1. Introduction
Following the complete sequencing and analysis of
the human genome, Snijders et al (1) assembled microarrays for genome-wide
measurement of DNA copy numbers and CNVs of the normal human genome
have been increasingly described (2). Genomic architecture has an important
role in CNVs (3). Pericentromeric
and sub-telomeric regions are rich in highly homologous duplicated
segments of DNA >1 Kb in length and with >90% sequence
similarity known as segmental duplications (SDs) (4). Non-allelic homologous recombination
(NAHR) may serve a more prominent role in larger CNVs and SDs than
smaller CNVs (5). SDs are also
referred to as low-copy number repeats (LCRs) (4). Harel and Lupski (6) defined LCRs as clusters of paralogous
sequences organized in hierarchical groups of direct and inversely
orientated sequences. LCRs vary in copy number and also mediate CNV
formation. Furthermore, LCR/SD pairs can contribute to initiate
NAHR and result in the formation of CNVs. NAHR events between
different chromatids give rise to duplications or deletions,
whereas NAHR events on the same chromatid give rise to deletions
(3).
Chromothripsis derived from chromosome
(chromo) shattering (thripsis) is a phenomenon that
involves complex chromosomal rearrangements (7). Marcozzi et al (7) reviewed a model of chromothripsis
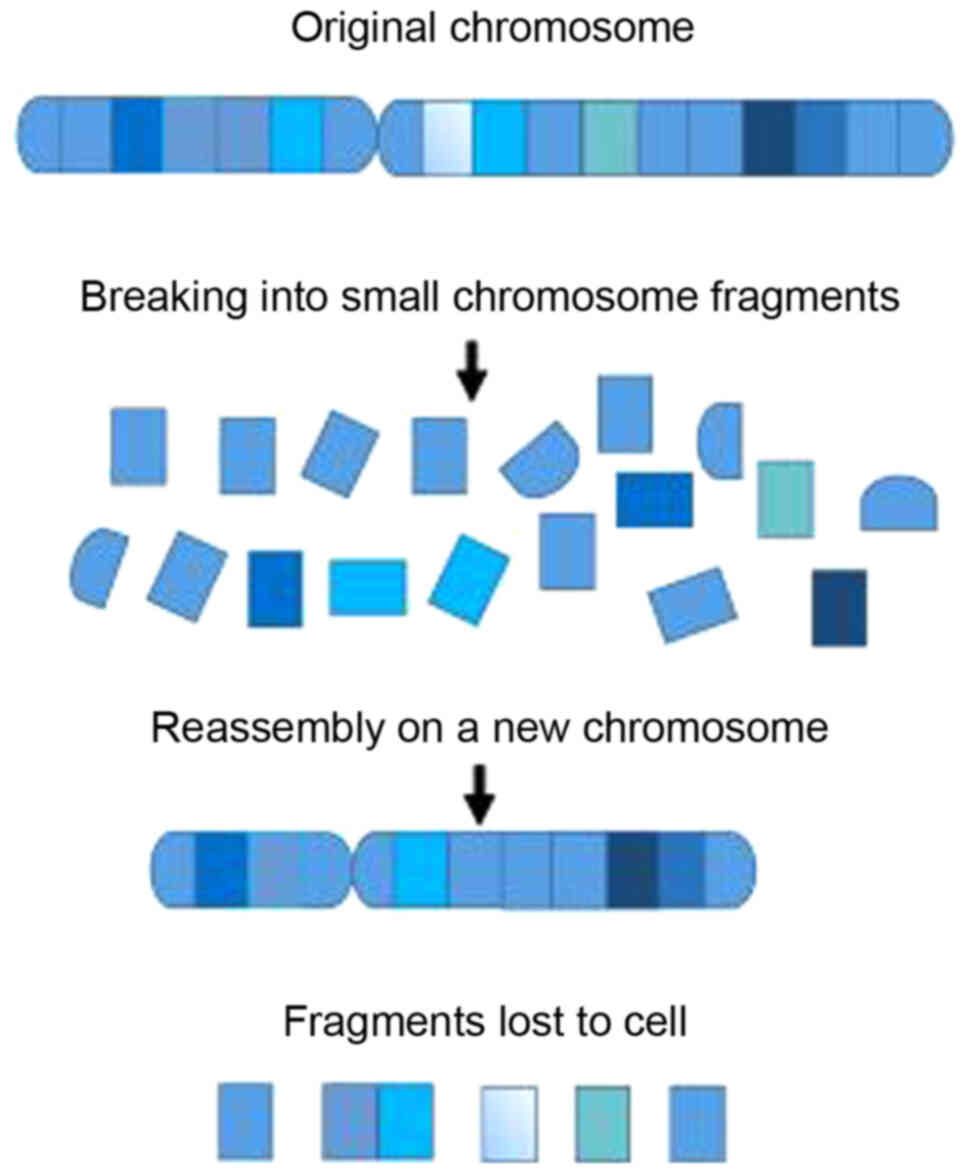
formation in which chromosomes are initially broken into small
chromosomal fragments, and subsequently, the chromosomal fragments
are reassembled into a new chromosome. However, the order and
orientation of the fragments are altered compared with the
structure of the original chromosome, and some of the chromosomal
fragments may be lost as they are not incorporated during the
reassembly process (7). Thus,
chromothripsis is characterized by extensive genomic rearrangements
and an oscillating pattern of DNA copy number levels of one or a
few chromosomes (8). Chromothriptic
breakpoints often occur in the vicinity of clusters of point
mutations, termed kataegis (from the Greek for ‘thunderstorm’)
(9), thus leading to the hypothesis
that kataegic regions of hypermutations may indicate, or even lead
to structural rearrangements (10).
