Introduction
Porphyrias are a group of metabolic disorders
affecting biosynthesis of heme; each specific subtype of Porphyria
is the result of a decrease in the activity of a specific enzyme
involved in the biosynthesis of heme (1,2). The
specific patterns of overproduction of heme precursors are
associated with characteristic clinical features; in particular,
porphyria cutanea tarda (PCT) is a hepatic cutaneous Porphyria
resulting from an acquired or inherited deficiency of the enzyme
Uroporphyrinogen decarboxylase (URO-D) (2-5).
PCT is present in two main forms: Type I, sporadic or acquired; and
type II, familial or hereditary. The clinical symptoms of PCT
include skin fragility, hyperpigmentation, bullae and
hypertrichosis. The onset of PCT is frequently associated with
different precipitating agents, primarily hepatotoxic drugs and
hepatotropic viral infection (6-8).
The prevalence of PCT varies worldwide from 1:5,000 (Czech Republic
and Slovakia) to 1:70,000 (Ireland) (9,10) and in
Argentina the prevalence is 1:20,000(2).
In Argentina, PCT patients have a high incidence
(16%) of human immunodeficiency virus (HIV) infection (11). However, since almost all HIV-infected
patients have additional risk factors for Porphyria manifestation,
it is still unclear whether HIV infection is a precipitating factor
for development of PCT. Despite this, several reports have
mentioned PCT being triggered after or during HIV therapy with
antiretroviral drugs, even in the absence of another precipitating
agent (12-14).
The human multidrug-resistance gene
(ABCB1/MDR1) encodes for the integral membrane
protein P-glycoprotein (P-gp), which is involved in the
energy-dependent transport of substances from the inside of cells
and/or from membranes to the outside space, acting as a pump that
effluxes a wide range of structurally diverse xenobiotics, such as
antiretroviral drugs, and protease and integrase inhibitors
(15-19).
According to the single nucleotide variant (SNV) database of the
National Center for Biotechnology Information, the human
ABCB1 coding region has >50 SNVs (ncbi.nlm.nih.gov/gene/5243). The most relevant amongst
these are: Exon 12 (rs1128503, c.1236C>T), exon 21 (rs2032582,
c.2677G>T/A) and exon 26 (rs1045642, c.3435 C>T), which
affect the expression and/or activity of P-gp, and therefore the
bioavailability of some drugs (20-22);
these three SNVs are the most common in Caucasian populations, and
are associated with an increased susceptibility of developing a
disease or to modify the effect of drugs used for therapy (17,23-26).
Glutathione S-transferases (GSTs) are a family of
enzymes belonging to the Phase II Drug Metabolizing System, which
catalyzes the synthesis of thioether conjugates between glutathione
and xenobiotics (27,28). These enzymes are also involved in the
detoxification of reactive oxygen species (ROS), environmental
carcinogens and steroid hormones, as well as in the metabolism of
chemotherapeutic agents (27,28).
Some genetic variants, including GSTT1 null, GSTM1
null and GSTP1 (rs1695, c.313A>G), are of clinical
importance because they alter the activity of GSTs, and may affect
the levels of hormones and xenobiotics. An increased susceptibility
of developing several different types of cancer (29,30),
liver failure due to alcoholism (31) and other diseases (32) has been associated with the presence
of non-wild-type variants. Singh et al (33) demonstrated the relationship between
the variants GSTM1, GSTT1 and GSTP1 and
hepatotoxicity, which was associated with antiretroviral therapy in
individuals with HIV.
Based on the above, the aim of the present study was
to evaluate the role of genetic variants in triggering PCT, and to
analyze the genetic basis of the association between PCT and
HIV.
Materials and methods
Subjects
The recruited cohorts consisted of Caucasian
individuals of both sexes. The individuals were stratified into
four groups: Control group (n=60, 32 males and 28 females, age
range 17-77 years, median age 38.5 years), individuals with a
negative diagnosis for both HIV and PCT; HIV group (n=35, 30 males
and 5 females, age range 20-53 years, median age 27 years),
patients infected with HIV; PCT group (n=40, 22 males and 18
females, age range 31-83 years, median age 49 years), patients with
acquired PCT without HIV (onset of PCT due to other triggering
factors); and PCT-HIV group (n=40, 36 males and 4 females, age
range 29-67 years, median age 44.2 years), patients diagnosed with
PCT and also infected with HIV. The exclusion criterion was:
Individuals of Control and HIV groups related to PCT patients.
Samples were collected from patients attending the
Research Center on Porphyrins and Porphyrias (CIPYP), Hospital de
Clínicas José de San Martín (Buenos Aires, Argentina) between March
2010 and December 2018. All individuals provided signed consent for
participation. The present study conformed with the guidelines
stated in the Declaration of Helsinki (34), and was approved by the Institutional
Research Ethics Committee of the CIPYP, National Scientific and
Technical Research Council, University of Buenos Aires,
Argentina.
Biological materials, DNA extraction
and genotyping
Genomic DNA was extracted from peripheral blood,
using the Illustra blood genomicPrep Mini Spin kit (Invitrogen;
Thermo Fisher Scientific, Inc.). PCR was performed using MyTaq HS
Red mix, 2x (Bioline); this kit includes the enzyme MyTaq HS DNA
Polymerase. Primers were designed using the SeqBuilder and
PrimerSelect programs (DNASTAR version 11.0; Lasergene).
ABCB1 gene variants
PCR-restriction fragment length polymorphism (RFLP)
was used to analyze the variants in exon 12 (c.1236C>T), 21
(c.2677G>T/A) and 26 (c.3435C>T), according to the protocols
described in previous studies (35-37).
Fig. 1 shows the representative
patterns of the genotypes of each SNV.
