Introduction
Acute myeloid leukemia (AML) is a major subtype of
leukemia resulting from the uncontrolled proliferation and
incomplete differentiation of the myeloid progenitor in the bone
marrow with possible spread of myeloblasts to the blood, liver or
spleen (1). According to the World
Health Organization (WHO), AML is diagnosed following ≥20%
myeloblasts in the blood or bone marrow smears, the characteristics
of which is of crucial importance in the classification of the
disease (2,3). The European Leukemia Net (ELN) has
updated their classification guidelines to match those of the WHO
to assist in consistent diagnosis, management and monitoring
(4). Of note, the genomic landscape
of AML is complicated resulting from different classes of genetic
mutations (5). In addition, it has
been reported that acquisition of additional mutations as a
consequence of genomic instability may contribute to the incidence
of AML, reviewed in (5). In ~50% of
AML patients, a normal karyotype is observed; however, advanced
sequencing techniques have assisted in identifying novel genetic
alterations that contribute to the progression of AML (6-8). In
this respect, RNA sequencing technology has been used to examine
the transcription rates of both wildtype and mutated alleles to
assess allelic imbalances in patients with AML (9). It has been reported that 99% of mutated
DNA is transcribed into RNA in animal model tumor cells (10). This suggests that RNA transcripts
reflect the genotype of tumor cells and may assist in understanding
the genetic landscape of various types of cancer, including AML,
and as such, is becoming an important factor with regards to
personalized medicine and clinical decision making.
Case report
The present study was reviewed and approved by the
Institutional Review Board of Imam Abdulrahman Bin Faisal
University (approval no. IRB #2017-03-147) and patient consent was
obtained prior to participation.
A 37 year old female presented with general
weakness, bone pain and Petechiae, suggestive of the
thrombocytopenia (Table I) for 2
weeks. The patient suffered from photosensitivity, arthralgia and
solar urticaria for the last 6 years, and was followed up by the
Dermatology Clinic. She also had a nephew who was diagnosed with
acute myeloid leukemia at the age of 6. Clinical examination showed
a well-developed, well-nourished and alert female. There was no
evidence of lymphadenopathy or organomegaly. She had scattered
Petechiae, but no purpura or ecchymosis. Laboratory analysis of
blood smears and complete blood counts revealed significant
relative leukocytosis and >20% blast counts. Of note, according
to the WHO definition, the presence of blast counts >20% is
