Introduction
Skin cancer is a malignant tumor that afflicts
individuals all over the world. Skin cancer is divided into
malignant melanoma and non-melanoma skin cancer (NMSC) (1,2). According
to global cancer statistics in 2018, there were 142,056 new cases
of NMSC, accounting for 5.8% of global cancer cases, and 65,155
NMSC-related deaths, accounting for 0.7% of global cancer
mortality. There were 779,723 new cases of skin malignant melanoma,
accounting for 1.6% of global cancer cases, and 60,712 skin
melanoma-related deaths, accounting for 0.6% of global cancer
mortality (3).
According to the 2015 Chinese Cancer Statistics
Report, the incidence of skin cancer in China was 8/1,000 and the
mortality rate was 3.22/1,000 (4,5). Compared
with other types of cancer, skin cancer has a higher recurrence
rate, with a 35% probability of recurrence in the first 3 years and
a 50% probability of recurrence in the first 5 years. Recurrent
skin cancer is usually the same sub-type as the original cancer
(6). Although the mortality rate of
skin cancer is extremely low and the cure rate is high, its high
incidence and recurrence rate constitute a significant economic
burden to health services. Furthermore, skin cancer located on the
head and other highly visible areas may affect a patients mental
wellbeing and quality of life (7,8).
Cutaneous metastases of malignant tumors can be
caused by malignant tumor cells traveling through the blood or
lymphatic system, tissue interstitial diffusion (tissue gap
diffusion), or surgical implantation (9). The higher the malignancy of the tumor,
the more likely it is to metastasize, and skin metastasis is often
the end-stage manifestation of a malignant tumor (10). Once metastasis occurs, the prognosis
of cancer is poor (11).
Unfortunately, skin metastasis may be the first clinical
manifestation of a malignant tumor (12). Therefore, a suitable prognosis serves
an important role in the recurrence and treatment of metastatic
skin cancer. The aim of the present study was to identify potential
prognostic biomarkers of metastatic skin cancer, which may be used
in a clinical setting, through data mining and analysis of skin
cancer prognosis genes.
With the development of gene chip technology and
next-generation sequencing technology (13), and the constant revision of the
viewpoints of individualized medical treatment and precision
medical care, understanding skin cancer from the perspective of the
genome and proposing more effective genetic biomarkers provides
more relevant information for drug development and clinical
decision-making (14-18).
Traditional statistical methods often yield unstable results and
excessive errors when applied to gene expression analysis (19). Furthermore, in high-throughput gene
expression experiments, the number of variables is generally much
higher than the sample size, which is called the Curse of
Dimensionality (20). Therefore, in
the present study, least absolute shrinkage and selection operator
(LASSO) was used to mine genomic data to minimize the instability
caused by high-dimensional data (21-27).
Materials and methods
Data collection
Using The Cancer Genome Atlas, data on skin cancer
from the Xena Functional Genomics Explorer (xenabrowser.net/datapages/) was obtained, including
the number of patients (n=481) and the number of genes assessed
(n=20,530). Among these, the number of primary samples (Primary
Tumor) was 105, the number of metastatic (Metastatic) samples was
368, and the number of other types of samples was 8 (28-32).
Data preprocessing
First, 219 samples with missing survival attribute
values were removed from 473 primary and metastatic samples and 255
patient samples were retained. These data were stratified into
primary tumor samples (n=72) and metastatic samples (n=183) by
matching the patients' samples in the gene expression spectrum.
Finally, using the ‘sample’ function in R version 3.5.2(33), the metastatic samples were randomly
divided into two groups; training samples (n=91) and test samples
(n=92).
Clinical data analysis
Several clinical factors affect the prognosis of
patients with skin cancer, so it is necessary for data analysis to
consider multiple attribute values in the sample as influencing
factors. Based on previous studies, clinical variables including
age, sex, history of radiotherapy, Tumor-Node-Metastasis
pathological stage (34) and cancer
status were used to analyze the related clinical factors in the
subsequent data mining analysis (35-38).
Analysis of gene data
First, the ‘Limma’ package version 3.42.2(39) in R was used to analyze the
differentially expressed genes of the 91 training set samples and
72 primary tumor samples. According to the adjusted P-value
(adj.P.val<0.001), 783 genes were considered to be
differentially expressed genes.
The ‘survival’ package (rdocumentation.org/packages/survival; version 3.1-8)
in R was used to perform Cox regression analysis of the
differentially expressed genes and the Cox coefficient, the hazard
ratio (HR) and the P-values of the Wald test of each gene were
calculated using the Kaplan-Meier method (40), and the genes that were significantly
associated with the survival of the patients (P<0.05) were
screened using the ‘survivalROC’ package (version number 1.0.3;
rdocumentation.org/packages/survivalROC).
Finally, the screened genes were analyzed again
using LASSO in the R package ‘glmnet’ (rdocumentation.org/packages/glmnet; version 3.0-2), to
obtain more critical genes. Through 10-fold cross validation, 20
risk genes, which were closely related to survival, were identified
(41-49).
Additionally, the Gene Ontology (GO) Cell Component Ontology Method
(50,51) was used to analyze pathway involved in
protein activity.
Prognostic index (PI) calculation
As an important indicator of the integration of risk
genes, a PI value can be determined for each patient with skin
cancer. The PI was obtained by linearly fitting the product of the
expression and the coefficient corrected by LASSO of each gene. The
formula for calculating the PI was: PI=β1
X1+β2X2+…+βiXi…+βnXn;
where Xi is the expression of the ith gene and
βi is the coefficient corrected by LASSO of the ith
gene.
Data validation
By extrapolating genes with a P<0.05, the 20 risk
genes obtained were verified using the test sample using the same
methods described above.
Statistical analysis of clinical
variables
From the clinical information of 255 patient
samples, the patients' age, sex, radiation therapy, pathological
T-stage and cancer status were taken as single variables. Cox
regression analysis was performed, and the P-value of the Log-rank
test and the HR value of each clinical factor were calculated.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinicopathological variables
Age, pathologic T-stage and cancer status were all
significantly associated with the recurrence of skin cancer,
suggesting that these three clinical factors may be used as
independent prognostic indicators (Table
I).
 | Table IClinicopathological characteristics
of the patients. |
Table I
Clinicopathological characteristics
of the patients.
|
Characteristics | Number of patients
(relapse) | P-value | HR (95% CI) |
|---|
| Age | | 0.030 | 1.669
(1.037-2.688) |
|
>57 | 123(35) | | |
|
≤57 | 132(36) | | |
| Sex | | 0.200 | 1.359
(0.8058-2.291) |
|
Male | 160(49) | | |
|
Female | 95(22) | | |
| Cancer status | | | |
|
Unknown | 3(1) |
7x10-10 | |
|
With
tumor | 160(18) | | 0.802
(0.1089-5.908) |
|
Tumor
free | 92(52) | | 0.1572
(0.0206-1.201) |
| Radiation
therapy | | 0.200 | 1.7204
(0.851-3.478) |
|
Unknown | 1 (0) | | |
|
Yes | 38(9) | | |
|
No | 216(62) | | |
| Pathological
T-stage | | 0.009 | 2.003
(1.175-3.412) |
|
T1-T3 | 165(49) | | |
|
T4 | 90(22) | | |
Genetic data analysis
After determining differential gene expression, 44
genes were considered significantly associated with the survival of
patients using survival analysis. Cox regression analysis and LASSO
analysis were performed, and 20 risk genes associated with skin
cancer were identified (Table
II).