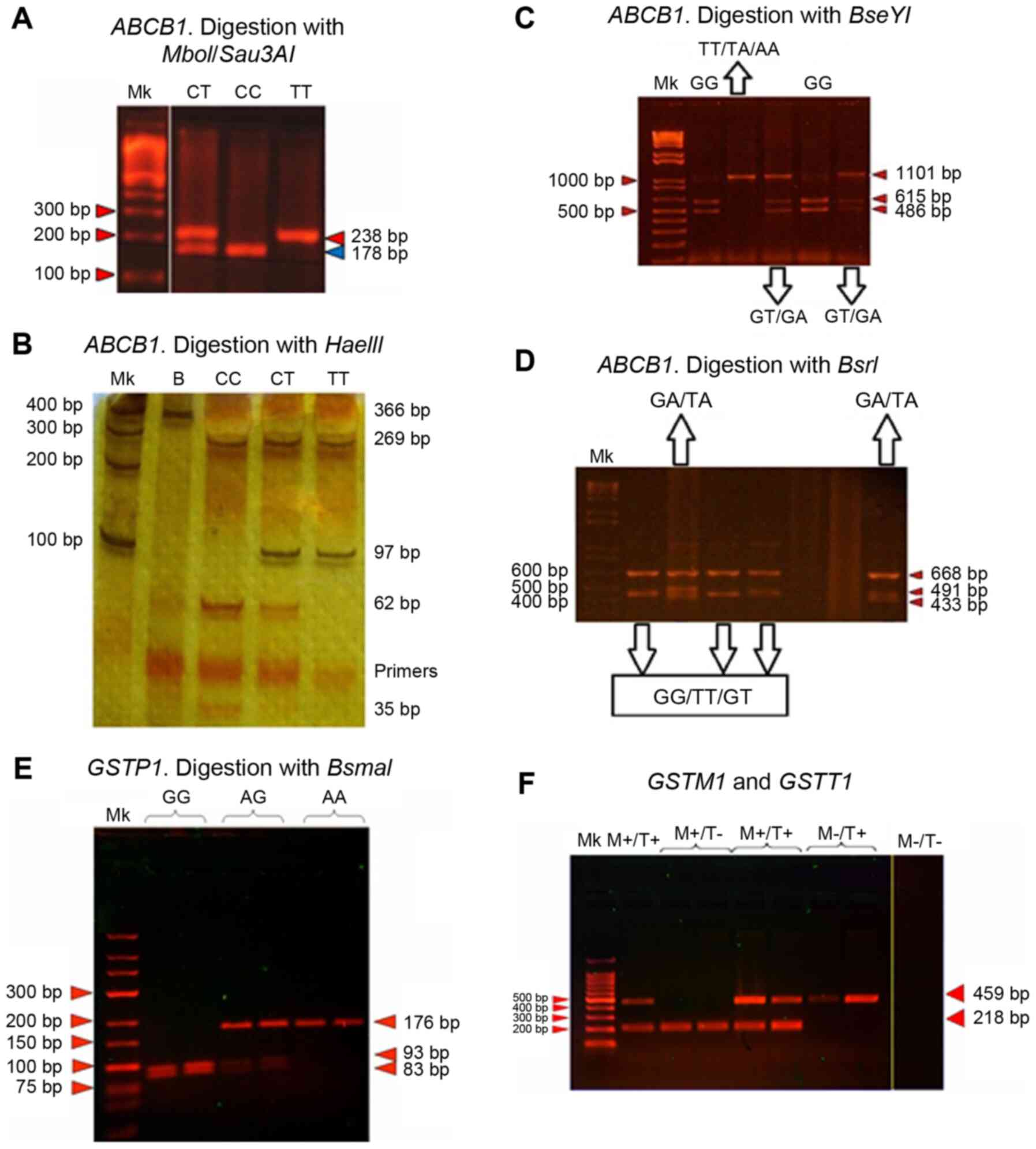 | Figure 1Genotyping band pattern of the
variants studied. Representative band pattern of (A-D) the
ABCB1 and (E and F) GST gene variants following
enzymatic digestion of the PCR products (panels A-E), or
PCR-multiplex amplification (panel F). (A) c.3435C>T (exon 26),
3% agarose gel in the presence of ethidium bromide (80 V, 45 min).
(B) c.1236C>T (exon 12), 12% polyacrylamide gel with 0.1% silver
staining (250 V, 80 min). (C and D) c.2677G>T/A (exon 21), 2%
agarose gel with ethidium bromide (80 V, 40 min). (E) GSTP1,
3% agarose gel with ethidium bromide (80 V, 60 min). (F)
GSTT1 and GSTM1, 2% agarose with ethidium bromide (80
V, 30 min). Mk, marker. |
To genotype the SNV c.3435C>T of exon 26, the
primers used were: ABCB1 3435 forward,
5'-GCTGGTCCTGAAGTTGATCTGTGAAC-3' and reverse,
5'-ACATTAGGCAGTGACTCGATGAAGGCA-3', which amplifies a 238 bp
fragment. The thermocycling conditions were: Initial denaturation
at 95˚C for 5 min; followed by 35 cycles at 94˚C for 30 sec, 61˚C
for 30 sec and 72˚C for 30 sec; and a final extension step of 72˚C
for 5 min. The PCR products were digested using the restriction
enzymes Sau3A1 or MboI. The wild-type allele (allele
C) has a cut-off site for these enzymes which generates two
fragments with lengths of 178 and 60 bp, whereas the T variant does
not possess this site (Fig. 1A).
To genotype the SNV c.1236C>T of exon 12, the
primers used were: ABCB1-15 forward,
5'-TATCCTGTGTCTGTGAATTGCC-3' and ABCB1-15 reverse
5'-CCTGACTCACCACACCAATG-3', which amplify a 366 bp fragment. The
thermocycling conditions were: Initial denaturation at 95˚C for 5
min; followed by 35 cycles at 94˚C for 30 sec, 60˚C for 30 sec and
72˚C for 30 sec; and a final extension step of 72˚C for 5 min. The
PCR product was digested with the enzyme HaeIII, which
yields three fragments of 269, 62 and 35 bp in the wild-type gene
(allele C). When the variant c.1236C>T was present, a
restriction digest site was abolished and only two fragments of 269
and 97 bp were obtained (Fig.
1B).
To genotype the SNV c.2677G>T/A of exon 21, the
primers used were 21F forward, 5'-GCTTTAGTAATGTTGCCGTGAT-3' and 21R
reverse, 5'-ATACCCCTAGCATTTTTCCATA-3', which amplify a 1,101 bp
fragment. The thermocycling conditions were: Initial denaturation
at 95˚C for 5 min; followed by 35 cycles at 94˚C for 1 min, 58˚C
for 30 sec and 72˚C for 2 min; and a final extension step of 72˚C
for 5 min. To evaluate the G and T alleles, the PCR products were
digested with the restriction enzyme BseYI; the pattern of
bands for the G allele consists of two bands of 615 and 486 bp,
whereas the cut-off site for the T allele is abolished, showing one
band of 1,101 bp (Fig. 1C). To
genotype the A allele, the PCR product was digested with the
restriction enzyme BsrI; the resulting pattern for the A
allele is three bands of 491, 433 and 177 bp, whereas that for the
G or T alleles is two bands of 668 and 433 bp (Fig. 1D).