2. CNVs
CNVs consist of duplications, deletions and
insertions of DNA sequences into an individual's genome that range
in size from 50 base pairs to millions of bases (11). They are structural variants that may
involve complex genomic rearrangements and seem to possess
additional mutations around their breakpoints (12). CNVs are seemingly frequent in the
human genome. Zarrei et al (4) constructed a CNV map of the human genome
and identified 11,742 CNV regions in the stringent map of healthy
individuals. Aberrant pairing between mismatched copies of the
segmental duplication and unequal crossing over in meiosis may be
involved in deletions. Additionally, interstitial duplications may
be due to inter- or intrachromosomal recombination between copies
of the segmental duplications (13).
Large LCRs are predisposed to DNA rearrangements, namely deletions,
duplications and inversions, via NAHR. NAHRs give rise to recurrent
structural variants, which share the same genomic content and size
in unrelated individuals (14).
Nonrecurrent structural variants which possess unique genomic
content and size at a given locus in unrelated individuals are
formed by other molecular mechanisms that include replicative and
non-replicative mechanisms, reviewed in (15). LCRs may possess a dual role in
structural variation: Mediating recurrent structural variants as
substrates for NAHR and then stimulating non-recurrent variants via
replication-based mechanisms (14).
Gu et al (16) suggested that
the high concentration of Alu elements in a specific region served
as a suitable substrate for formation of CNVs and Alu-Alu-mediated
mechanisms contribute considerably to the formation of complex
CNVs.
The formation of structural variants may be
associated with the genomic architecture. For example, NAHR- and
nonhomologous-mediated CNVs are associated with different timings
of DNA replication: Hotspots of NAHR-mediated events were enriched
in early-replicating regions, whereas nonhomologous hotspots were
enriched in late-replicating regions (17).
Recently, Hattori et al (18) reported a case with several features
of the multiple de novo CNVs (mdnCNVs), including multiple
rearrangements in perizygotic cells, non-recurrent rearrangements,
and rearrangements that were present in one chromosomal arm but
were not present at the inter-chromosomal translocation (18). According to Liu et al
(19), the timeframe of the mdnCNV
phenomenon predicts that the dnCNVs which occur prior to the embryo
reaching the 2-cell stage are constitutional by default.
CNVs remain a major challenge with regard to
clinical interpretation. The American College of Medical Genetics
established standards and guidelines for interpretation and
reporting of postnatal constitutional CNVs (20). There are three main categories of
significance: Pathogenic, benign and uncertain clinical
significance. The last category is subdivided into; likely
pathogenic, likely benign and uncertain clinical significance (no
sub-classification). The interpretation of the clinical relevance
of CNV is complex but necessary to the practice of medicine.
3. Chromothripsis
Chromothripsis, chromoanasynthesis and chromoplexy
are collectively termed chromoanagenesis (from the Greek
chromo for chromosome and anagenesis for rebirth).
For a recent review see Zepeda-Mendoza and Morton, 2019(21). In the present review, only
chromothripsis will be discussed.