 | Table IIPrognostic genes. |
Table II
Prognostic genes.
| Gene symbol | Name | Univariate HR (95%
CI) | Coefficient | P-value | LASSO HR (95%
CI) |
|---|
| TMEM45B | Transmembrane
protein 45B | 0.899
(0.822-0.982) | -0.086 |
1.88x10-2 | 0.918
(0.864-0.975) |
| CDKN1B | Cyclin-dependent
kinase inhibitor 1B (p27, Kip1) | 0.928
(0.886-0.972) | -0.029 |
1.55x10-3 | 0.971
(0.914-1.032) |
| PKHD1L1 | Polycystic kidney
and hepatic disease 1 (autosomal recessive)-like 1 | 0.938
(0.886-0.993) | -0.010 |
2.73x10-2 | 0.990
(0.932-1.052) |
| PLCL2 | Phospholipase
C-like 2 | 0.961
(0.929-0.994) | -0.006 |
2.14x10-2 | 0.994
(0.936-1.056) |
| CKMT1A | Creatine kinase,
mitochondrial 1A | 1.024
(1.000-1.049) | 0.001 |
4.90x10-2 | 1.001
(0.942-1.063) |
| SPINK5 | Serine peptidase
inhibitor, Kazal type 5 | 1.027
(1.002-1.052) | 0.007 |
3.31x10-2 | 1.008
(0.948-1.070) |
| CST6 | Cystatin E/M | 1.027
(1.000-1.054) | 0.011 |
4.67x10-2 | 1.011
(0.952-1.074) |
| FABP5 | Fatty acid binding
protein 5 (psoriasis-associated) | 1.037
(1.007-1.067) | 0.007 |
1.53x10-2 | 1.007
(0.948-1.070) |
| PTK6 | Protein tyrosine
kinase 6 | 1.047
(1.010-1.084) | 0.029 |
1.11x10-2 | 1.030
(0.969-1.094) |
| TXNDC17 | Thioredoxin domain
containing 17 | 1.047
(1.002-1.095) | 0.004 |
4.02x10-2 | 1.004
(0.945-1.066) |
| LTB4R | Leukotriene B4
receptor | 1.050
(1.007-1.096) | 0.018 |
2.39x10-2 | 1.018
(0.958-1.082) |
| FAM100A | Family with
sequence similarity 100, member A | 1.050
(1.008-1.094) | 0.015 |
1.90x10-2 | 1.016
(0.956-1.079) |
| KRT10 | keratin 10 | 1.057
(1.025-1.091) | 0.016 |
4.33x10-4 | 1.016
(0.956-1.080) |
| PPP1R13L | Protein phosphatase
1, regulatory subunit 13 like | 1.059
(1.010-1.109) | 0.000 |
1.65x10-2 | 1.000
(0.941-1.063) |
| DLX3 | Distal-less
homeobox 3 | 1.063
(1.025-1.102) | 0.008 |
9.40x10-4 | 1.008
(0.949-1.071) |
| KPRP | Keratinocyte
proline rich protein | 1.064
(1.015-1.115) | 0.044 |
9.68x10-3 | 1.045
(0.984-1.111) |
| VSIG10L | V-set and
immunoglobulin domain containing 10 like | 1.066
(1.011-1.123) | 0.021 |
1.87x10-2 | 1.021
(0.961-1.085) |
| MYOM3 | Myomesin 3 | 1.073
(1.015-1.134) | 0.040 |
1.25x10-2 | 1.041
(0.979-1.106) |
| NMU | Neuromedin U
receptor 1 | 1.092
(1.042-1.144) | 0.072 |
2.22x10-4 | 1.075
(1.012-1.142) |
| PIGW |
Phosphatidylinositol glycan anchor
biosynthesis, class W | 1.113
(1.044-1.188) | 0.002 |
1.08x10-3 | 1.002
(0.943-1.064) |
Gene PI analysis
Through linear fitting of the product of expression
and regression coefficient of the 20 genes in each sample, the PI
of each patient was calculated, and the patients were sorted from
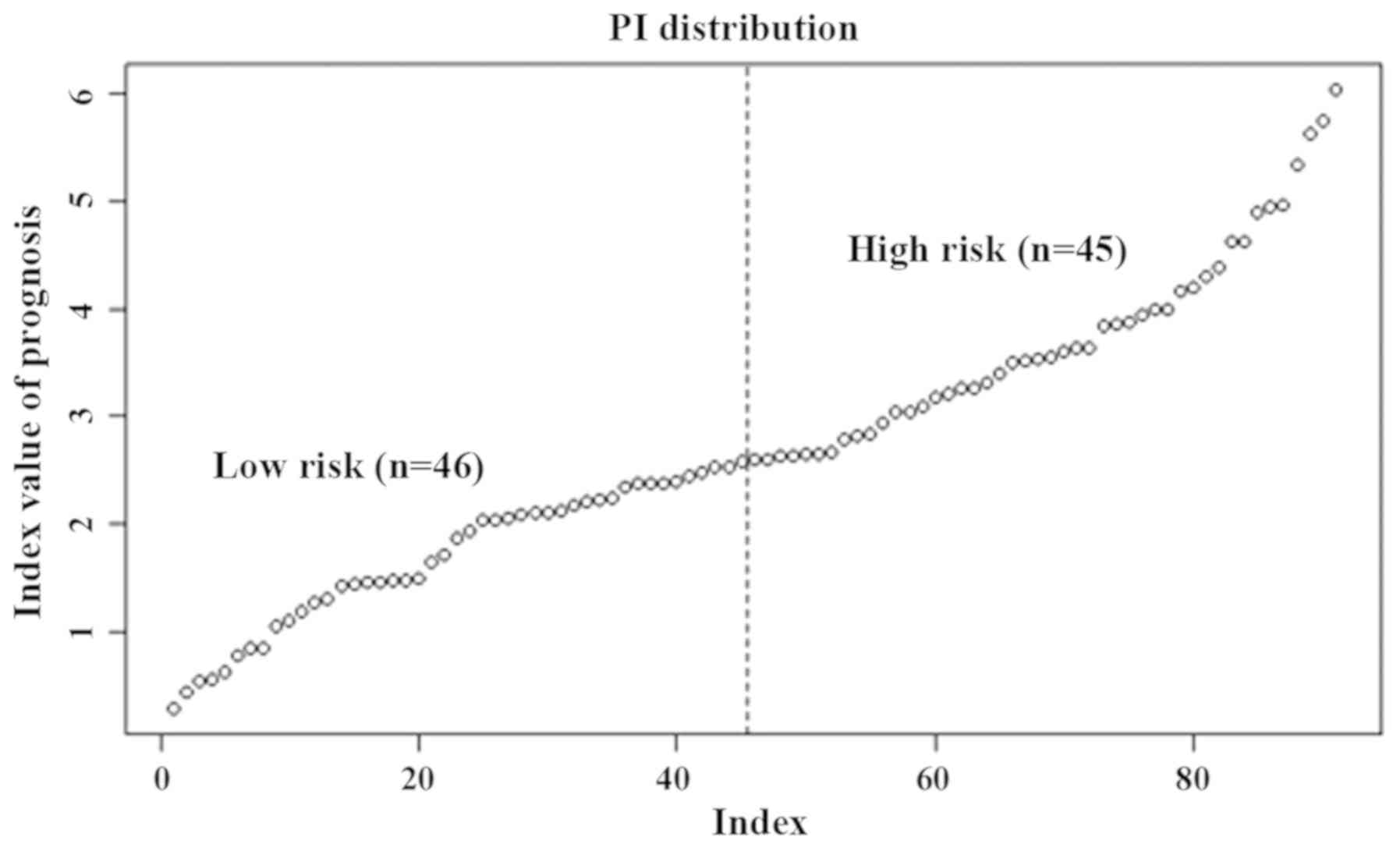
lowest to highest according to their PI value. Based on the median
PI value, the patients were divided into high-risk and low-risk
groups (Fig. 1). The lower the PI
value, the lower the risk of recurrent survival, and the higher the
PI value, the higher the risk of recurrent survival.