confirmatory of an AML diagnosis (11). According to the definition of the
French-American-British classification system (2), blast counts >30% are confirmatory of
a diagnosis of AML (3). Blood smears
were assessed on admission along with complete blood counts, and
the blast count was 40%, which was confirmed in subsequent blood
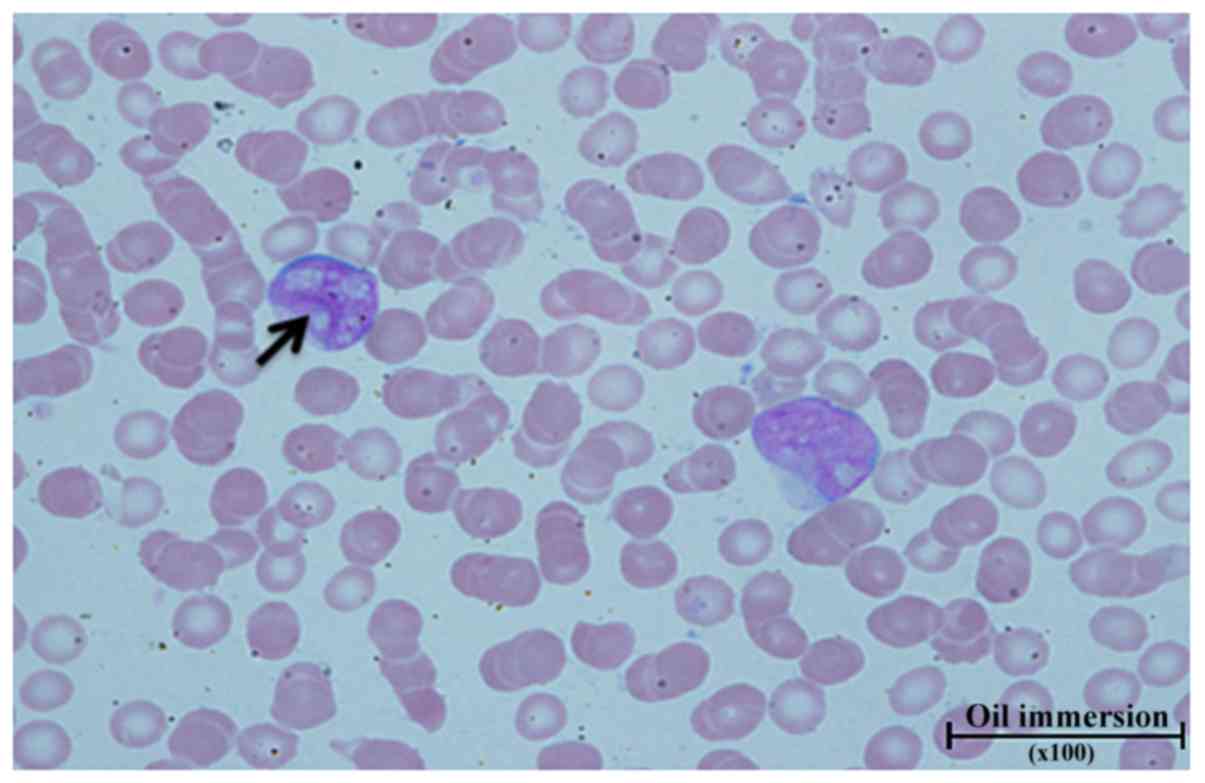
samples, with blasts counts of 30 and 41%. Blast cells were large
with a high nuclear to cytoplasmic ratio, fine opened chromatin and
irregular nuclear outlines, and some cells showed inclusion of
Auer-rods. Additionally, several cells showed fine chromatin with
2-3 nucleoli. Some blasts showed convoluted cribriform nuclei
(Fig. 1). Morphological blood sample
analysis and blood counts (Table I)
suggested a diagnosis of AML. The hematopathologist recommended
bone marrow analysis, for karyo-and immunotyping.
 | Table IComplete and differential blood
counts. |
Table I
Complete and differential blood
counts.
| Parameter (reference
range) | 1st check | 2nd check | 3rd
check |
|---|
| Red blood cells
(4.2-5.5), Mil/ul | 4.51 | 3.9 | 4.14 |
| Hemoglobin (12-16),
g/dl | 11.5 | 10 | 10.6 |
| Hematocrit (37-47),
% | 35 | 29.9 | 31.5 |
| Red blood cell
distribution width (11.5-14.5) | 17.7 | 18.1 | 17.6 |
| Mean corpuscular
volume (80-94), fl | 77.6 | 76.7 | 76.1 |
| Mean corpuscular
hemoglobin (27-32), pg | 25.6 | 25.6 | 25.5 |
| Mean corpuscular
hemoglobin concentration (32-36), g/dl | 33 | 33.4 | 33.5 |
| White blood cells
(4-11), k/ul | 8.6 | 7.9 | 9.1 |
| Corrected white
blood cell (4-11), k/ul | 8.4 | - | - |
| Platelet (140-450),
k/ul | 62 | 45 | 47 |
| Mean platelet
volume (7.2-11.1), fl | 8.4 | 8.8 | 8.1 |
| Segmented (38-79),
% | 15 | 23 | 22 |
| Band lymphocytes
(0-3), % | 8 | 2 | 2 |
| Lymphocytes
(12-15), % | 23 | 23 | 22 |
| Monocytes (0-10),
% | 2 | - | 4 |
| Atypical
lymphocytes (0), % | 9 | - | - |
| Metamyelocytes (0),
% | 1 | 3 | - |
| Myelocytes (0),
% | 1 | 3 | 2 |
| Promyelocytes (0),
% | 1 | - | 1 |
| Eosinophils (0-8),
% | - | 2 | - |
| Basophiles (0-1),
% | - | 5 | 6 |
| Blasts (0), % | 40 | 30 | 41 |
| Nucleated red blood
cells (0) | 2 | - | - |
The patient was then transferred to the King Fahd
Specialist Hospital, wherein molecular analysis, bone marrow
analysis, karyotyping and immunophenotyping were performed. The
cytogenetics report (Table II) of
bone marrow aspirates indicated the presence of an abnormal tumoral
clone with trisomy 8 (47, XX, +8[18]/46, XX[1]); this karyotype was
confirmed by fluorescence in situ hybridization, which
showed 88% trisomy 8 (Data not shown); a karyotype abnormality that
is associated with a moderate risk of AML according to ELN
(12).
 | Table IICytogenetics report. |
Table II
Cytogenetics report.