GST variants
To study the GSTM1 and GSTT1 variants,
the presence or absence of deletion was evaluated using multiplex
PCR; to study the c.313A>G of GSTP1, PCR-RFLP was used.
Fig. 1E and F shows the characteristic patterns of the
different genotypes of each variant.
The thermocycling conditions of multiplex PCR were:
Initial denaturation at 95˚C for 5 min; followed by 35 cycles at
94˚C for 30 sec, 60˚C for 30 sec and 72˚C for 30 sec; and a final
extension step of 72˚C for 5 min. In the case of GSTM1, the
primers used were: Forward, 5'-GAACTCCCTGAAAAGCTAAAGC-3' and
reverse, 5'-TTGGGCTCAAATATACGGTGGA-3', resulting in an
amplification product of 218 bp. For GSTT1, the primers were
forward, 5'-TTCCTTACTGGTCCTCACATCTC-3' and reverse,
5'-TCACCGGATCATGGCCAGCA-3'. The band patterns were: Two bands of
218 and 459 bp for M+/T+, one band at 218 bp for M+/T-, one band at
459 bp for M-/T+, and the absence of bands for M-/T- (Fig. 1F). It is necessary to consider that
this methodology only allows for discrimination between the absence
and presence, but not differentiating between heterozygous and
non-zero homozygous genotypes, in which the existence of one or two
alleles is indistinguishable. Thus, samples with double deletion
were amplified again to confirm this aspect.
To genotype the variant c.313A>G of GSTP1,
the primers used were: Forward, 5'-ACCCCAGGGCTCTATGGGAA-3' and
reverse, 5'-TGAGGGCACAAGAAGCCCCT-3', obtaining a product of 176 bp.
The thermocycling conditions were: Initial denaturation at 95˚C for
5 min; followed by 35 cycles at 94˚C for 30 sec, 62˚C for 30 sec
and 72˚C for 30 sec; and a final extension step of 72˚C for 5 min.
The PCR product was digested with the restriction enzyme
BsmAI, and three possible patterns were obtained according
to the genotype of the samples: For the homozygous wild type (AA),
one band of 176 bp; for the heterozygous genotype (AG), three bands
of 176, 93 and 83 bp (AG); and for the homozygous mutant genotype
(GG), two bands of 93 and 83 bp (Fig.
1E).
The specific run conditions for analysis of the
variants and alleles varied and are described in the figure legend
for each specific condition.
Data management and statistical
analysis
The results were evaluated using a χ² using VCCStats
Beta 3.0 (institutodemetodologia.net/propios-c5xu). The
frequencies of alleles and genotypes were calculated by directly
counting. Haplotype analysis was performed using SNPStats
(snpstats.net/start.htm) (38). The association with the risk of
developing PCT was estimated using the odds ratios (ORs) and 95%
confidence intervals (CIs). P<0.05 was considered to indicate a
statistically significant difference.
To compare the variant frequencies in the present
study with that reported for other human populations, a literature
search was performed and used to develop a quantitative systematic
review (meta-analysis) following some of the guidelines stated in
the PRISMA (39). The search was
performed using the following terms: ‘ABCB1’, ‘MDR1’,
‘GST’, ‘GSTT1’, ‘GSTM1’, ‘GSTP1’,
‘genetic variants’, ‘rs1045642’, ‘rs2032582’, ‘rs1128503’ and
‘rs1695’ in PubMed (pubmed.ncbi.nlm.nih.gov) and SciELO (scielo.org/es) in October 2020. The language used was
generally English, although searches in Spanish (using SciELO) were
also performed to avoid the bias of using only one language. In
addition, the references in the studies deemed relevant were also
assessed. The criteria used were as follows: i) Studies in humans;
ii) investigation of the genetic variants analyzed in the present
work (ABCB1 and GST) that included a control group
(without any associated pathologies); and iii) studies published
since 2000. The data were extracted from each study (19 in total)
to construct comparative tables, which included the following
information: Name of the first author, and population and allelic
(ABCB1 and GSTP1 variants) or genotypic (GSTM1
and GSTT1 variants) frequency of the control group. The
frequencies are expressed as ranges, using the maximum and minimum
values found in all studies as the extremes. The data from the
literature search was also compared with the values in the 1000
Genomes Browser (browser.1000genomes.org; August 2020).
Results
Allelic and genotypic frequencies of
the ABCB1 gene
The allelic and genotypic frequencies of the
ABCB1 gene were calculated and compared between the groups
(Table I). For the c.3435C>T
variant, the frequency of the T allele in both groups with PCT (PCT
and PCT-HIV) was significantly higher than in the Control or HIV
individuals. When c.1236 C>T SNV was evaluated, the frequency of
the T allele in the PCT group was higher than in the other groups,
whereas no differences were detected between the Control, HIV and
PCT-HIV groups. The evaluation of the SNV c.2677G>T/A included
the analysis of two non-wild-type alleles (A and T); the frequency
of the A allele was similar in all the groups evaluated, whereas
that of the T allele in the PCT-HIV group was significantly higher
than in Control individuals and HIV and PCT patients.
 | Table IAllelic and genotypic frequencies for
c.3435 C>T, c.1236 C>T and c.2677G>T/A variants of the
ABCB1 gene. |
Table I
Allelic and genotypic frequencies for
c.3435 C>T, c.1236 C>T and c.2677G>T/A variants of the
ABCB1 gene.
| | Allelic
frequency |
Genotypic
frequency |
|---|
| | c.3435 C>T | c.1236 C>T | c.2677G>T/A | c.3435 C>T | c.1236 C>T | c.2677G>T/A |
|---|
| Groups | C | T | C | T | G | T | A | CC | CT | TT | CC | CT | TT | GG | GT | TT | TA/GA |
|---|
| Control, n=60 | 0.6 | 0.36 | 0.7 | 0.33 | 0.5 | 0.45 | 0 | 33 | 61 | 5.3 | 41 | 51 | 8 | 33 | 40 | 22.5 | 5.0/0 |
| HIV, n=35 | 0.5 | 0.46 | 0.6 | 0.39 | 0.6 | 0.37 | 0 | 23 | 63 | 14.3 | 26 | 71 | 2.9 | 40 | 43 | 14.3 | 2.9/0 |
| PCT, n=40 | 0.5 | 0.52a | 0.4 | 0.59a | 0.5 | 0.48 | 0 | 16 | 63 | 20.9a | 6.9 | 69 | 24.1a | 22 | 54 | 19.5 | 2.4/2.4 |
| PCT-HIV, n=40 | 0.5 | 0.55a | 0.7 | 0.35 | 0.4 | 0.61a | 0 | 18 | 55 | 27.3a | 33 | 59 | 7.7 | 5.9 | 62 | 29.4a | 2.9/0 |
When the genotypic frequency was analyzed,
c.3435C>T was more common in the polymorphic variant (TT) in
both PCT groups (PCT and PCT-HIV; both P<0.05) compared with the
Control group. For the c.1236C>T variant, only the PCT group had
an increased frequency (P<0.05) when compared with the other
groups. In the case of c.2677G>T/A SNV, the frequency of
genotypes that included the presence of A (TA and GA) was similar
between the groups, but the TT genotype was higher in the PCT-HIV
group compared with the HIV and PCT groups (P<0.05).