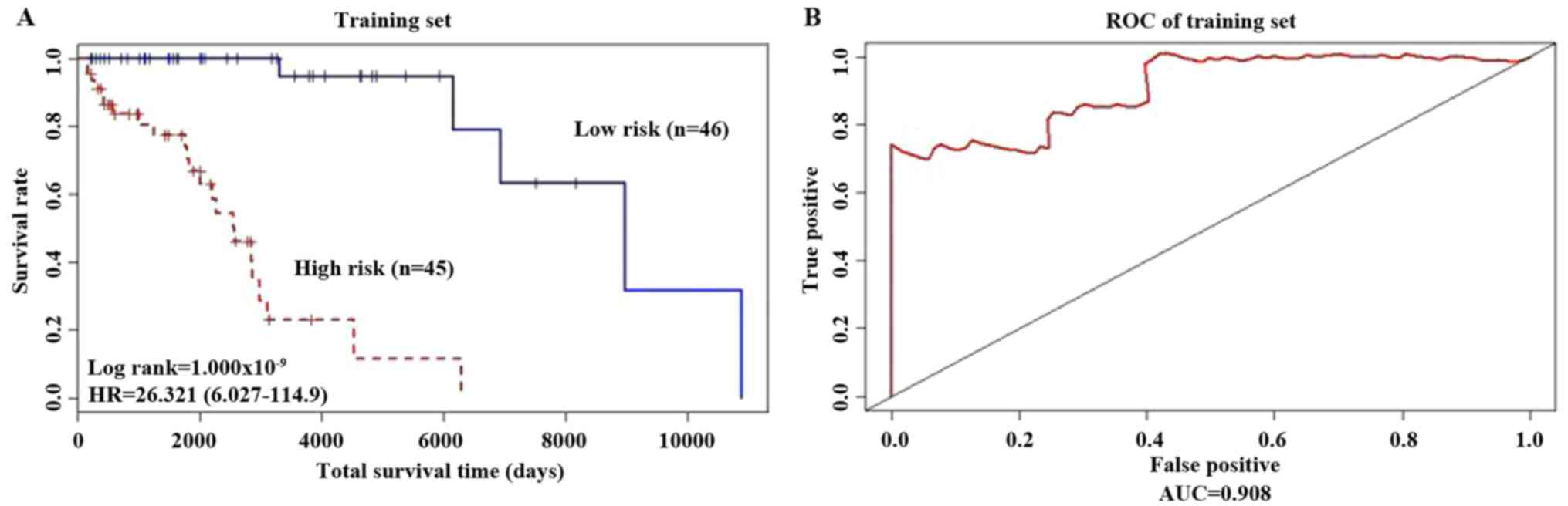
Using the Kaplan Meier method, the survival curves
for the two groups of patients, are presented in Fig. 2A; patients considered low risk
exhibited significantly longer overall survival times (P<0.001;
HR=26.321). Using the 5-year survival rates, a Receiver Operating
Characteristic (ROC) curve was drawn (Fig. 2B) and analysis was performed using the
‘survivalROC’ package. The advantages and disadvantages of the
model constructed using the 20 gene biomarkers were determined
based on the Area Under the Curve (AUC) value. The results showed
that AUC was equal to 0.908 (AUC >0.5 indicates a suitable
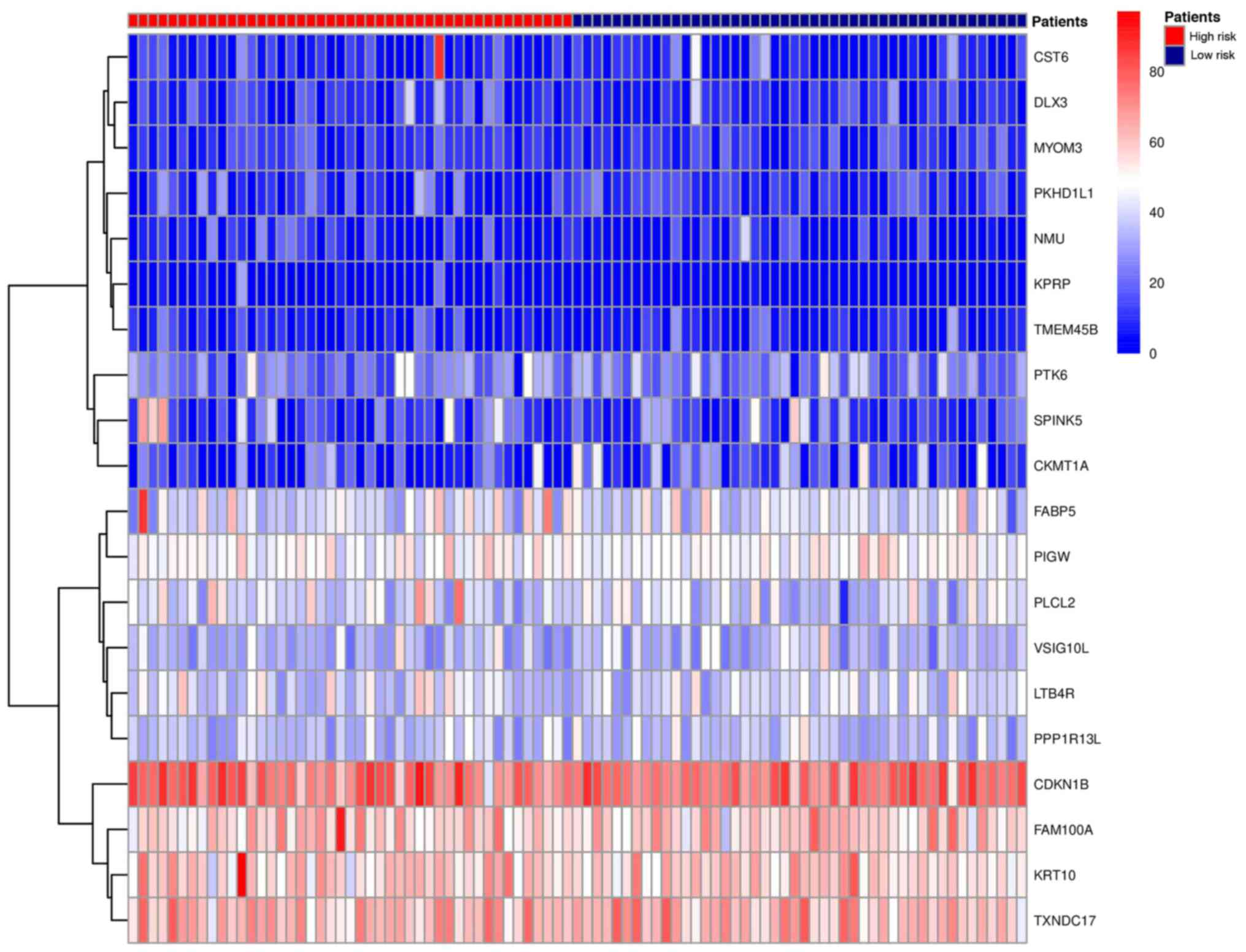
model). The heat map of risk gene expression profiles in high-risk
and low-risk patients were plotted (Fig.