Stephens et al (22) characterized a phenomenon during
cancer development, which they termed chromothripsis. The
chromosome or chromosomal region was segmented/broken into 10-100s
of pieces, some of which were subsequently stitched back together
by the endogenous DNA repair mechanisms, such as
microhomology-mediated break repair and/or nonhomologous
end-joining (NHEJ). The result was a mosaic patchwork of genomic
fragments (22). The database
ChromothripsisDB (23) is a resource
for mining the existing knowledge of chromothripsis (24). Fig. 1
shows a scheme of the process of chromothripsis in a single
chromosome. Ionizing radiation may contribute in part to
chromothripsis (25). Mladenov et
al (26) developed a model of
double stranded break (DSB) clustering which permitted direct
analysis of the consequences of determined configurations of DSB
clusters in cells. Their results suggested that DSB clusters
constitute the first-line DSB-processing pathways of canonical-NHEJ
and homologous recombination repair. Consequently, there is an
increase in the contribution of alternative end-joining and the
formation of chromosomal aberrations. The authors were thus able to
hypothesize a mechanism for the damage caused by high linear energy
transfer radiation and the genomic rearrangements associated with
chromothripsis (26). One of the
mechanisms that may underlie chromothripsis involves the
segmentation and breakdown of chromosomes in micronuclei where
isolated chromosomes or chromosome arms undergo significant local
DNA breakage and rearrangement (27). The chromosomes from ruptured
micronuclei are reincorporated into daughter nuclei (28). Maciejowski et al (29) suggested another mechanism: Telomere
fusions during telomere crisis, giving rise to anaphase bridges
that persist and develop into chromatin bridges. Several steps
occur after and at the end of clonal descendants derived from
telomere crisis in cells displaying chromothripsis and kataegis
(29). Kataegis is associated with
chromothripsis (9). Chromothripsis
breakpoints may result in the presence of clusters of base
substitutions with close proximity (kataegis), displaying the
C>T and C>G signature at TpC dinucleotides, which are
associated with mutagenesis mediated by apolipoprotein B mRNA
editing catalytic polypeptide-like family (29). Tubio and Estivill (30) reported that chromothripsis may be
caused by aborted programmed cell death (apoptosis). Several
genotoxic agents such as radiation, nutrient deprivation, infection
or oxygen shortage resulted in higher-order fragmentation of
chromatin and apoptosis in a cell population. However, one or even
a small number of cells may not complete apoptosis and survive.
These surviving cells may incorrectly repair their DNA, giving rise
to rearrangements characteristic of chromothripsis (30). Using an approach based on complex
alterations after selection and transformation, Mardin et al
(31) reported an association
between telomere stability and hyperploidy with chromothripsis
(31). Ivkov and Bunz (32) established a model for the suppression
of chromothripsis by p53, where the mutational loss of p53 gives
rise to chromothripsis.
Kloosterman et al (33) reported evidence of local shattering
of chromosomes followed by NHEJ leading to the formation of complex
constitutional rearrangements involved in congenital defects, and
suggested the possibility of constitutional chromothripsis
(33). Analysis of 10 constitutional
complex chromosomal rearrangements demonstrated that chromothripsis
rearrangements may result from chromosome breakage by multiple DSBs
(34). The authors found that in two
patients the rearrangements were confined to a single chromosome,
but in one patient multiple chromosomes were involved. These
rearrangements gave rise to deletions or to copy neutral
rearrangements, and clusters of DSBs were observed. The authors
concluded that a common mechanism involved in chromothripsis
rearrangements associated with developmental malformations may be
the chromosome shattering and nonhomologous or microhomology
mediated repair mechanisms (34).
Nazaryan-Petersen et al (35)
demonstrated that constitutional chromothripsis may be driven by
L1-Mediated Retro-transposition and Alu/Alu Homologous
Recombination. Masset et al (36) suggested a mechanism through which
shattered chromosomes are reassembled primarily by NHEJ giving rise
to complex rearranged chromosomes with or without copy-number
losses, as illustrated by Fukami et al (37). Additionally, microhomology-mediated
break-induced replication (MMBIR) may be implicated in
chromothripsis (37).
The number of breaks is lower for chromothripsis
rearrangements observed in the germline cells compared with
chromothripsis in cancer genomes (34). These lower numbers of breaks and copy
number changes in congenital chromothripsis may be due to different
molecular mechanisms occurring in the development of the disease,
as well as to selection in a developing embryo. It is possible that
congenital chromothripsis rearrangements possess a similar
architecture as simple reciprocal translocation, which, involves
two breaks and subsequent formation of two derivative chromosomes
(38). Constitutional chromothripsis
may thus be a more complex variant of a simple reciprocal
translocation (38).