| Test name | Results |
|---|
| Cytogenetics
(CBFB)-FISH | 100% normal
CBFB | Normal |
| Cytogenetics
MLL-FISH | 100% normal
MLL | Normal |
| Chromosomal
analysis neoplastic study | 47, XX, +8[18]/46,
XX[1] | Abnormal |
| Cytogenetics
PML/RARA-FISH | 100 normal
PML/RARA | Normal |
| Cytogenetics
RUNX1/RUNX1T1-FISH | 88% trisomy
RUNX1T1 | Abnormal |
| Cytogenetics
BCR/ABL1-FISH | 100 BCR/ABL1
normal | Normal |
Post treatment bone marrow aspirates and biopsy from
the right posterior iliac crest were assessed. The biopsy was a
single core (0.9 cm in length) in which the cellular area showed
<10% cellularity with stromal edema and serous degeneration that
was deemed to be therapy-related. A mixture of lymphocytes,
macrophages, plasma cells and a few megakaryocytes were observed in
the cellular area. There was no increase in blast cells as
indicated by the negative immunostaining of CD117. Reticulin
staining showed scattered linear reticulin indicating bone marrow
fibrosis grading of MF-0, which is indicative of a normal
architecture (13). The bone marrow
aspirates showed marked suppression of trilineage hematopoiesis
with a relative increase in lymphocytes and macrophages.
Differential counts showed 7% erythroid precursors, 2% blast cells,
2% segments, 86% lymphocytes and 2% plasma cells. The complete
blood count, which was performed on the same day showed marked
leucopenia, neutropenia and thrombocytopenia. The differential
blood counts indicated that lymphocytes accounted for 91.8% of the
count, monocytes 4.9% and basophils 3.3%. A case of AML showing
hypocellular marrow with 2% blasts was recommended for bone marrow
re-evaluation. The patient then underwent Allogenic stem cell bone
marrow transplantation, as recommended as a consolidation treatment
for patients with trisomy 8 AML (12). Bone marrow aspirates and biopsy were
assessed again 5 months later; the marrow aspirates were
aparticulated and diluted, almost mimicking the peripheral blood,
with no blast cells observed upon scanning. The biopsy was 1.4 cm
in length, in which the cellular area examined showed
intertrabecular hemorrhage with crushing and tissue loss. A
cellularity of 10% (marked hypocellular marrow) was also reported
with trilineage hematopoiesis and no blast cells were observed.
Flow cytometry analysis showed 0.1% positivity for the gated
population of CD45dim/CD34. The patient passed away in April
2019.
In the present case report, the transcriptomic
landscape was assessed in this patient at the time of AML diagnosis
by performing whole genome RNA sequencing (14,15).
This was followed by gene expression quantification using the RSEM
software package (16).
Bioinformatics analysis and the software used are described in the
supplementary materials and methods. The bioinformatics data showed
a marked number of differentially expressed genes when compared
with a matched female control profile; >2,500 differentially
upregulated genes (log2 fold change ≥1, probability of
≥0.8 (where the P-value was used to calculate the probability of
gene expression) and 252 downregulated genes (log2 fold
change ≤-1, probability ≥0.8) (Fig.
2). The volcano plots (NOISeq method) (17) in Fig.
2B also shows the number of non-differentially expressed genes
(14,087; log2 fold change <1 and >-1, probability
of <0.8). Genes were sorted according to the log2
fold change and the probability of being upregulated. Genes with a
log2 fold change <5 and a probability <80% were
excluded from subsequent analysis. In this respect, log2
(fold change) was calculated as follows: Log[(gene expression in
AML/gene expression in control)/2], where gene expression in
AML/gene expression in control=the actual fold change. The
following formula was used to convert log2 (fold change
values) to the actual fold change: Actual fold change=2log2
(fold change). Gene ontology analysis was used during the
filtration process, which resulted in identification of possible
crucial hits that may be of potential importance in disease
progression and in clinical management of the patient. ADGRA3, Rh
associated glycoprotein (RhAG), succinate receptor 1 (SUCNR1) and
transmembrane-4 L-six family member-1 (TM4SF1) are all cell
membrane proteins that were found to be upregulated in this patient
compared with the corresponding control, with log2 fold
change and gene expression probability values of 5.25, 91.2%; 7.2,
93%; 9, 97%; and 8.6, 97%, respectively. Table III shows the log2 (fold
change), the converted fold change values and the gene expression
probabilities. The results of RNA sequencing were further validated
by quantitative (q)PCR. The sequences of the primers are presented
in Table SI. The results confirmed
the significant upregulation of RHAG, SUCNR1 and TM4SF1, with
2-DDCq values of 50.6, 266.5 and 392,476.7respectively
(Fig. S1). These values show the
number of times each gene is expressed more in the AML case
compared with the control sample.