3). The high-risk group was clearly distinguished from the
low-risk group. This indicated that the models constructed by these
20 gene biomarkers performed well.
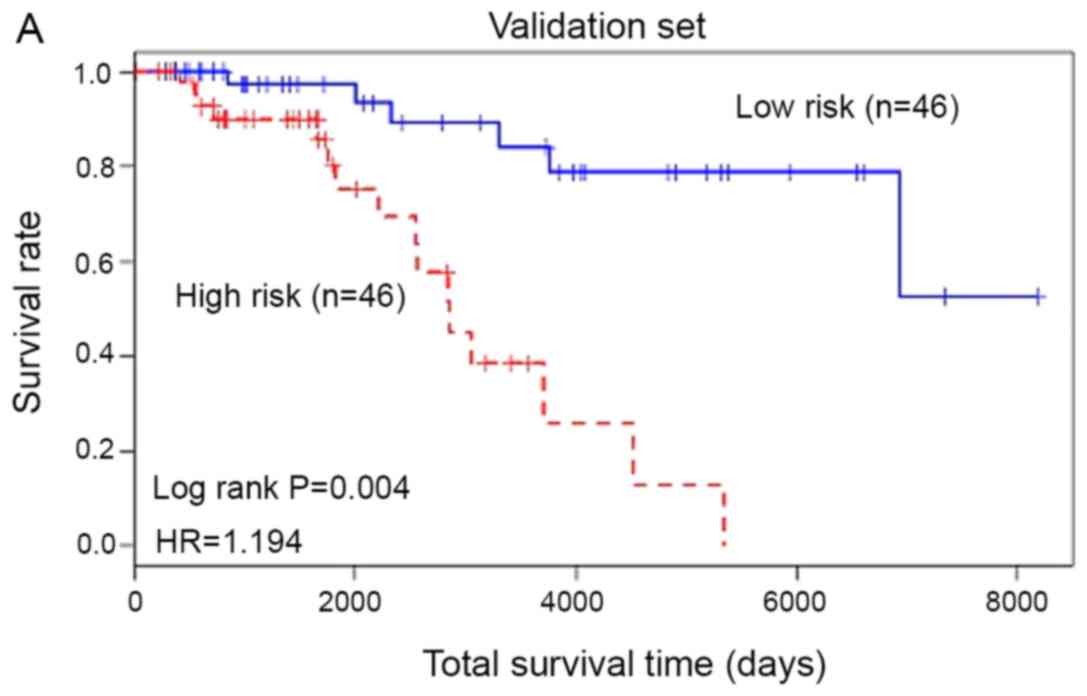
Based on the experimental results, the 20 gene
prognostic biomarkers could be used to significantly classify the
patients with skin cancer in the training samples into two groups:
High risk and low risk. In order to further verify the accuracy of
the test results, the 20 genetic biomarkers were used to validate
the test samples. As shown in Fig. 4,
these genetic biomarkers could still classify patients with skin
cancer in the test sample into high-risk and low-risk categories
(P=0.004, HR=1.194). The AUC of the ROC curve was 0.855.