The mutation rate is greater in spermatogenesis than
in oogenesis (39). In the course of
spermatogenesis, chromothripsis can arise due to environmental
stimuli such as ionizing radiation or the generation/presence of
free radicals which act as initiators of DNA damage (39). Constitutional chromothripsis may be
due to an imbalance between DSB formation and repair in meiosis
(40). The rearrangements observed
by Kloosterman et al (34)
were present on paternal chromosomes. This finding highlights the
vulnerability of spermatogenesis to DNA damage and show that
spermatogenesis is a critical stage in the genesis of congenital
chromothripsis. Failure in DNA repair pathways in oocytes with the
potential occurrence of abortive apoptosis or replicative stress
may also trigger chromothripsis (39).
In addition to the cases of congenital disease
referred to above, multiple other cases with abnormal phenotypes
and constitutional chromothripsis have been described (41-43).
Chromothripsis in healthy females was described by
de Pagter et al (44) who
demonstrated that the human genome can tolerate chromothripsis
rearrangements, disrupting multiple protein-coding genes with a
normal phenotype. Additionally, Bertelsen et al (45) showed that constitutional
chromothripsis may occur over several generations and was not
always associated with an abnormal phenotype. Chiang et al
(46) showed that chromothripsis
occurred in the germline where it resulted in a karyotypically
balanced state with chromosomal balanced abnormalities such as
inversions (46).
An increasing number of reports of chromothriptic
events in patients with congenital diseases (33,34,41-43),
in embryos (39), but also in
healthy individuals (44,45) suggest that chromothripsis is
considerably more frequent than expected (7). These studies are important for the
evaluation of patients with congenital diseases as well as in human
health.
4. CNVs and constitutional
chromothripsis
Clustered CNVs detected by chromosomal microarray
analysis (CMA) are frequently reported as constitutional
chromothripsis (47). Pettersson
et al (48) presented two
different rearrangements on chromosome 5p in a mother and her
daughter, initially classified as simple CNVs. Using a combination
of microarray analysis and massive parallel whole-genome
sequencing, it was shown that, due to unequal crossing-over during
meiosis, there was an evolution from a chromothriptic rearrangement
in the mother to another complex rearrangement involving both
deletions and duplications in her daughter (48). Slamova et al (49) studied the case of a boy with
developmental and growth delay in whom karyotyping showed a
seemingly balanced de novo complex rearrangement of 4
chromosomes. Microarray analysis detected two paternal de
novo deletions and subsequent whole-genome mate-pair sequencing
confirmed the chromothriptic nature of the rearrangement (49).
Chromothripsis results in CNVs and other structural
changes (50). As referred to above,
rearrangements in chromothripsis were associated with Alu/Alu NAHR.
Due to the high copy number of the Alu elements, these elements are
prone to NAHR events which have resulted in benign and pathogenic
genomic deletions, duplications and inversions (35). Similarly, the presence of repeated
sequences, such as segmental duplications or Alu sequences, were
frequently observed at the break points of chromothripsis, similar
to CNVs (50).
Nazaryan-Petersen et al (47) studied 21 clustered CNV carriers with
congenital developmental disorders, intellectual disability or
autism. Using whole genome sequencing to study the structures of
the rearrangement first investigated by CMA, they identified a
total of 83 breakpoint junctions (BPJs). Their results indicated 8
cases with deletions that frequently had additional structural
rearrangements, such as insertions and inversions typical to
chromothripsis, 7 cases with duplications, and 6 cases with
combinations of duplications and deletions showing interspersed
duplications and BPJs enriched with microhomology. Some
rearrangements also indicated both a breakage-fusion-bridge cycle
process and haltered formation of a ring chromosome, and 2 cases
showed rearrangements mediated by Alu and long interspersed nuclear
elements (LINE). The authors concluded that various mechanisms may
be involved in the formation of clustered CNVs: Replication
independent canonical NHEJ and alt-NHEJ, microhomology-mediated
break-induced replication (MMBIR)/fork stalling and template
switching, and breakage-fusion-bridge cycle and Alu- and
LINE-mediated pathways. They suggested that 7 cases were
chromothripsis and 10 cases were chromoanasynthesis events
(47). The primary difference
between chromoanasynthesis and chromothripsis is the presence of
copy gains such as duplication, triplication, in addition to
deletions and copy-neutral chromosomal regions (7).