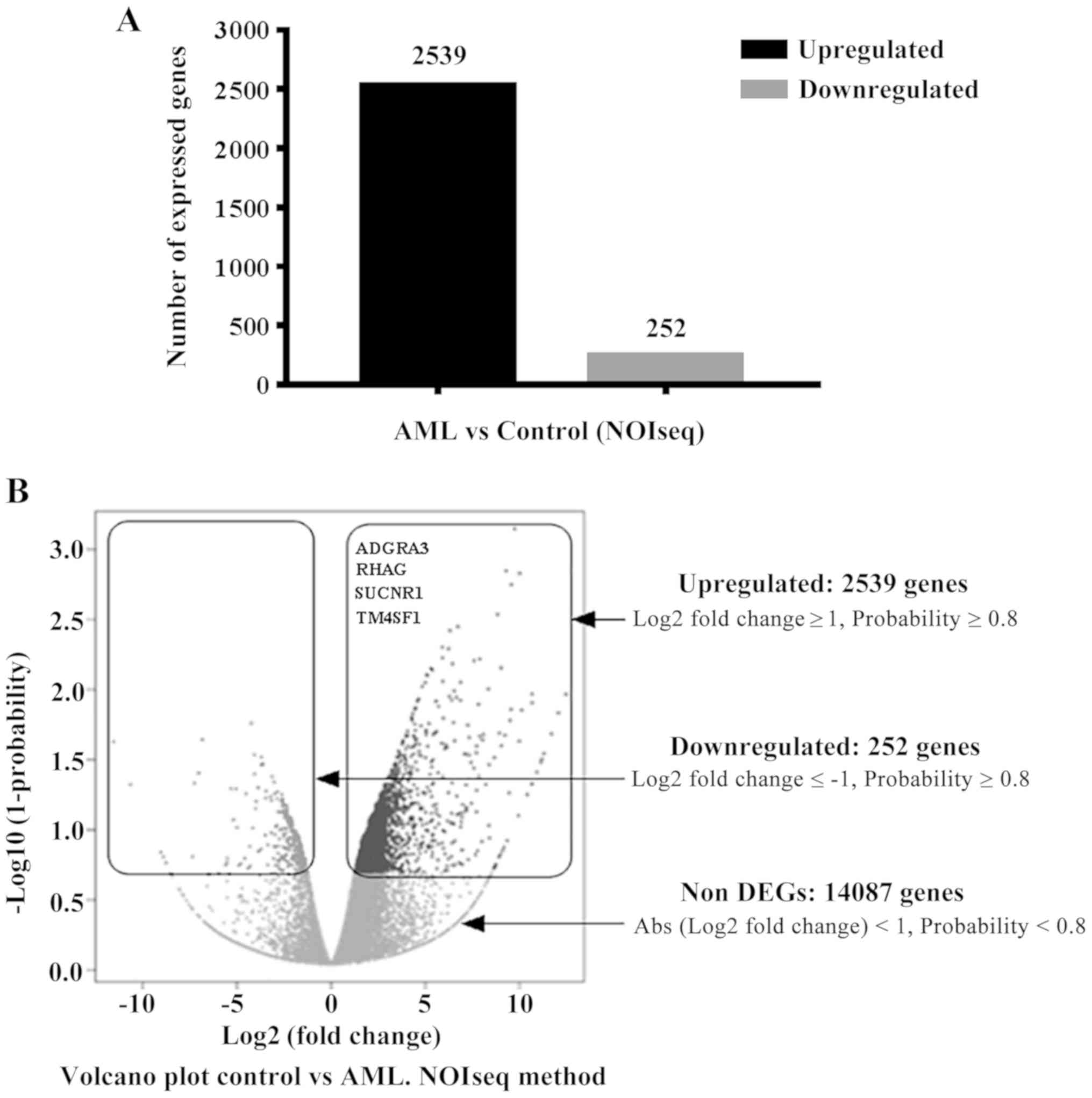 | Figure 2Differentially expressed genes in the
patient with AML compared with a healthy matched-control. (A) A
total of 2,539 upregulated and 252 downregulated genes were
identified in the patient with AML. (B) Volcano plot showing the up
and downregulated genes, and the criteria used to define whether a
gene was significantly dysregulated. Amongst the overexpressed
genes, ADGRA3, RHAG, SUCNR1 and TM4SF1 were further considered as
potential targets in AML. The log2 fold change and the
probability of gene expression for each gene compared with the
corresponding control were: ADGRA3 (5.25, 91.2%), RHAG (7.2, 93%),
SUCNR1 (9, 97%) and TM4SF1 (8.6, 97%), respectively. |
 | Table IIIGenes with potential significance in
acute myeloid leukemia. |
Table III
Genes with potential significance in
acute myeloid leukemia.