Prognostic gene function analysis
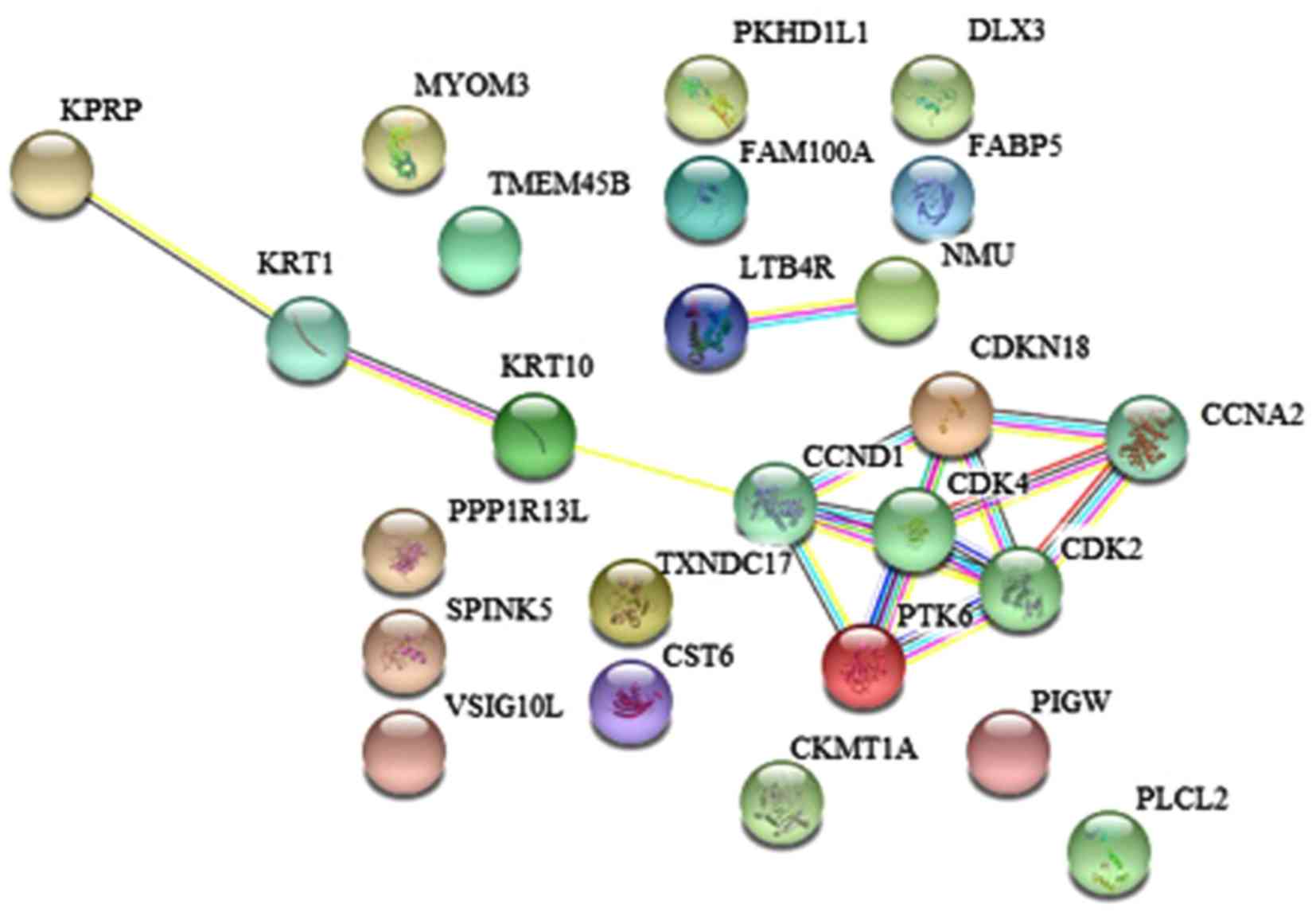
The online genetic analysis tool STRING (52) was used to analyze and study the
association between the identified gene biomarkers and their
associated protein synthesis pathways (Fig. 5; Table
III) Gene Ontology (GO) cellular component ontology analysis
identified a pathway of which involved a cyclin-dependent protein
kinase holoenzyme complex, which included three proteins (CCND1,
CDK2 and CDK4).
 | Table IIIGO biological processes associated
with the prognostic genes. |
Table III
GO biological processes associated
with the prognostic genes.
| Pathway ID | Pathway
description | Gene count | False discovery
rate | Matching
proteins |
|---|
| GO.0010948 | Negative regulation
of cell cycle process | 5 | 0.0219 | CCNA2, CCND1, CDK2,
CDK4, CDKN1B |
| GO.0031100 | Organ
regeneration | 4 | 0.0219 | CCNA2, CCND1, CDK2,
CDK4 |
| GO.0044773 | mitotic DNA Damage
checkpoint | 4 | 0.0219 | CCNA2, CCND1, CDK2,
CDKN1B |
| GO.0071156 | Regulation of cell
cycle arrest | 4 | 0.0219 | CCND1, CDK2, CDK4,
CDKN1B |
| GO.1901990 | Regulation of
mitotic cell cycle phase transition | 5 | 0.0219 | CCNA2, CCND1, CDK2,
CDK4, CDKN1B |
| GO.0032355 | Response to
estradiol | 4 | 0.0238 | CCNA2, CCND1, CDK2,
CDKN1B |
| GO.0010389 | Regulation of G2/M
transition of mitotic cell cycle | 3 | 0.0277 | CCNA2, CCND1,
CDK4 |
| GO.0044772 | Mitotic cell cycle
phase transition | 5 | 0.0277 | CCNA2, CCND1, CDK2,
CDK4, CDKN1B |
| GO.0097305 | Response to
alcohol | 5 | 0.0277 | CCNA2, CCND1, CDK2,
CDK4, CDKN1B |
| GO.1901991 | Negative regulation
of mitotic cell cycle phase transition | 4 | 0.0287 | CCNA2, CCND1, CDK2,
CDKN1B |
| GO.0000082 | G1/S transition of
mitotic cell cycle | 4 | 0.0326 | CCND1, CDK2, CDK4,
CDKN1B |
| GO.0048513 | Organ
development | 11 | 0.0345 | CCNA2, CCND1, CDK2,
CDK4, CDKN1B, DLX3, KRT1, KRT10, PLCL2, PPP1R13L, SPINK5 |
| GO.0060429 | Epithelium
development | 7 | 0.0457 | CCND1, CST6, DLX3,
FABP5, KRT10, PTK6, SPINK5 |
| GO.0048545 | Response to steroid
hormone | 5 | 0.0482 | CCNA2, CCND1, CDK2,
CDK4, CDKN1B |
Performance of the biomarkers in
clinical subtypes
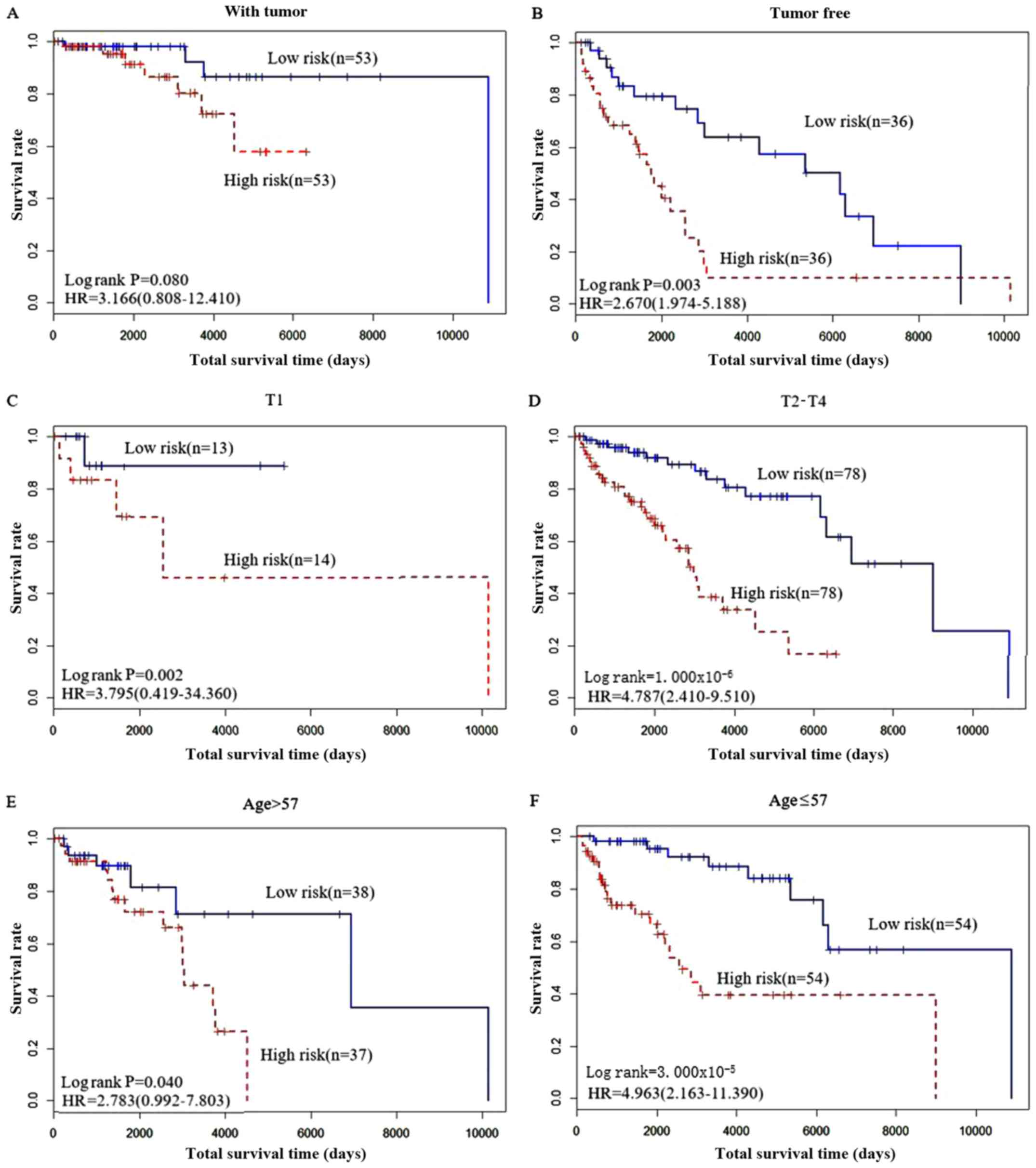
Among the clinical factors, pathological T stage (T1
and T2-4), cancer status (with tumor and tumor free) and age
(>57 years vs. ≤57 years) were all significantly associated with
the recurrence status of patients with skin cancer (Fig. 6). Therefore, these 20 genes should to
be considered in different clinical types to determine which
clinical state they are more suitable for.
The results in Fig. 6
shows the predictive effect of these 20 gene biomarkers. Patients
who were tumor free had improved survival compared with those with
tumor. Similarly, patients with a T-stage of T1-3 exhibited
improved survival compares with patients classed as T4, and
patients ≤57 years old had improved survival compared with patients
>57 years old.
Discussion
In the present study, a variety of statistical
analysis methods were used (including LASSO regression,
single-factor survival analysis, multi-factor Cox proportional
hazards regression model and ROC curve analysis) to identify
differences in the gene expression profiles of patients with skin
cancer. A supervised cluster analysis method was used and 20
prognostic genes from the training samples were identified and
verified against a test sample. The results showed that these 20
genes can stratify patients with skin cancer as high-risk and
low-risk, which shows the feasibility of the mining method used in
the present study.
From a biological point of view, the 20 prognostic
genes identified in the present study successfully divided the
patients with cancer according to the risk of recurrence, which may
have important reference value for the treatment of recurrence of
metastatic skin cancer, and may influence future clinical studies
of skin cancer and drug development.