Recently, Hattori et al (18) reported a case of a patient with
transient neonatal diabetes mellitus and multiple congenital
malformations who possessed a simple tandem duplication on
chromosome 6q, a simple balanced inversion on chromosome 14q, two
tandem inversions with a deletion on chromosome 2q, an inverted
duplication with a deletion on chromosome 13q, and catastrophic
rearrangements on chromosome 21q. The substantial genomic changes
on chromosomes 2q and 21q were indicative of chromothripsis, and
the eventual rearrangement of 13q may have also resulted from
chromothripsis. The rearrangements on chromosomes 6q and 13q were
are likely created initially during premeiotic mitosis in a
testicular germ cell, and thereafter modified by physiological
homologous recombination during meiosis I, whereas simple
rearrangements on 6q and 14q are likely the result of NHEJ or
replication-based errors. It is expected that these five
chromosomal aberrations are not independent, but reflect a specific
mutagenic event. The case in (18)
had multiple de novo CNVs. Breakpoints of the rearrangements
were indicative of replication-based errors, NHEJ and
chromothripsis (18).
It thus seems possible that multiple de novo
CNVs and constitutional chromothripsis may occur in the human
genome probably as a result of the same mutagenic event. Table I summarizes the published studies on
CNVs and constitutional chromothripsis and outlines the primary
features and disease cases already described in the literature.
These studies are important for understanding the development of
the human embryo and thus in health and human disease.
 | Table IStudies on copy number variations and
constitutional chromothripsis. |
Table I
Studies on copy number variations and
constitutional chromothripsis.
| Author, year | Phenotype | Copy number
variationsa | Chromothripsis | (Refs.) |
|---|
| Bertelsen et
al, 2016 | Normal |
hg19xg.[chr3:[pter_135827611::137890282_138510036
inv::138510037_142218722::135827614_137735948]::chr5:
118834146_qter]; g.[chr5:pter_118834138::chr3:
[137845987_137890201inv::142218723_qter]] An ~109-kb deletion on
3q22.3 was detected. | Chromothripsis
transmitted through three generations in 11 healthy carriers | (45) |
| Pettersson et
al, 2018 | Normal |
5p13.2(31820212-32131586)x1
5p13.2(36418846-36521666)x1 5p13.2(37072236-37092106)x1
5p13.2(37577701-37742275)x1 5q13.2(70150001-70220000)x1 | Chromothriptic
rearrangement | (48) |
| Pettersson et
al, 2018 | Developmental
delay |
5p13.2(31820212-32131586)x1
5p13.2(36521666-37072247)x3 5p13.2(37092106-37577669)x3 | Complex
rearrangement that evolved from a chromothriptic rearrangement in
the mother | (48) |
| Slamova et
al, 2018 | Developmental and
growth delay | Two de novo
deletions of 0.7 and 2.5 Mb at two of the breakpoints in 1q24.3 and
6q24.1-q24.2, respectively | Chromothriptic
rearrangement | (49) |
| Nazaryan-Petersen
et al, 2018 | Liver
malformation | arr[GRCh37]
5p15.1(16715952_16736553x1, 16758650_16771432x1)
NC_000005.9:g.[16715952_16736553del;16736554_16758649inv;16758650_16771432del] | Chromothripsis | (47) |
| Nazaryan-Petersen
et al, 2018 | Speech delay,
autism | arr[GRCh37]
7q11.22q11.23(70610154_72399292x 1,74050199_74834365x1) dn
NC_000007.14:g.[70609300_72422999del;72423000_74047984inv;74047986_74049000del] | Chromothripsis | (47) |
| Nazaryan-Petersen
et al, 2018 | Developmental
delay, speech delay, visual abnormality, craniosynostosis | arr[GRCh37] 11q14.3
(89843044_91294308)x1 mat
NC_000011.9:g.[89543002_89640782del;89640783_89766001inv;89766002_91339106del] | Chromothripsis | (47) |
| Nazaryan-Petersen
et al, 2018 | Speech delay, ADHD,
autism | arr[GRCh37]
17p13.3(2173896_2414920)x1 pat
NC_000017.10:g.[2220422_2484969del;2484970_2617882inv;2617882_2649613del] | Chromothripsis | (47) |
| Nazaryan-Petersen
et al, 2018 | Developmental
delay, speech delay | arr[GRCh37]
21q22.3(43427355_44858483x1,45803409_48095807x1) dn
NC_000021.8:g.[43414907_44797114del; 44797115_44797221inv;
44797222_45781000del; 45781001_45781001inv;45781002_
48101999del] | Chromothripsis | (47) |
| Nazaryan-Petersen
et al, 2018 | Developmental
delay, speech delay, growth retardation | arr[GRCh37]
4q31.3q34.1(155165258_158705411x1,
161300937_166372343x1,171349346_174403566x1) dn
NC_000004.11:g.[154997276_155050346del;155164913_158707725del;158707726_171342995
inv;161297891_166374443del;171342996_174401004del] | Chromothripsis | (47) |
| Nazaryan-Petersen
et al, 2018 | Infantile spasms,
hypotonia | arr[GRCh37]
7q11.23q21.11(75063222_77310662x1,
77629679_77770664x1,78236090_79911425x1, 82687283_82746799x1)dn