Haplotype analysis was performed on the three
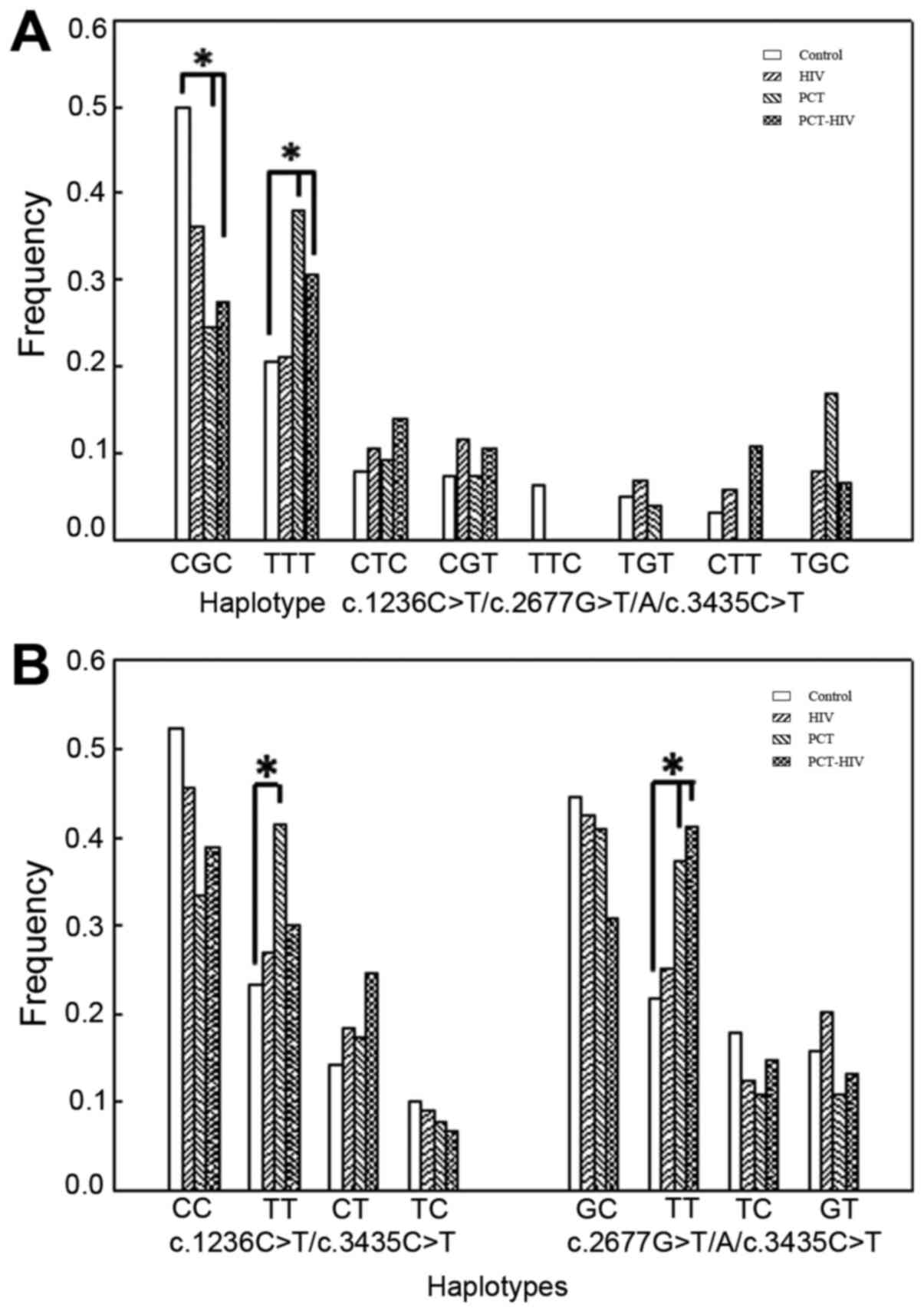
ABCB1 SNVs (Fig. 2A). The
results indicated that the CGC and TTT haplotype frequencies were
>20% in all the groups. In the PCT and PCT-HIV groups, the TTT
haplotype was present at a higher frequency than in the Control and
HIV groups, and the OR values indicated that it was a risk
haplotype for the onset of the disease [PCT group: OR=12.70 (CI,
1.98-81.31), P<0.01; PCT-HIV group: OR=4.64 (CI, 1.16-18.57),
P<0.05]. An opposite relationship was observed for the wild-type
haplotype CGC, which showed a high frequency in the Control and HIV
groups (P<0.05).
Since the results suggested a possible role of the
c.3435C>T variant in the initiation of PCT, a paired haplotype
analysis was performed (Fig. 2B). In
the PCT group, the TT frequency was increased (P<0.05) for the
combination c.1236C>T/c.3435C>T, indicating that this
combination is a risk haplotype [OR=6.53 (CI, 1.72-24.70),
P<0.01]. When the c.2677G>T/A/c.3435C>T pair was
evaluated, there was a higher frequency of the TT haplotype for
both PCT populations (PCT and PCT-HIV; P<0.05) when compared
with the Control and HIV groups; the OR values revealed a risk
haplotype for PCT vs. Control [OR=2.32 (CI, 0.81-6.64), P<0.05]
and PCT-HIV vs. Control [OR=3.61 (CI, 1.25-10.32), P<0.05].