| Gene name | Log2
(fold change) | Actual fold
change | Probability of
expression, % |
|---|
| ADGRA3 | 5.25 | 38.05 | 91.2 |
| RhAG | 7.2 | 147.03 | 93 |
| SUCNR1 | 9 | 512 | 97 |
| TM4SF1 | 8.6 | 388.02 | 97 |
To assess the genetic variations in this patient
compared with the reference genome, single nucleotide polymorphism
(SNP) and mutation analyses were performed. Briefly, data analyses
revealed that the mapping ratio with the reference gene sequences
was 75%, a total of 15,763 genes were identified in this patient.
Of those, 1,251 novel coding transcripts were reported for this
patient. In total, 10,444 novel transcripts were generated,
including the 1,251 novel gene transcripts that require further
characterization, 1,160 novel non-coding transcripts and 8,033
novel splicing variants of known genes. Genome analysis toolkit
(18) was used for
insertion/deletion and SNP analyses. Transition events that
involved an interchange of purine nucleotides [adenine (A) and
guanine (G)] or pyrimidine nucleotides [cytosine (C) and thymine
(T)] accounted for 113,011 events in total. There were 43,161
transversion events involving the transition of purine nucleotides
to pyrimidine nucleotides or vice versa. Transversion events
observed included A-C, A-T, C-G and G-T. The transition and
transversion events are summarized in Table SII). The results revealed intronic
mutations in fms-like tyrosine kinase 3 (FLT3), mutant variants of
which are present in ~30% of AML cases according to a recent report
(19). Another important mutation
observed in the present study was in the NRAS gene, which is a
common genetic signature observed in AML patients (20). In the present study, two exonic and
one intronic mutation of NRAS were observed. In addition, intronic
mutations were also identified in nucleophosmin 1 gene (NPM1).
According to Heath et al (21), NPM1 was frequently mutated in ~one
third of AML patients and its mutated forms were associated with
improved prognosis (21). In this
regard, it has been reported that mutations in AML belong to two
main classes: i) class I, which consists of mutations in genes that
can stimulate proliferation and cell survival (such as mutations in
Ras, JAK2 and FLT3) and ii) class II, where mutations affect cell
differentiation and the apoptotic machinery (such as mutations in
MLL, CEBPA and NPM1). A third class of mutations was also reported,
which included mutations that affect epigenetic modifications (such
as mutations in ASXL1 and DNMT 3a) (5).
Discussion
AML is characterized by a heterogeneous genomic
landscape resulting from numerous genetic alterations, making
disease stratification and management complicated (5). RNA transcripts may assist in
understanding the transcription rates of wild type and mutated
genes that may serve a role in sustaining the disease process and
outcomes (9,10). By mining the RNA-sequencing
bioinformatics data, 4 genes (ADGRA3, RHAG, SUCNR1 and TM4SF1) that
were highly expressed in the patient with AML were identified. The
4 genes encoded cell surface proteins that are important in
conveying extracellular signaling cascades. Thus, it was proposed
that these genes may serve as potentially novel biomarkers of AML,
pending further validation in additional patients, and in
vitro and in vivo experimental models of AML. ADGRA3,
also known as GPR125 is an adhesion G-protein-coupled membrane
receptor, which mediates downstream signaling pathways by
activation of G proteins (22). With
respect to the role of ADGRA3 in AML, it has been reported that its
protein product, GPR125, is downregulated in AML, in contrast to
the results of the present study, suggesting that the exact role of
this gene in AML requires further study (23). It has been recently reported that
upregulation of GPR125 has prognostic value in colorectal cancer
(24). In addition, the genomic data
commons (GDC) data portal (25)
contains 302 reports of different ADGRA3 mutations and 392 cases of
copy number variations have been reported in 27 and 28 projects,
respectively, indicating the role of ADGRA3 gene in neoplastic
disorders of lung, bronchus, uterus, cervix, bladder, mature B cell
lymphoma and others (26). SUCNR1
was significantly upregulated in the patient with AML in the
present study. To the best of our knowledge, the role of SUCNR1 in
AML has not yet been explored. SUCNR1 is a sensor of oxidative
stress, which may assist in the cells ability to sense and manage
excessive oxidative stress (27). In
this regard, succinate is considered an oncometabolite, which
promotes tumorigenesis by promoting angiogenesis in experimental
models as well as a transgenic zebrafish cancer model (28). Furthermore, single nucleotide
variants of SUCNR1 gene have been reported; however, the