NC_000007.14:g.[74942506_77216338delins[77754229_77756619inv;77770732_78236952inv;78265840_82690202inv];77226982_77226980del;77226981_77626463
inv;77626464_77626462del;77626463_78265840inv;
78265841_82754313del] | Chromothripsis | (47) |
| Hattori et
al, 2019 | Transient neonatal
diabetes mellitus and multiple congenital malformations | 46,XY,der (6) add (6)(q23.3),der (13) add (13)(q12.1),der (14) add (14)(q31),der (21) del(q11.2) add(q11.2) Two tandem
inversions with a deletion on 2q Catastrophic rearrangements on
21q | Chromothripsis or
chromoanasynthesis. Chromothripsis was particularly likely. | (18) |
5. Conclusion
Several studies have highlighted the importance of
CNVs in human health and pathology. Likewise constitutional
chromothripsis described shortly after cancer chromothripsis has
become increasingly important in the study of birth defects and has
also been observed in healthy subjects. Constitutional
chromothripsis may coexist with multiple de novo CNVs.
Future studies are required to further clarify the relationship
between CNVs and constitutional chromothripsis. This work is of
great interest not only to researchers but also to clinicians who
should consider how these phenomena are involved from a clinical
perspective.
For a more complete overview of chromothripsis, the
book Chromothripsis. Methods and Protocols is recommended (51).
Acknowledgements
Not applicable.
Funding
Funding
The present review was supported by Fundação da
Ciência e Tecnologia (FCT, Portugal; grant nos. UID/BIM/00009/2013
and UID/BIM/00009/2016).
Availability of data and materials
Not applicable.
Authors' contributions
AB and ASR wrote and revised the manuscript. JR
proposed the subject and reviewed the manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Snijders AM, Nowak N, Segraves R,
Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B,
Kimura K, et al: Assembly of microarrays for genome-wide
measurement of DNA copy number. Nat Genet. 29:263–264.
2001.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Database of Genomic Variants: A curated
catalogue of human genomic structural variation. dgv.tcag.ca/. Accessed September 24, 2019.
|
|
3
|
Chen L, Zhou W, Zhang L and Zhang F:
Genome architecture and its roles in human copy number variation.
Genomics Inform. 12:136–144. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zarrei M, MacDonald JR, Merico D and
Scherer SW: A copy number variation map of the human genome. Nat
Rev Genet. 16:172–183. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Conrad DF, Pinto D, Redon R, Feuk L,
Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et
al: Origins and functional impact of copy number variation in the
human genome. Nature. 464:704–712. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Harel T and Lupski JR: Genomic disorders
20 years on-mechanisms for clinical manifestations. Clin Genet.
93:439–449. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Marcozzi A, Pellestor F and Kloosterman
WP: The genomic characteristics and origin of chromothripsis.
Methods Mol Biol. 1769:3–19. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang CZ, Spektor A, Cornils H, Francis
JM, Jackson EK, Liu S, Meyerson M and Pellman D: Chromothripsis
from DNA damage in micronuclei. Nature. 522:179–184.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nik-Zainal S, Alexandrov LB, Wedge DC, Van
Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J,
Stebbings LA, et al: Mutational processes molding the genomes of 21
breast cancers. Cell. 149:979–993. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Luijten MNH, Lee JXT and Crasta KC:
Mutational game changer: Chromothripsis and its emerging relevance
to cancer. Mutat Res. 777:29–51. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bickhart DM (ed): Copy Number Variants.
Methods and Protocols. Humana Press, New York, NY, pp10-206,
2018.
|
|
12
|
Dhokarh D and Abyzov A: Elevated variant
density around SV breakpoints in germline lineage lends support to
error-prone replication hypothesis. Genome Res. 26:874–881.
2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Emanuel BS and Shaikh TH: Segmental
duplications: An ‘expanding’ role in genomic instability and
disease. Nat Rev Genet. 2:791–800. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Carvalho CM and Lupski JR: Mechanisms
underlying structural variant formation in genomic disorders. Nat
Rev Genet. 17:224–238. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hastings PJ, Lupski JR, Rosenberg SM and
Ira G: Mechanisms of change in gene copy number. Nat Rev Genet.