Among the 20 prognostic genes identified in the
present study, several are closely associated with skin cancer and
metastatic skin cancer, including DLX3, PTK6 and CST6 genes. For
example, physical interaction between DLX3 and P53 on P21 promoter
can enhance the expression of P21(53). Increasing DLX3 expression in
keratinocytes produces a G1-S blockade associated with P53
signature transcriptional profiles (54).
Deletion of DLX3 promotes a mitogenic phenotype
associated with constitutive activation of ERK (55). The loss of DLX3 expression in human
skin cancer suggests that a DLX3-P53 interaction may be a primary
regulatory axis of epidermis differentiation and that DLX3 may be a
regulator of skin cancer development. Protein tyrosine kinase 6
(PTK6) is expressed in ~70% of cases of triple-negative breast
cancer, in which it serves an important role in promoting
metastatic lung colonization and survival (56). PTK6 inhibits the inhibition of
metastasis of triple-negative breast cancer via SNAIL-dependent
regulation of E-cadherin expression (57). Epigenetic changes associated with
upregulation of CST6 gene expression may be accompanied by
metastatic diffusion of primary tumor sites, and current studies
have shown that methylation-dependent epigenetic silencing of CST6
represents an important mechanism for loss of CST6 during the
development and/or progression to metastasis (58). Other gene prognostic biomarkers
identified in the present study have potential research value and
need further exploration and research.
Among these, several genes, including PLCL2, serine
protease inhibitor Kazal type-5 (SPINK5) and KRT10 are associated
with skin diseases. Inosine strongly enhances proliferation of
human C32 melanoma cells through the PLCL2 and PI3K pathways
(59). SPINK5 serves a crucial role
in the timing of desquamation of the skin (60). Biallelic KRT10 mutations result in
skin fragility caused by self-improving epidermolytic ichthyosis
(61). Several other genes, such as
transmembrane protein 45B (TMEM45B), CDKN1B, CKMT1A, FABP5 and
PPP1R13L have also been shown to be associated with several types
of cancer. TMEM45B has been shown to be abnormally expressed in
gastric tumors and serves an important role in gastric
tumorigenesis (62). The CCND1-A870G
and CDKN1B-C79T polymorphisms are associated with breast cancer
risk (63). The
n335586/miR-924/CKMT1A axis contributes to migration and invasion
of hepatocellular carcinoma cells (64). FABP5 serves an important role in the
carcinogenesis and metastasis of cervical cancer, and may be a
novel predictor for prognostic assessment of patients with cervical
cancer (65). Polymorphisms of ERCC1,
CD3EAP and PPP1R13L in chromosomal region 19q13.2-3 have previously
been shown to exhibit a synergistic effect on apoptosis and DNA
repair pathways (66).
In summary, the 20 gene biomarkers identified based
on the LASSO algorithm can effectively predict the risk of patients
with skin cancer and may be more convenient as a model of
prognosis.
Acknowledgements
Not applicable.
Funding
This work was supported by the Key Project of China
Ministry of Education for Philosophy and Social Science: Big Data
Driven Risk Research on City's Public Safety (grant no. 16JZD023)
and the Fundamental Research Funds for the Central Universities:
Big Data Driven Risk Pre-Warning Research on City's Public Safety
(grant no. 17LZUJBWZD012).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
GL analyzed and interpreted the patient data was a
major contributor in writing the manuscript. CL analyzed the data.
HZ made substantial contributions to acquisition of data. ZZ was a
major contributor in interpreting the data and in writing the
manuscript. YS made substantial contributions to analysis and
interpretation of data. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Diepgen TL and Mahler V: The epidemiology
of skin cancer. Br J Dermatol. 146 (Suppl 61):S1–S6.