involvement of this gene was excluded from being implicated in the
familial inheritance of gastric and rectal cancer (29). Importantly, a role of SUCNR1 in
hematopoietic progenitor cell development was reported (30). The activated SUCNR1 receptor
potentiates proliferation of hematopoietic precursors as well as
protecting erythroleukemic cells from starvation-caused cell death
(31). It has been reported that the
proliferative effect of SUCNR1 in hematopoietic progenitor cells is
mediated via the ERK1/2 pathway (32). In addition, 106 somatic mutations and
copy number variants, gain and loss, have also been reported in
multiple projects reporting these types of genotypic variants in
SUCNR1 in breast, lung, bladder, uterus, ovary, liver and other
types of cancer (25). Additionally,
SCUNR1 overexpression was reported in ankle tissue samples in an
adjuvant arthritis model (33),
where arthralgia (joint pain) is a common symptom; this may reflect
the universal metabolic role of SCUNR1 in multiple types of cells,
including blood cells in the present study. In this regard,
accumulation of succinate in multiple types of cancer has been
reported, including in ovarian and thyroid cancer (34), renal and gastric carcinoma (35), and familial pheochromocytoma
(36). Accumulation of succinate in
cancer cells promotes oncogenesis by promoting angiogenesis via
activation of STAT3 and ERK signaling through SUCNR1 (28,37).
However, the possibility that the upregulation of SCUNR1 in the
patient presented in this case report was due to arthralgia can be
excluded, as the transcriptomic profile was based on RNA
transcripts extracted from blood cells, not synovial fluid samples
nor the patient's chondrocytes. RhAG is an ammonia transporter,
which has previously been associated with cancer based on its
downregulated expression in esophageal cancer cell lines compared
with normal esophageal cells (38).
In contrast to the previous study, in the present study, RHAG
expression was upregulated in the patient with AML, and thus may
possess prognostic value as a biomarker of AML. According to the
GDC data portal, 160 somatic mutations in the RHAG gene have been
reported in patients from 21 different projects, suggesting a role
in neoplasms of the bronchus and lung, adenocarcinomas of the
colon, melanomas and other types of neoplastic diseases. Finally,
the present study identified TM4SF1 as a potential crucial hit in
AML. TM4SF1 is a transmembrane protein with oncogenic properties in
lung cancer, where it has been shown to participate in cell
migration and tumor metastasis (39). In addition, TM4SF1 was also reported
to promote metastasis in pancreatic cancer (40). To the best of our knowledge the role
of TM4SF1 has not yet been reported in AML.
The cell surface protein products of the identified
genes found to be dysregulated may sustain the external cell
signaling mechanisms in AML, and thus should be further studied to
determine their diagnostic and prognostic value in patients with
AML. However, the present study has some limitations. First, the
expression of these dysregulated genes was not validated at the
protein level. Thus, the roles of these four genes will be further
verified as a continuation of the present study using in
vitro and in vivo models of AML. Additionally, the
differences in transcriptomic profiles were concluded from
comparisons with a single healthy female control. A larger sample
size of both patients and controls are required to establish more
generalizable and convincing results.
In conclusion, RNA-sequencing bioinformatics data
revealed the overexpression of a large number of differentially
transcribed genes in this patient. Data mining led to
identification of ADGRA3, RHAG, SUCNR1 and TM4SF1 as potential
candidate genes for further investigation to validate their exact
roles in AML.
Supplementary Material
qPCR analysis of RHAG, SUCNR1 and
TM4SF1 expression. Quantitative validation of the RNA sequencing
results by qPCR confirmed the apparent gene expression changes
observed for RHAG, SUCNR1 and TM4SF1 in the AML case when compared
with the corresponding control. β-actin was used as the
housekeeping control. Samples were run in triplicate sets and
repeated independently twice. qPCR, quantitative PCR; RhAG, Rh
associated glycoprotein; SUCNR1, succinate receptor 1; TM4SF1,
transmembrane-4 L-six family member-1; AML, acute myeloid
leukemia.
Sequences of the primers.
Summary of coding and non-coding
transcripts in the patient with acute myeloid leukemia.
Transition and transversion events in
the patient with acute myeloid leukemia.
Acknowledgements
Not applicable.
Funding
This work was funded by the Deanship of Scientific
Research at Imam Abdulrahman Bin Faisal University (grant no.