10:551–564. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Gu S, Yuan B, Campbell IM, Beck CR,
Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, et
al: Alu-mediated diverse and complex pathogenic copy-number
variants within human chromosome 17 at p13.3. Hum Mol Genet.
24:4061–4077. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Koren A, Polak P, Nemesh J, Michaelson JJ,
Sebat J, Sunyaev SR and McCarroll SA: Differential relationship of
DNA replication timing to different forms of human mutation and
variation. Am J Hum Genet. 91:1033–1040. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hattori A, Okamura K, Terada Y, Tanaka R,
Katoh-Fukui Y, Matsubara Y, Matsubara K, Kagami M, Horikawa R and
Fukami M: Transient multifocal genomic crisis creating
chromothriptic and non-chromothriptic rearrangements in prezygotic
testicular germ cells. BMC Med Genomics. 12(77)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu P, Yuan B, Carvalho CMB, Wuster A,
Walter K, Zhang L, Gambin T, Chong Z, Campbell IM, Coban Akdemir Z,
et al: An organismal CNV mutator phenotype restricted to early
human development. Cell. 168:830–842.e7. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kearney HM, Thorland EC, Brown KK,
Quintero-Rivera F and South ST: Working Group of the American
College of Medical Genetics Laboratory Quality Assurance Committee.
American college of medical genetics standards and guidelines for
interpretation and reporting of postnatal constitutional copy
number variants. Genet Med. 13:680–685. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zepeda-Mendoza CJ and Morton CC: The
iceberg under water: Unexplored complexity of chromoanagenesis in
congenital disorders. Am J Hum Genet. 104:565–577. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Stephens PJ, Greenman CD, Fu B, Yang F,
Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA,
et al: Massive genomic rearrangement acquired in a single
catastrophic event during cancer development. Cell. 144:27–40.
2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chromothripsis DB: A curated database of
chromothripsis. http://cgma.scu.edu.cn/ChromothripsisDB. Accessed
September 2, 2019.
|
|
24
|
Cai H: ChromothripsisDB: A curated
database for the documentation, visualization, and mining of
chromothripsis data. Methods Mol Biol. 1769:279–289.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Morishita M, Muramatsu T, Suto Y, Hirai M,
Konishi T, Hayashi S, Shigemizu D, Tsunoda T, Moriyama K and
Inazawa J: Chromothripsis-like chromosomal rearrangements induced
by ionizing radiation using proton microbeam irradiation system.
Oncotarget. 7:10182–10192. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mladenov E, Saha J and Iliakis G:
Processing-challenges generated by clusters of dna double-strand
breaks underpin increased effectiveness of High-LET radiation and
chromothripsis. Adv Exp Med Biol. 1044:149–168. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Crasta K, Ganem NJ, Dagher R, Lantermann
AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D and
Pellman D: DNA breaks and chromosome pulverization from errors in
mitosis. Nature. 482:53–58. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Leibowitz ML, Zhang CZ and Pellman D:
Chromothripsis: A new mechanism for rapid karyotype evolution. Annu
Rev Genet. 49:183–211. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Maciejowski J, Li Y, Bosco N, Campbell PJ
and de Lange T: Chromothripsis and kataegis induced by telomere
crisis. Cell. 163:1641–1654. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tubio JM and Estivill X: Cancer: When
catastrophe strikes a cell. Nature. 470:476–477. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
31
|
Mardin BR, Drainas AP, Waszak SM,
Weischenfeldt J, Isokane M, Stütz AM, Raeder B, Efthymiopoulos T,
Buccitelli C, Segura-Wang M, et al: A cell-based model system links
chromothripsis with hyperploidy. Mol Syst Biol.
11(828)2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ivkov R and Bunz F: Pathways to
chromothripsis. Cell Cycle. 14:2886–2890. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kloosterman WP, Guryev V, van Roosmalen M,
Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B,
Hochstenbach R, Poot M and Cuppen E: Chromothripsis as a mechanism
driving complex de novo structural rearrangements in the germline.
Hum Mol Genet. 20:1916–1924. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kloosterman WP, Tavakoli-Yaraki M, van
Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L,
Vergult S, Giardino D, Hansson K, et al: Constitutional
chromothripsis rearrangements involve clustered double-stranded DNA
breaks and nonhomologous repair mechanisms. Cell Rep. 1:648–655.