Analysis of the frequencies of GSTT1,
GSTM1 and GSTP1
Results of the frequencies of GSTT1,
GSTM1 and GSTP1 are shown in Table II. The genotypic frequencies of the
presence or absence of GSTT1 showed that the frequency of
the homozygous null genotype was increased in the PCT-HIV group
when compared with the HIV group, although the differences were not
statistically significant. In the case of GSTM1, the effect
was opposite to that described for GSTT1; the PCT-HIV group
presented a significantly lower frequency for the null genotype in
homozygosis when compared with the HIV group.
 | Table IIGSTT1, GSTM1 and
GSTP1 (c.313 A>G) frequencies. |
Table II
GSTT1, GSTM1 and
GSTP1 (c.313 A>G) frequencies.
| | GSTT1 | GSTM1 | c.313 A>G
(GSTP1) |
|---|
| | Genotypic
frequency | Allelic | Genotypic |
|---|
| Groups | +/+, +/- | -/- | +/+, +/- | -/- | A | G | AA | AG | GG |
|---|
| Control, n=60 | 91.67 | 8.33 | 58.33 | 41.67 | 0.58 | 0.42 | 29 | 58 | 13 |
| HIV, n=35 | 93.33 | 6.67 | 46.67 | 53.33 | 0.53 | 0.47 | 28 | 50 | 22 |
| PCT, n=40 | 89.47 | 10.53 | 63.16 | 36.84 | 0.55 | 0.45 | 25 | 60 | 15 |
| PCT-HIV, n=40 | 85.71 | 14.29b | 67.86 | 32.14a | 0.54 | 0.46 | 27 | 54 | 19 |
When the variant c.313A>G of GSTP1 was
analyzed, no significant differences in the allelic frequencies
were observed between the different groups. An almost equivalent
distribution was observed between both alleles (A and G), with a
slightly higher prevalence of the wild-type variant. The genotypic
frequency showed no significant differences between the groups
studied, although the presence in heterozygosis (AG) was 2-fold
higher than the homozygote genotype (GG).
Considering that the allelic and genotypic
frequencies of the variant c.313A>G of GSTP1 showed no
appreciable differences between the groups studied, only the
combinations of GSTM1 and GSTT1 were further
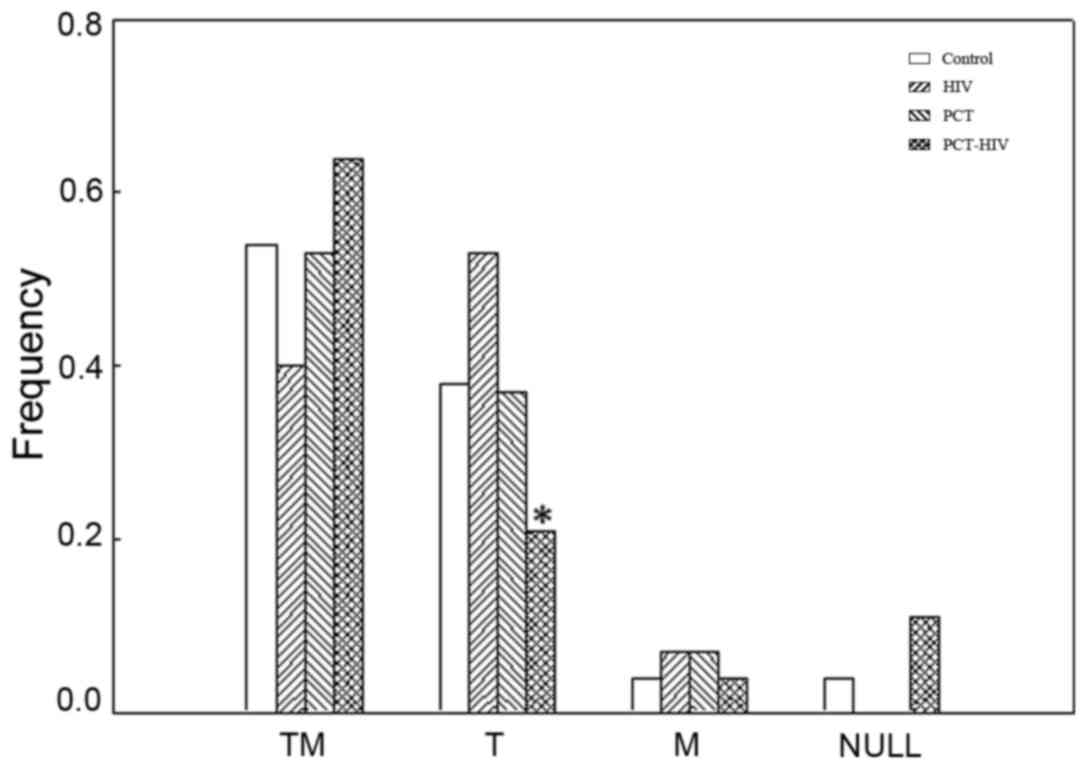
evaluated (Fig. 3). The genotype
presence for both genes (TM), either in homozygosis or in
heterozygosis, represented a frequency >40% for all the groups
studied. The frequency of individuals with the presence of
GSTT1 in at least one of the alleles and absence in
homozygosis of GSTM1 (T) was lower in the PCT patients
infected with HIV (PCT-HIV) compared with HIV infected individuals.
Regarding the presence of GSTM1, either in homozygosis or
heterozygosis, in the absence of GSTT1 (M), all the groups
studied had a similar frequency (<10%). It is noteworthy that no
HIV or PCT patients with the null genotype (absence of both genes
in homozygosis) were detected in the population analyzed.