2017-099-CAMS).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
OSEM conceived and designed the study, and wrote the
manuscript. AMAA collected the clinical data and wrote the
manuscript. AA collected the clinical data and revised the
manuscript. KA performed the bioinformatics analysis. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was reviewed and approved by the
Institutional Review Board at Imam Abdulrahman Bin Faisal
University (approval no. IRB # 2017-03-147).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hasserjian RP: Acute myeloid leukemia:
Advances in diagnosis and classification. Int J Lab Hematol.
35:358–366. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposals for the
classification of the acute leukaemias. French-American-British
(FAB) co-operative group. Br J Haematol. 33:451–458.
1976.PubMed/NCBI View Article : Google Scholar
|
|
3
|
DiNardo CD, Garcia-Manero G, Pierce S,
Nazha A, Bueso-Ramos C, Jabbour E, Ravandi F, Cortes J and
Kantarjian H: Interactions and relevance of blast percentage and
treatment strategy among younger and older patients with acute
myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Am J
Hematol. 91:227–232. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Döhner H, Estey E, Grimwade D, Amadori S,
Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA,
et al: Diagnosis and management of AML in adults: 2017 ELN
recommendations from an international expert panel. Blood.
129:424–447. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lagunas-Rangel FA, Chávez-Valencia V,
Gómez-Guijosa MÁ and Cortes-Penagos C: Acute myeloid
leukemia-genetic alterations and their clinical prognosis. Int J
Hematol Oncol Stem Cell Res. 11:328–339. 2017.PubMed/NCBI
|
|
6
|
Martelli MP, Sportoletti P, Tiacci E,
Martelli MF and Falini B: Mutational landscape of AML with normal
cytogenetics: Biological and clinical implications. Blood Rev.
27:13–22. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rubnitz JE, Gibson B and Smith FO: Acute
myeloid leukemia. Hematol Oncol Clin North Am. 24:35–63.
2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Takahashi S: Current findings for
recurring mutations in acute myeloid leukemia. J Hematol Oncol.
4(36)2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Batcha AMN, Bamopoulos SA, Kerbs P, Kumar
A, Jurinovic V, Rothenberg-Thurley M, Ksienzyk B, Philippou-Massier
J, Krebs S, Blum H, et al: Allelic imbalance of recurrently mutated
genes in acute myeloid leukaemia. Sci Rep. 9(11796)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Castle JC, Loewer M, Boegel S, Tadmor AD,
Boisguerin V, de Graaf J, Paret C, Diken M, Kreiter S, Türeci O and
Sahin U: Mutated tumor alleles are expressed according to their DNA
frequency. Sci Rep. 4(4743)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vardiman JW, Harris NL and Brunning RD:
The world health organization (WHO) classification of the myeloid
neoplasms. Blood. 100:2292–2302. 2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hemsing AL, Hovland R, Tsykunova G and
Reikvam H: Trisomy 8 in acute myeloid leukemia. Expert Rev Hematol.
12:947–958. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Thiele J, Kvasnicka HM, Facchetti F,
Franco V, van der Walt J and Orazi A: European consensus on grading
bone marrow fibrosis and assessment of cellularity. Haematologica.
90:1128–1132. 2005.PubMed/NCBI
|
|
14
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12(323)2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tarazona S, Furió-Tarı P, Nueda MJ, Ferrer
A and Conesa A: NOISeq: Differential expression in RNA-seq 2013.
https://bicoductor.statistik.tu-dortmund.de/packages/3.7/bior/vignettes/NOISwq/inst/doc/NOISeq.pdf.
|
|
18
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A mapreduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Daver N, Schlenk RF, Russell NH and Levis
MJ: Targeting FLT3 mutations in AML: Review of current knowledge
and evidence. Leukemia. 33:299–312. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Berman JN, Gerbing RB, Alonzo TA, Ho PA,
Miller K, Hurwitz C, Heerema NA, Hirsch B, Raimondi SC, Lange B, et
al: Prevalence and clinical implications of NRAS mutations in
childhood AML: A report from the Children's oncology group.
