of mutations in different genes within larger DNA sequences
(15).
Ishige et al reported that 25% of the
mutations in exon 2 were in codon 12 and 4% in codon 13(6). Neumann et al (16) also reported that 80% of the mutations
in exon 2 were in codon 12. In the present study, the prevalence of
KRAS mutations was 34%, mainly in exon 2. The mutations on
exon 2 were 12% in codon 13, and 54% in codon 12. Although findings
of those studies lacked concordance, they did agree that codon 12
at exon 2 is the most common site of mutation in KRAS for
colorectal cancer.
High-resolution melting has been used as an
alternative strategy for sensitive detection of DNA sequence
variations in different genes (17).
This method is based on the different DNA melting temperatures for
specific targets. This method cannot, however, be used to identify
changes in the DNA sequence, although wild-type sequences and
mutant sequences correlate with specific, and different, melting
curves. In CRC, HRM has been evaluated to validate its routine use
compared with both direct sequencing and NGS in various studies
(18-23).
The HRM assay with DNA concentrations ranging from
20 to 100 ng, were able to evaluate the exon 2 of KRAS with
a sensitivity and specificity of 83.3 and 96.6%, respectively,
compared to NGS. Demonstrating a high concordance between the two
methods used (ĸ = 0.816). Negru et al reported concordance
of approximately 99% between HRM and DNA sequencing for exon
2(20). The large capacity for
detecting mutations in exon 2 of KRAS in our study was
similar to that in other studies with different populations
(17,24-26).
The short number of samples is a limitation of this
study. However, this study proved that this technique had a high
sensitivity and specificity that may be validated in a high number
of tumors. This assay, different from the others that was carried
out in formalin-fixed paraffin-embedded (FFPE) tissue, was
elaborate in fresh tumor samples that can increase the sensitivity
of detection of mutations. A possible explanation is a possible DNA
degeneration during the preparation and maintenance of the archived
samples in FFPE. Another possible advantage when compared to other
published articles is that the sequencing of KRAS mutation
was by NGS and not by Sanger or pyrosequencing, which both have a
low sensitivity.
Two advantages had to be considered in the diagnosis
of KRAS mutations in CRC by HRM. This method is less
expensive than NGS and may be an alternative method to diagnose
KRAS mutation This is particularly important in view of the
growing need to identify mutant profiles for different target genes
in many types of cancer. Another advantage of HRM is the time spent
for performance of the test, i.e., approximately 3.5 h compared to
NGS, which requires some days.
In conclusion, in our cohort, HRM had a high index
of comparison with NGS, which was the gold standard for detecting
DNA mutations of KRAS in colorectal cancer.
Acknowledgements
We would like to acknowledge the Surgical Department
and Pathology Department from Federal University of São Paulo and
Fundação de Amparo a Pesquisa do Estado de São Paulo for aiding in
implementing this project.
Funding
The study was supported by the CAPES Coordination
for the Improvement of Higher Education Personnel and Sao Paulo
Research Foundation grant number 13/19268-3.
Availability of data and materials
The datasets generated and analyzed in the present
study are included in this published article.
Authors' contributions
RRM contributed to performance of the experiments of
HRM, statistical analysis and writing the article. TDS contributed
to acquisition of tumor samples, extraction of DNA and DNA
sequencing. NMF contributed to the concept, design and drafted the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was conducted in accordance with the
Resolution 466 of December 08, 2012, of the National Health Council
of Brazil. Ethical approval for this study was obtained from the
Ethics and Clinical Research Committee Coordenadoria de Ensino e
Pesquisa do Hospital São Paulo-HU/UNIFESP. CAAE: 55446116000005505.
Patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soeriomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
National Cancer Institute: Comprehensive
Cancer Information. https://www.inca.gov.br/sites/ufu.sti.inca.local/files//media/document//estimativa-incidencia-de-cancer-no-brasil-2018.
Accessed June 29th, 2019.
|
|
3
|
Genetics Home Reference KRAS gene. US
National Library of Medicine. https://ghr.nlm.nih.gov/gene/KRAS.
Accessed June 30th, 2019.
|
|
4
|
Ciombor KK and Bekaii-Saab T: A
comprehensive review of sequencing and combination strategies of
targeted agents in metastatic colorectal cancer. Oncologist.
23:25–34. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ye LC, Liu TS, Ren L, Wei Y, Zhu DX, Zai
SY, Ye QH, Yu Y, Xu B, Qin XY, et al: Randomized controlled trial
of cetuximab plus chemotherapy for patients with KRAS
wild-type unresectable colorectal liver-limited metastases. J Clin
Oncol. 31:1931–1938. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ishige T, Itoga S, Sato K, Kitamura K,
Nishimura M, Sawai S, Matsushita K, Suzuki K, Ota S, Miyauchi H, et
al: High-throughput screening of extended RAS mutations
based on high-resolution melting analysis for prediction of
anti-EGFR treatment efficacy in colorectal carcinoma. Clin Biochem.
47:340–343. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yokota T: Are KRAS/BRAF mutations potent
prognostic and/or predictive biomarkers in colorectal cancers?