Distribution of combined ABCB1, GSTM1
and GSTT1 genotypes
Using the variants studied for ABCB1,
GSTM1 and GSTT1, the risk alleles of the individuals
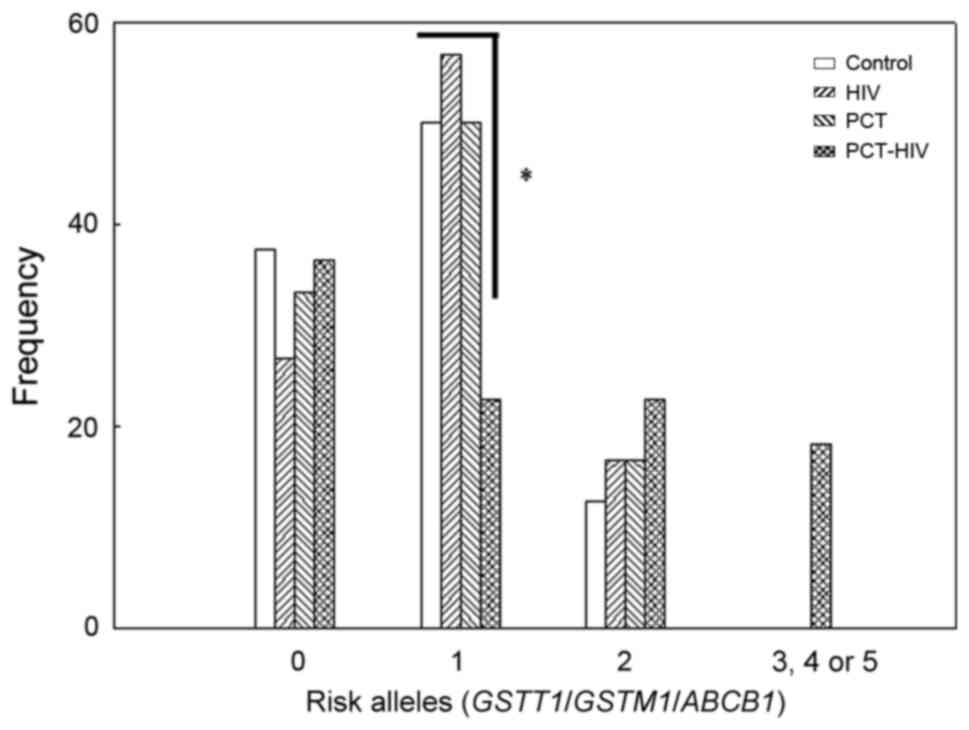
in each group were determined (Fig.
4). The absence of TT for all ABCB1 SNVs plus the
presence of GSTT1 plus GSTM1 was considered as 0 risk
alleles; the presence of only one non-wild type variant in
homozygosis for ABCB1 SNVs or absence of GSTT1 or
GSTM1 was considered as 1 risk allele; and the cases in
which 2-5 of the variants were not wild-type in homozygosis was 2-5
risk alleles, respectively. The results showed that the PCT-HIV
group had the highest proportion of 2 risk alleles and the lowest
proportion of 1 risk allele. Moreover, this group was the only
group that had individuals with 3-5 risk alleles.
Discussion
The results of the present study showed that the
variants of the ABCB1 gene may influence the initiation of
PCT. It is important to note that Porphyrias are multifactorial
diseases, and that PCT in particular can be triggered by
alcoholism, estrogens, drug abuse, iron overload or hepatotropic
viral infections, through different mechanisms of alterations of
heme metabolism (2,4,12,13). In
our previous studies (12) and in
those from other authors (40,41), the
clinical symptomatology and biochemical alterations commonly
observed in PCT patients are similar to those seen in HIV patients
who develop PCT. Moreover, no differences were observed in terms of
response to treatments for PCT (hydroxychloroquine alone or
combined with phlebotomies).
When evaluating the c.3435C>T variant in
ABCB1, its allelic (T) and genotypic (TT) frequencies in the
PCT and PCT-HIV groups were significantly higher compared with the
Control group, suggesting that the role of this variant in
triggering PCT may not be exclusively associated with HIV infection
or antiretroviral therapy. The nucleotide position analyzed is
located in the second ATP binding domain and although the change is
synonymous, there are studies that confirm that it can affect the
folding of the protein, insertion to the membrane, the translation
process, and the interaction with ATP and substrates/inhibitors
(16,42-44).
At the hepatic level, alterations in P-gp activity may increase
cellular toxicity due to the inefficiency in export of substances
and metabolites, resulting in hepatotoxicity and oxidative stress
caused by an increase in ROS (17,45). In
this context, the administration of substances or drugs that are
metabolized in the liver in individuals with the TT genotype for
c.3435C>T SNV may promote and contribute to the inhibition of
hepatic URO-D and, consequently, to the onset of PCT.
Regarding the SNV c.1236C>T, both allelic (T) and
genotypic (TT) frequencies were significantly higher in PCT
individuals than in the other groups, and this variant may be
associated with triggering the development of PCT as an inducer of
hepatotoxicity, independent of HIV infection and antiretroviral
treatment. The findings of Fung and Gottesman (16) suggest that the primary impact of this
change lies in the presence of a rare codon (GGT instead of GGC)
which can lead to a pause or slowdown of ribosomal function, and to
a decrease in the activity and/or protein levels of P-gp.
When evaluating the results obtained for the
c.2677G>T/A variant, the fact that the frequencies of the A
allele and those of genotypes TA/GA were similar in all the groups
studied, suggests that there is no evidence to associate this
variant with the initiation of PCT in individuals, regardless of
HIV infection status. The frequencies of the T allele and the TT
genotype were significantly higher for the PCT-HIV group than for
the other groups, especially for the patients infected with HIV.
This result suggests that the c.2677G>T/A variant may influence
initiation of PCT in HIV-infected individuals, possibly through a
mechanism that involves antiretroviral therapy based on the fact
that anti-HIV drugs are substrates of P-gp and genetic variants
alter the expression and activity of the transporter (16,17).
Although in the nucleotide position studied there is no functional
domain (intracellular loop), biochemical evidence has confirmed
that the change in alanine to serine or threonine may alter the
transport of drugs, due to irregularities in the ATPase activity of
P-gp (46). The P-gp transporter is
a key determinant of the bioavailability and penetration of
protease inhibitors used as antiretroviral therapies. Taking into
account that the deficiencies in drug transporters may increase the
risk of hepatotoxicity, a possible explanation for the high
incidence of PCT in the Caucasian population of HIV infected
patients in Argentina (1:370) compared with the prevalence of PCT
in this country (1:20,000) may be linked to the high presence of
this variant and the consequent context of hepatotoxicity resulting
from the suboptimal transport of antiretrovirals by P-gp, favoring
the inhibition of URO-D.
The analysis of haplotypes of the three SNVs of the
ABCB1 gene, the significant increase in TTT in both PCT
groups compared with Control and HIV individuals, and the inverse
relationship in the wild-type haplotype highlight the potential
role of ABCB1 variants in initiation of PCT. On the other
hand, the analysis of the SNV pairs c.1236C>T and c.3435C>T
indicated that the frequency of the haplotype TT was significantly
higher in the PCT individuals, demonstrating the possible influence
of this SNV combination on the development of PCT. The variant
c.3435C>T has been reported to be of great relevance in the
predisposition to various pathologies, such as thyroid cancer,
early-onset Parkinson's disease and methotrexate-induced adverse
events in rheumatoid arthritis, amongst others (21,47,48).