2002.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lai V, Cranwell W and Sinclair R:
Epidemiology of skin cancer in the mature patient. Clin Dermatol.
36:167–176. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Pilgrim W, Hayes R, Hanson DW, Zhang B,
Boudreau B and Leonfellner S: Skin Cancer (Basal Cell Carcinoma,
Squamous Cell Carcinoma, and Malignant Melanoma): New cases,
treatment practice, and health care costs in New Brunswick, Canada,
2002-2010. J Cutan Med Surg. 18:320–331. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in china,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Karagas MR, Stukel TA, Greenberg ER, Baron
JA, Mott LA and Stern RS: Risk of subsequent basal cell carcinoma
and squamous cell carcinoma of the skin among patients with prior
skin cancer. Skin cancer prevention study group. JAMA.
267:3305–3310. 1992.PubMed/NCBI
|
|
7
|
Jerant AF, Johnson JT, Sheridan CD and
Caffrey TJ: Early detection and treatment of skin cancer. Am Fam
Physician. 62:357–368, 375-376, 381-382. 2000.PubMed/NCBI
|
|
8
|
Gauci J and Muscat G: A local perspective
on basal cell carcinoma: Frequency of subsequent skin tumours.
Malta Med J. 1:46–55. 2017.
|
|
9
|
Gillner J, Kirchberg K, Korge B,
Hunzelmann N, Krieg T and Scharffetter-Kochanek K: Cutaneous
metastases from a leiomyosarcoma of the testicular tunica
albuginea. Hautarzt. 51:41–45. 2000.PubMed/NCBI View Article : Google Scholar : (In German).
|
|
10
|
Hoyt BS and Cohen PR: Cutaneous scrotal
metastasis: Origins and clinical characteristics of visceral
malignancies that metastasize to the scrotum. Int J Dermatol.
52:398–405. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bremnes RM, Veve R, Hirsch FR and Franklin
WA: The E-cadherin cell-cell adhesion complex and lung cancer
invasion, metastasis, and prognosis. Lung Cancer. 36:115–124.
2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhen Y, Wu C and Jiang J: Analysis of
clinicopathological features and prognostic factors in cutaneous
metastases of malignant tumors. Chin Clin Oncol. 19:152–155.
2014.
|
|
13
|
Niu JX, Meng XK and Ren JJ: Studied
microRNA gene expression in human hepatocellular carcinoma by
microRNA microarray techniques. World J Gastroenterol.
21:12605–12611. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hou X, Peng JX, Hao XY, Cai JP, Liang LJ,
Zhai JM, Zhang KS, Lai JM and Yin XY: DNA methylation profiling
identifies EYA4 gene as a prognostic molecular marker in
hepatocellular carcinoma. Ann Surg Oncol. 21:3891–3899.
2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pivarcsi A and Sonkoly E: Skin cancer
associated microRNAs. US Patent 12/994,734. Filed June 4, 2009;
issued May 5, 2011.
|
|
16
|
Glavač D and Ravnik-Glavač M: Essential
role of microRNA in skin physiology and disease. Adv Exp Med Biol.
888:307–330. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Leffell DJ: The scientific basis of skin
cancer. J Am Acad Dermatol. 42 (Suppl):S18–S22. 2000.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Thyagarajan A, Shaban A and Sahu RP:
MicroRNA-directed cancer therapies: Implications in melanoma
intervention. J Pharmacol Exp Ther. 364:1–12. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu Z, Chen D, Xu Y and Liu J: Logistic
support vector machines and their application to gene expression
data. Int J Bioinform Res Appl. 1:169–182. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yongchao GE, Sealfon SC and Speed TP:
Multiple testing and its applications to microarrays. Stat Methods
Med Res. 18:543–563. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Okimoto G, Zeinalzadeh A, Wenska T, Loomis
M, Nation JB, Fabre T, Tiirikainen M, Hernandez B, Chan O, Wong L
and Kwee S: Joint analysis of multiple high-dimensional data types
using sparse matrix approximations of rank-1 with applications to
ovarian and liver cancer. BioData Min. 9(24)2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Xiong J, Xiong K and Bing Z: Clinical and
RNA expression integrated signature for urothelial bladder cancer
prognosis. Cancer Biomark. 21:535–546. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tang RX, Chen WJ, He RQ, Zeng JH, Liang L,
Li SK, Ma J, Luo DZ and Chen G: Identification of a RNA-Seq based
prognostic signature with five lncRNAs for lung squamous cell
carcinoma. Oncotarget. 8:50761–50773. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gao X, Wu Y, Yu W and Li H: Identification
of a seven-miRNA signature as prognostic biomarker for lung
squamous cell carcinoma. Oncotarget. 7:81670–81679. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
van Malenstein H, Gevaert O, Libbrecht L,
Daemen A, Allemeersch J, Nevens F, Van Cutsem E, Cassiman D, De
Moor B, Verslype C and van Pelt J: A seven-gene set associated with
chronic hypoxia of prognostic importance in hepatocellular
carcinoma. Clin Cancer Res. 16:4278–4288. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Nakazawa H, English D, Randell PL,
Nakazawa K, Martel N, Armstrong BK and Yamasaki H: UV and skin
cancer: Specific p53 gene mutation in normal skin as a biologically
relevant exposure measurement. Proc Natl Acad Sci USA. 91:360–364.
1994.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Linck L, Liebig J, Völler D, Eichner N,
Lehmann G, Meister G and Bosserhoff A: MicroRNA-sequencing data
analyzing melanoma development and progression. Exp Mol Pathol.
105:371–379. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Goldman M, Craft B, Swatloski T, Ellrott
K, Cline M, Diekhans M, Ma S, Wilks C, Stuart J, Haussler D and Zhu
J: The UCSC Cancer Genomics Browser: Update 2013. Nucleic Acids
Res. 41 (Database Issue):D949–D954. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cline MS, Craft B, Swatloski T, Goldman M,
Ma S, Haussler D and Zhu J: Exploring TCGA Pan-Cancer Data at the
UCSC Cancer Genomics Browser. Sci Rep. 3(2652)2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Goldman M, Craft B, Swatloski T, Cline M,
Morozova O, Diekhans M, Haussler D and Zhu J: The UCSC cancer
genomics browser: Update 2015. Nucleic Acids Res. 43 (Database
Issue):D812–D817. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Turner N, Ware O and Bosenberg M: Genetics
of metastasis: Melanoma and other cancers. Clin Exp Metastasis.
35:379–391. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kong J, Zhang Y and Zhao B: A
clinicopathological analysis of 104 cases with metastatic tumors of
the skin. Chin J Diagnostic Pathol. 2:211–213. 1995.
|
|
33
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing,
Vienna, 2006.
|
|
34
|
Loh KC, Greenspan FS, Gee L, Miller TR and
Yeo PP: Pathological Tumor-Node-Metastasis (pTNM) staging for
papillary and follicular thyroid carcinomas: A retrospective
analysis of 700 patients. J Clin Endocrinol Metab. 82:3553–3562.