2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Nazaryan-Petersen L, Bertelsen B, Bak M,
Jønson L, Tommerup N, Hancks DC and Tümer Z: Germline
chromothripsis driven by L1-mediated retrotransposition and Alu/Alu
homologous recombination. Hum Mutat. 37:385–395. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Masset H, Hestand MS, Van Esch H,
Kleinfinger P, Plaisancié J, Afenjar A, Molignier R, Schluth-Bolard
C, Sanlaville D and Vermeesch JR: A distinct class of
chromoanagenesis events characterized by focal copy number gains.
Hum Mutat. 37:661–668. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fukami M, Shima H, Suzuki E, Ogata T,
Matsubara K and Kamimaki T: Catastrophic cellular events leading to
complex chromosomal rearrangements in the germline. Clin Genet.
91:653–660. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kloosterman WP and Cuppen E:
Chromothripsis in congenital disorders and cancer: Similarities and
differences. Curr Opin Cell Biol. 25:341–348. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pellestor F and Gatinois V: Potential role
of chromothripsis in the genesis of complex chromosomal
rearrangements in human gametes and preimplantation embryo. Methods
Mol Biol. 1769:35–41. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Poot M and Haaf T: Mechanisms of origin,
phenotypic effects and diagnostic implications of complex
chromosome rearrangements. Mol Syndromol. 6:110–134.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Macera MJ, Sobrino A, Levy B, Jobanputra
V, Aggarwal V, Mills A, Esteves C, Hanscom C, Pereira S,
Pillalamarri V, et al: Prenatal diagnosis of chromothripsis, with
nine breaks characterized by karyotyping, FISH, microarray and
whole-genome sequencing. Prenat Diagn. 35:299–301. 2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gamba BF, Richieri-Costa A, Costa S,
Rosenberg C and Ribeiro-Bicudo LA: Chromothripsis with at least 12
breaks at 1p36.33-p35.3 in a boy with multiple congenital
anomalies. Mol Genet Genomics. 290:2213–2216. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
van Heesch S, Simonis M, van Roosmalen MJ,
Pillalamarri V, Brand H, Kuijk EW, de Luca KL, Lansu N, Braat AK,
Menelaou A, et al: Genomic and functional overlap between somatic
and germline chromosomal rearrangements. Cell Rep. 9:2001–2010.
2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
de Pagter MS, van Roosmalen MJ, Baas AF,
Renkens I, Duran KJ, van Binsbergen E, Tavakoli-Yaraki M,
Hochstenbach R, van der Veken LT, Cuppen E and Kloosterman WP:
Chromothripsis in healthy individuals affects multiple
protein-coding genes and can result in severe congenital
abnormalities in offspring. Am J Hum Genet. 96:651–656.
2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Bertelsen B, Nazaryan-Petersen L, Sun W,
Mehrjouy MM, Xie G, Chen W, Hjermind LE, Taschner PE and Tümer Z: A
germline chromothripsis event stably segregating in 11 individuals
through three generations. Genet Med. 18:494–500. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chiang C, Jacobsen JC, Ernst C, Hanscom C,
Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger
SR, et al: Complex reorganization and predominant non-homologous
repair following chromosomal breakage in karyotypically balanced
germline rearrangements and transgenic integration. Nat Genet.
44:390–397, S1. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Nazaryan-Petersen L, Eisfeldt J,
Pettersson M, Lundin J, Nilsson D, Wincent J, Lieden A, Lovmar L,
Ottosson J, Gacic J, et al: Replicative and non-replicative
mechanisms in the formation of clustered CNVs are indicated by
whole genome characterization. PLoS Genet.
14(e1007780)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Pettersson M, Eisfeldt J, Syk Lundberg E,
Lundin J and Lindstrand A: Flanking complex copy number variants in
the same family formed through unequal crossing-over during
meiosis. Mutat Res. 812:1–4. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Slamova Z, Nazaryan-Petersen L, Mehrjouy
MM, Drabova J, Hancarova M, Marikova T, Novotna D, Vlckova M,
Vlckova Z, Bak M, et al: Very short DNA segments can be detected
and handled by the repair machinery during germline chromothriptic
chromosome reassembly. Hum Mutat. 39:709–716. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Liu P, Erez A, Nagamani SC, Dhar SU,
Kołodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang
F, Withers MA, et al: Chromosome catastrophes involve replication
mechanisms generating complex genomic rearrangements. Cell.
146:889–903. 2011.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Pellestor F (ed): Chromothripsis. Methods
and Protocols. Humana Press, Montpellier, Hérault, pp11-367,
2018.
|