Anticancer Agents Med Chem. 12:163–171. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tosi F, Magni E, Amatu A, Mauri G,
Bencardino K, Truini M, Veronese S, De Carlis L, Ferrari G,
Nichelatti M, et al: Effect of KRAS and BRAF
mutations on survival of metastatic colorectal cancer after liver
resection: A systematic review and meta-analysis. Clin Colorectal
Cancer. 16:e153–e163. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Brudvik KW, Kopetz SE, Li L, Conrad C,
Aloia TA and Vauthey JN: Meta-analysis of KRAS mutations and
survival after resection of colorectal liver metastases. Br J Surg.
102:1175–1183. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Douillard JY, Oliner KS, Siena S,
Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham
D, Jassem J, et al: Panitumumab-FOLFOX4 treatment and RAS
mutations in colorectal cancer. N Engl J Med. 369:1023–1034.
2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Venook A, Niedzwiecki D, Innocenti F, et
al: Impact of primary (1°) tumor location on overall survival (OS)
and progression-free survival (PFS) in patients (pts) with
metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405
(Alliance). J Clin Oncol. 34:3504. 2016. View Article : Google Scholar
|
|
12
|
Hutchison CA III: DNA sequencing: Bench to
bedside and beyond. Nucleic Acids Res. 35:6227–6237.
2007.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wojdacz TK, Dobrovic A and Hansen LL:
Methylation-sensitive high-resolution melting. Nat Protoc.
3:1903–1908. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Van Krieken JH, Rouleau E, Ligtenberg MJ,
Normanno N, Patterson SD and Jung A: RAS testing in metastatic
colorectal cancer: Advances in Europe. Virchows Arch. 468:383–396.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kothari N, Schell MJ, Teer JK, Yeatman T,
Shibata D and Kim R: Comparison of KRAS mutation analysis of
colorectal cancer samples by standard testing and next-generation
sequencing. J Clin Pathol. 67:764–767. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Neumann J1, Zeindl-Eberhart E, Kirchner T
and Jung A: Frequency and type of KRAS mutations in routine
diagnostic analysis of metastatic colorectal cancer. Pathol Res
Pract. 205:858–862. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Simi L, Pratesi N, Vignoli M, Sestini R,
Cianchi F, Valanzano R, Nobili S, Mini E, Pazzagli M and Orlando C:
High-resolution melting analysis for rapid detection of
KRAS, BRAF, and PIK3CA gene mutations in
colorectal cancer. Am J Clin Pathol. 130:247–253. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Er TK, Chang YS, Yeh KT, Chang TJ and
Chang JG: Comparison of two different screening methods for the
KRAS mutation in colorectal cancer. Clin Lab. 56:175–186.
2010.PubMed/NCBI
|
|
19
|
Karim B, Florence C, Kamel R, Nadia K,
Ines O, Raja M, Sarra BJ, Florent S and Amel BA: KRAS mutation
detection in Tunisian sporadic coloractal cancer patients with
direct sequencing, high resolution melting and denaturating high
performance liquid chromatography. Cancer Biomark. 8:331–340.
2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Negru S, Papadopoulou E, Apessos A,
Stanculeanu DL, Ciuleanu E, Volovat C, Croitoru A, Kakolyris S,
Aravantinos G, Ziras N, et al: KRAS, NRAS and
BRAF mutations in Greek and Romanian patients with
colorectal cancer: A cohort study. BMJ Open.
4(e004652)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Mancini I, Santucci C, Sestini R, Simi L,
Pratesi N, Cianchi F, Valanzano R, Pinzani P and Orlando C: The use
of COLD-PCR and high-resolution melting analysis improves the limit
of detection of KRAS and BRAF mutations in colorectal
cancer. J Mol Diagn. 12:705–711. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Borràs E, Jurado I, Hernan I, Gamundi MJ,
Dias M, Martí I, Mañé B, Arcusa A, Agúndez JA, Blanca M, et al:
Clinical pharmacogenomic testing of KRAS, BRAF and
EGFR mutations by high resolution melting analysis and
ultra-deep pyrosequencing. BMC Cancer. 11(406)2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Oati Ashtiani Z, Mehrsai AR, Pourmand MR
and Pourmand GR: High resolution melting analysis for raid
detection of PIK3CA gene mutations in bladder cancer. A
mutated target for cancer therapy. Urol. 15:26–31. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ma ES, Wong CL, Law FB, Chan WK and Siu D:
Detection of KRAS mutations in colorectal cancer by
high-resolution melting analysis. J Clin Pathol. 62:886–891.
2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Porcelli B, Frosi B, Terzuoli L, Arezzini
L, Pagani R, Civitelli S, Tanzini G, Orlando C, Pazzagli M,
Marziliano N, et al: Melting temperature analysis as quantitative
method for detection of point mutations. Clin Chem Lab Med.
39:501–504. 2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Karbalaie Niya MH, Basi A, Koochak A,
Safarnezhad Tameshkel F, Rakhshani N, Zamani F, Imanzade F, Rezvani
H, Adib Sereshki MM and Sohrabi MR: Sensitive melting analyses for
screening of KRAS and BRAF mutations in Iranian human metastatic
colorectal cancers. Asian Pac J Cancer Prev. 17:5147–5152.
2016.PubMed/NCBI View Article : Google Scholar
|