Regarding the results obtained for the haplotypes of the
combination c.2677G>T/A/c.3435C>T, a significant increase in
TT was detected for the two PCT groups compared with that observed
in the Control and HIV groups, indicating that this haplotype may
influence development of PCT mediated by both antiretroviral
therapy and other risk factors.
Based on the results of the present study, it can be
concluded that the decrease in the expression of ABCB1
and/or the activity of P-gp, and its role as a predisposing factor
in triggering PCT, requires a synergistic combination of changes,
altering the molecular and protein structure of the transporters of
drugs and xenobiotics.
Since HIV and PCT patients usually present with
liver damage, and PCT is a hepatic Porphyria (2,4,49-52),
it was of interest to extend this work to study the influence of
variants of GST, a marker enzyme involved in cellular
detoxification. When the GSTM1 variant was genotyped, the
frequency of the null genotype was significantly lower in the
HIV-PCT group, suggesting that the presence of this gene could
predispose an individual to development of PCT in HIV-infected
patients. Regarding GSTT1, the frequency of null homozygotes
in PCT-HIV individuals was increased, although this result was not
statistically significant; this could be attributed to the fact
that the absence of elements of the cellular detoxification system
can cause an increase in hepatotoxicity, leading to the onset of
PCT in individuals with antiretroviral treatment.
The fact that the null genotype frequencies in
homozygosis for GSTM1 and GSTT1 showed opposite
results indicates the existence of an opposite mechanism and
biological implications in the influence of the triggering of the
acquired PCT in HIV-infected individuals, without neglecting the
multifactorial nature of the pathology.
It was hypothesized that the variant c.313A>G
(GSTP1) would have some influence on the development of PCT,
taking into account that the mutation is located in the active
protein site and thus causes suboptimal catalytic activity and,
therefore, lower cellular detoxification capacity of xenobiotics
and even ROS (26). The allelic and
genotypic frequencies between the different groups were similar,
although this was probably due to multiple factors, one of which
may be that GSTP1 is not primarily expressed in the
liver.
It was considered appropriate to evaluate the
combination of variants corresponding to the GSTT1 and
GSTM1 genes, excluding the GSTP1 gene, which was
similar in all the groups studied. The combination of both genes
(GSTT1 and GSTM1) showed there were no PCT or HIV
patients with absence in homozygosis of both genes. Moreover, this
null genotype for GSTT1 and GSTM1 showed a tendency
to be increased in the PCT-HIV group, but the results were not
statistically different; this condition may predispose individuals
to an increased risk of hepatotoxicity, but to a lesser degree than
other variants/alleles, that, in combination with other factors,
may lead to the development of PCT. The absence in homozygosis of
GSTM1 and the presence of at least one allele of
GSTT1 was significantly lower in the PCT-HIV group, which is
consistent with that observed for GSTT1 and GSTM1
individually; the condition described could represent a combination
that decreases the risk of triggering PCT. It is known that the
cDNAs encoded by GSTM1 and GSTM2 share a significant
amount of sequence identity (~99%) and that following elimination
of GSTM1, GSTM2 is overexpressed (53). Thus, GSTM2 may exhibit more
efficient detoxification activity regarding the conjugation of
antiretrovirals than GSTM1.
Based on the above analysis, it was concluded that
the development of PCT in HIV-infected individuals may have a
genetic basis regarding GST enzymes via a combination of different
genotypes in the GSTT1 (absence) and GSTM1 (presence)
genes.
When the variants of the ABCB1 and GST
genes were evaluated as a whole, only PCT-HIV individuals possessed
≥2 risk alleles. This aspect provides strong evidence that non-wild
type variants of these genes contribute to the triggering of PCT in
HIV-infected individuals, possibly due to inefficient transport of
antiretrovirals and thus increased liver toxicity. External and/or
genetic factors that predispose an individual to hepatotoxicity
promote the inhibition of URO-D, increasing the probability of the
onset of the disease (2,4).
The GST variants analyzed in the present study are
related to an increase in oxidative stress markers and ROS in blood
samples in individuals exposed to toxic factors or with other
pathologies increasing ROS levels (54-56).
In this context, HIV-infected individuals carrying GST variants may
result in high hepatic toxicity under antiretroviral treatment
and/or other triggering factors related to drug metabolism and
cellular detoxification.
The allelic frequencies of ABCB1 found in the
Control group were compared with those reported for other countries
(Table III). This comparison
showed differences between various regions of the world. For
example, individuals of African descent were considerably more
likely to possess wild-type variants for the three SNVs compared
with other ethnicities (57,58). For the c.3435C>T SNV, the mutant
variants in the present study was significantly higher than that
observed for the African population (57,58). The
T frequency of the c.2677C>T variant was significantly higher in
the Argentine population than in African individuals (57,58) and
in some ethnic groups of Chile (20); for variant A, no significant
differences were observed between groups. Regarding the
c.1236C>T SNV, the frequency of the T variant in African
individuals (57,58) was significantly lower than that found
in the present study; in contrast, the frequencies for Mapuche
(20) and Asian (57,58)
populations were significantly higher than those found in the
present study. Results of other studies in Caucasians showed no
notable differences with the results of the present study (15,58). The
bibliographic data are consistent with that provided by the 1000
Genome Browser.
 | Table IIIAllelic frequencies of c.3435C>T,
c.1236C>T and c.2677G>T/A variants of ABCB1 gene in
different populations. |
Table III
Allelic frequencies of c.3435C>T,
c.1236C>T and c.2677G>T/A variants of ABCB1 gene in
different populations.