1997.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lezcano C, Shoushtari AN, Ariyan C,
Hollmann TJ and Busam KJ: Primary and metastatic melanoma with NTRK
fusions. Am J Surg Pathol. 42:1052–1058. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rhee JS, Matthews BA, Neuburg M, Logan BR,
Burzynski M and Nattinger AB: The skin cancer index: Clinical
responsiveness and predictors of quality of life. Laryngoscope.
117:399–405. 2007.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Morgan M, McCreedy R, Simpson J and Hay
RJ: Dermatology quality of life scales: A measure of the impact of
skin diseases. Br J Dermatol. 136:202–206. 1997.PubMed/NCBI
|
|
38
|
Rogers EM, Connolly KL, Nehal KS, Dusza
SW, Rossi AM and Lee E: Comorbidity scores associated with limited
life expectancy in the very elderly with nonmelanoma skin cancer. J
Am Acad Dermatol. 78:1119–1124. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Nagy A, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep.
8(9227)2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zaorsky NG, Lee CT, Zhang E and Galloway
TJ: Skin CanceR Brachytherapy vs External beam radiation therapy
(SCRiBE) meta-analysis. Radiother Oncol. 126:386–393.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Riker AI, Enkemann SA, Fodstad O, Liu S,
Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al: The
gene expression profiles of primary and metastatic melanoma yields
a transition point of tumor progression and metastasis. BMC Med
Genomics. 1(13)2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lawrence MS, Stojanov P, Polak P, Kryukov
GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH,
Roberts SA, et al: Mutational heterogeneity in cancer and the
search for new cancer-associated genes. Nature. 499:214–218.
2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wiltgen M, Gerger A and Smolle J: Tissue
counter analysis of benign common nevi and malignant melanoma. Int
J Med Inform. 69:17–28. 2003.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Esteva A, Kuprel B, Novoa RA, Ko J,
Swetter SM, Blau HM and Thrun S: Dermatologist-level classification
of skin cancer with deep neural networks. Nature. 542:115–118.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Esteva A, Kuprel B, Novoa RA, Ko J,
Swetter SM, Blau HM and Thrun S: Corrigendum: Dermatologist-level
classification of skin cancer with deep neural networks. Nature.
546(686)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Masood A and Al-Jumaily AA: Computer aided
diagnostic support system for skin cancer: A review of techniques
and algorithms. Int J Biomed Imaging. 2013(323268)2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chang P, Bing Z, Tian J, Zhang J, Li X, Ge
L, Ling J, Yang K and Li Y: Comprehensive assessment gene
signatures for clear cell renal cell carcinoma prognosis. Medicine
(Baltimore). 97(e12679)2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View Article : Google Scholar
|
|
51
|
The Gene Ontology Consortium. The Gene
Ontology Resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Panno ML, Giordano F, Mastroianni F,
Morelli C, Brunelli E, Palma MG, Pellegrino M, Aquila S, Miglietta
A, Mauro L, et al: Evidence that low doses of Taxol enhance the
functional transactivatory properties of p53 on p21 waf promoter in
MCF-7 breast cancer cells. FEBS Lett. 580:2371–2380.
2006.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Palazzo E, Kellett MD, Cataisson C, Bible
PW, Bhattacharya S, Sun HW, Gormley AC, Yuspa SH and Morasso MI: A
novel DLX3-PKC integrated signaling network drives keratinocyte
differentiation. Cell Death Differ. 24:717–730. 2017.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Jiang L, Campagne C, Sundström E, Sousa P,
Imran S, Seltenhammer M, Pielberg G, Olsson MJ, Egidy G, Andersson
L and Golovko A: Constitutive activation of the ERK pathway in
melanoma and skin melanocytes in Grey horses. BMC Cancer.
14(857)2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Palazzo E, Kellett M, Cataisson C, Gormley
A, Bible PW, Pietroni V, Radoja N, Hwang J, Blumenberg M, Yuspa SH
and Morasso MI: The homeoprotein DLX3 and tumor suppressor p53
co-regulate cell cycle progression and squamous tumor growth.
Oncogene. 35:3114–3124. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Ito K, Park SH, Nayak A, Byerly JH and
Irie HY: PTK6 Inhibition suppresses metastases of triple-negative
breast cancer via SNAIL-dependent E-cadherin regulation. Cancer
Res. 76:4406–4417. 2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Rivenbark AG, Livasy CA, Boyd CE, Keppler
D and Coleman WB: Methylation-dependent silencing of CST6 in
primary human breast tumors and metastatic lesions. Exp Mol Pathol.
83:188–197. 2007.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Soares AS, Costa VM, Diniz C and Fresco P:
Inosine strongly enhances proliferation of human C32 melanoma cells
through PLC-PKC-MEK1/2-ERK1/2 and PI3K pathways. Basic Clin
Pharmacol Toxicol. 116:25–36. 2015.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Le NA, Katsuyama M, Demura M, Tanii H,
Katsuyama H and Saijoh K: Regulation of serine protease inhibitor
Kazal type-5 (SPINK5) gene expression in the keratinocytes. Environ
Health Prev Med. 19:307–313. 2014.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Frommherz L, Küsel J, Zimmer A, Fischer J
and Has C: Skin fragility caused by biallelic KRT10 mutations: An
intriguing form of self-improving epidermolytic ichthyosis. Br J
Dermatol. 182:780–785. 2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Shen K, Yu W, Yu Y, Liu X and Cui X:
Knockdown of TMEM45B inhibits cell proliferation and invasion in
gastric cancer. Biomed Pharmacother. 104:576–581. 2018.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Canbay E, Eraltan IY, Cercel A, Isbir T,
Gazioglu E, Aydogan F, Cacina C, Cengiz A, Ferahman M, Zengin E and
Unal H: CCND1 and CDKN1B polymorphisms and risk of breast cancer.
Anticancer Res. 30:3093–3098. 2010.PubMed/NCBI
|
|
64
|
Fan H, Lv P, Mu T, Zhao X, Liu Y, Feng Y,
Lv J, Liu M and Tang H: LncRNA n335586/miR-924/CKMT1A axis
contributes to cell migration and invasion in hepatocellular
carcinoma cells. Cancer Lett. 429:89–99. 2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wang W, Chu HJ, Liang YC, Huang JM, Shang
CL, Tan H, Liu D, Zhao YH, Liu TY and Yao SZ: FABP5 correlates with
poor prognosis and promotes tumor cell growth and metastasis in
cervical cancer. Tumor Biol. 37:14873–14883. 2016.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Chae YS, Kim JG, Kang BW, Lee SJ, Jeon HS,
Park JS, Choi GS and Lee WK: PPP1R13Lvariant associated with
prognosis for patients with rectal cancer. J Cancer Res Clin Oncol.
139:465–473. 2013.PubMed/NCBI View Article : Google Scholar
|