Leukemia. 25:1039–1042. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Heath EM, Chan SM, Minden MD, Murphy T,
Shlush LI and Schimmer AD: Biological and clinical consequences of
NPM1 mutations in AML. Leukemia. 31:798–807. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Spiess K, Bagger SO, Torz LJ, Jensen KHR,
Walser AL, Kvam JM, Møgelmose ASK, Daugvilaite V, Junnila RK,
Hjortø GM and Rosenkilde MM: Arrestin-independent constitutive
endocytosis of GPR125/ADGRA3. Ann N Y Acad Sci. 1456:186–199.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Maiga A, Lemieux S, Pabst C, Lavallée VP,
Bouvier M, Sauvageau G and Hébert J: Transcriptome analysis of G
protein-coupled receptors in distinct genetic subgroups of acute
myeloid leukemia: Identification of potential disease-specific
targets. Blood Cancer J. 6(e431)2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu Y, Chen W, Gong L, Ke C, Wang H and Cai
Y: Elevated G-protein receptor 125 (GPR125) expression predicts
good outcomes in colorectal cancer and inhibits wnt/β-catenin
signaling pathway. Med Sci Monitor. 24:6608–6616. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Institute NC: Genomic data commons data
portal. https://portal.gdc.cancer.gov/genes/ENSG00000152990.
J 2020.
|
|
26
|
Institute NC: Genomic data commons data
portal. https://portal.gdc.cancer.gov/genes/ENSG00000198829.
J 2020.
|
|
27
|
Louer EMM, Lores-Motta L, Ion AM, Den
Hollander AI and Deen PMT: Single nucleotide polymorphism
rs13079080 is associated with differential regulation of the
succinate receptor 1 (SUCNR1) gene by miRNA-4470. RNA Biol.
16:1547–1554. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mu X, Zhao T, Xu C, Shi W, Geng B, Shen J,
Zhang C, Pan J, Yang J, Hu S, et al: Oncometabolite succinate
promotes angiogenesis by upregulating VEGF expression through
GPR91-mediated STAT3 and ERK activation. Oncotarget. 8:13174–13185.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Thutkawkorapin J, Picelli S, Kontham V,
Liu T, Nilsson D and Lindblom A: Exome sequencing in one family
with gastric- and rectal cancer. BMC Genet. 17(41)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Macaulay IC, Tijssen MR, Thijssen-Timmer
DC, Gusnanto A, Steward M, Burns P, Langford CF, Ellis PD,
Dudbridge F, Zwaginga JJ, et al: Comparative gene expression
profiling of in vitro differentiated megakaryocytes and
erythroblasts identifies novel activatory and inhibitory platelet
membrane proteins. Blood. 109:3260–3269. 2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ariza AC, Deen PMT and Robben JH: The
succinate receptor as a novel therapeutic target for oxidative and
metabolic stress-related conditions. Front Endocrinol (Lausanne).
3(22)2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hakak Y, Lehmann-Bruinsma K, Phillips S,
Le T, Liaw C, Connolly DT and Behan DP: The role of the GPR91
ligand succinate in hematopoiesis. J Leukoc Biol. 85:837–843.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chen H, Pan T, Liu P, Wang P and Xu S:
Baihu Jia Guizhi decoction improves rheumatoid arthritis
inflammation by regulating succinate/SUCNR1 metabolic signaling
pathway. Evid Based Complement Alternat Med.
2019(3258572)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bardella C, Pollard PJ and Tomlinson I:
SDH mutations in cancer. Biochim Biophys Acta. 1807:1432–1443.
2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ricketts C, Woodward ER, Killick P, Morris
MR, Astuti D, Latif F and Maher ER: Germline SDHB mutations and
familial renal cell carcinoma. J Natl Cancer Inst. 100:1260–1262.
2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Baysal BE, Ferrell RE, Willett-Brozick JE,
Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE,
Rubinstein WS, Myers EN, et al: Mutations in SDHD, a mitochondrial
complex II gene, in hereditary paraganglioma. Science. 287:848–851.
2000.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhao T, Mu X and You Q: Succinate: An
initiator in tumorigenesis and progression. Oncotarget.
8:53819–53828. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chen BS, Xu ZX, Xu X, Cai Y, Han YL, Wang
J, Xia SH, Hu H, Wei F, Wu M and Wang MR: RhCG is downregulated in
oesophageal squamous cell carcinomas, but expressed in multiple
squamous epithelia. Eur J Cancer. 38:1927–1936. 2002.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ye L, Pu C, Tang J, Wang Y, Wang C, Qiu Z,
Xiang T, Zhang Y and Peng W: Transmembrane-4 L-six family member-1
(TM4SF1) promotes non-small cell lung cancer proliferation,
invasion and chemo-resistance through regulating the
DDR1/Akt/ERK-mTOR axis. Respir Res. 20(106)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yang JC, Zhang Y, He SJ, Li MM, Cai XL,
Wang H, Xu LM and Cao J: TM4SF1 promotes metastasis of pancreatic
cancer via regulating the expression of DDR1. Sci Rep.
7(45895)2017.PubMed/NCBI View Article : Google Scholar
|