| | | c.3435C>T | c.2677G>T/A | c.1236C>T | |
|---|
| First author,
year | Ethnicity | C | T | G | T | A | C | T | (Refs.) |
|---|
| Wielandt et
al, 2004 | Chilean | | | | | | | | (20) |
| | Mestizo | 0.67 | 0.33 | 0.65 | 0.26 | 0.09 | 0.59 | 0.41 | |
| | Mapuche | 0.65 | 0.35 | 0.69 | 0.16a | 0.15 | 0.4 | 0.6a | |
| | Pascuense | 0.75 | 0.25 | 0.78 | 0.15a | 0.07 | 0.7 | 0.3 | |
| Hoffmeyer et
al, 2000 | Caucasian | 0.46-0.48 | 0.52-0.54 | 0.53-0.61 | 0.39-0.43 | 0.02-0.04 | 0.54-0.60 | 0.40-0.46 | (15) |
| Milojkovic et
al, 2011 | | | | | | | | | (58) |
| Mhaidat et
al, 2011 | Asian | 0.38-0.53 | 0.47-0.62 | 0.36-0.62 | 0.36-0.42 | 0.02-0.22 | 0.35-0.44 |
0.56-0.65a | (57) |
| Milojkovic et
al, 2011 | | | | | | | | | (58) |
| Komoto et
al, 2006 | | | | | | | | | (59) |
| Mhaidat et
al, 2011 | African | 0.83-0.84 |
0.16-0.17a | 0.89-0.96 |
0.04-0.11a | Not determined | 0.85-0.86 |
0.14-0.15a | (57) |
| Milojkovic et
al, 2011 | | | | | | | | | (58) |
| Present study | Argentinian | 0.64 | 0.36 | 0.52 | 0.5 | 0.03 | 0.67 | 0.33 | - |
The frequencies obtained for the variants of the
GST genes with other populations were also compared
(Table IV). No significant
differences were detected between our results and another study
performed in Argentina (59) or
those reported for Brazilian, Caucasian, Asian and African
populations (61-73),
and were consistent with the 1000 Genome Browser, except for the
Asian population, where the database reported a larger range
compared to that found in other studies (0.78-0.90 for variant
A).
 | Table IVAllelic frequencies of GSTP1
(c.313A>G) and genotypic frequencies of GSTM1 and
GSTT1 in different populations. |
Table IV
Allelic frequencies of GSTP1
(c.313A>G) and genotypic frequencies of GSTM1 and
GSTT1 in different populations.
| | | GSTM1 | GSTT1 | GSTP1 (c.313
A>G) | |
|---|
| First author,
year | Ethnicity | +/+, +/- | -/- | +/+, +/- | -/- | A | G | (Refs.) |
|---|
| Rossini et
al, 2002 | Brazillian | 54 | 46 | 87 | 13 | 0.69 | 0.31 | (61) |
| Pinheiro et
al, 2017 | | | | | | | | (62) |
| Weich et al,
2017 | Caucasian | 44-58 | 45-58 | 73-88 | 12-27 | 0.64-0.71 | 0.29-0.36 | (60) |
| Klusek et
al, 2018 | | | | | | | | (63) |
| Srivastava et
al, 2018 | | | | | | | | (64) |
| Stamenkovic et
al, 2018 | | | | | | | | (65) |
| ThekkePurakkal
et al, 2019 | | | | | | | | (66) |
| Srivastava et
al, 2018 | Asian | 35-58 | 20-65 | 49-94 | 6-51 | 0.74 | 0.26 | (64) |
| Musavi et
al, 2019 | | | | | | | | (67) |
| Zehra et al,
2018 | | | | | | | | (68) |
| Saravani et
al, 2019 | | | | | | | | (69) |
| Farmohammadi et
al, 2020 | | | | | | | | (70) |
| Oshodi et
al, 2017 | African | 45-89 | 11-55 | 53-88 | 12-47 | 0.65-0.72 | 0.28-0.35 | (71) |
| Srivastava et
al, 2018 | | | | | | | | (64) |
| Idris et al,
2020 | | | | | | | | (72) |
| Rebai et al,
2020 | | | | | | | | (73) |
| Present study | Argentinian | 58 | 42 | 92 | 8 | 0.58 | 0.42 | - |
In conclusion, based on the ethnic diversity
observed in individuals from different regions of the world
compared with the results of the present study, it is important to
emphasize that each individual possesses a particular combination
of allelic variants which leads to specific biological
inter-individual differences. Therapies and drugs, such as
antiretrovirals may be metabolized in slightly different ways
between individuals, and thus may exhibit slightly different
effects or a per individual basis. Therefore, there are individuals
to whom certain substances are innocuous and others to whom the
doses may be excessive and cause metabolic damage. The observation
that there are combinations of variants and haplotypes that could
trigger PCT in HIV-infected individuals highlights the possibility
in which chronic therapy with antiretrovirals causes collateral
damage, favoring the triggering of this pathology. The study of
genetic variants and their impact on drug metabolism must be
considered to improve personalized medical therapy, according to
the genetic profile of each patient. Pharmacogenetics will optimize
the efficiency of xenobiotic action, avoiding harmful effects that
lead to collateral damage.
The genetic variants analyzed in the present study,
together with other linked genes or marker parameters of liver
damage, may improve evaluation of the status of HIV-infected
patients, thus providing a powerful therapeutic tool when
administering treatments for the background disease to prevent the
triggering of PCT or to reduce its impact, protecting the hepatic
status via administration of antioxidants.
This is the first study to investigate the possible
role of variants of GST and ABCB1 in the development
of PCT in HIV-infected individuals and suggests that variants in
genes that encode for proteins involved in the removal of
xenobiotics and in the Phase II Drug Metabolizing System may have
an influence on development of PCT in HIV-infected individuals.
Acknowledgements
The authors want to make a special mention to the
memory of Dr Alcira Batlle, our mentor and founder of CIPYP, who
dedicated her life researching Porphyrias. We would also like to
thank Dr Alcira Batlle for her contribution in the development of
this research, and MD Hector Muramatsu and Mrs Victoria Castillo
for their technical assistance with patients.
Funding
This work was supported by grants from the
University of Buenos Aires (UBACYT 2014-2017; 01/Q839 and 01/Q287),
UBACYT 2018 (20020170100609BA), and CONICET (PIP 0528; CONICET),
Argentina.
Availability of data and materials
All data generated or analyzed during the present
study is included in the published article.
Authors' contributions
PAP, JRZ, VAM and JVL designed the methodology used,
as well as validated and analyzed the data. VEP, JRZ, AMB and MVR
conceptualized the study. PAP, JRZ, VAM and AMB wrote and edited
the manuscript. VAM and JRZ supervised the study. All authors made
substantial contributions to the writing of the manuscript as well
as read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional
Research Ethics Committee of the CIPYP, National Scientific and
Technical Research Council, University of Buenos Aires (Buenos
Aires, Argentina). Patients provided signed informed consent for
participation in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Batlle A: Porfirias humanas. Signos y
tratamientos. En Porfirias y porfirinas. Aspectos clínicos,
bioquímicos y biología molecular. Editorial Federación Bioquímica
de la Provincia de Buenos Aires, La Plata, ISSN 0325-2957. Acta
Bioquím Clín Latinoam Supl. 3:37–69. 1997.(In Spanish).
|
|
2
|
Rossetti MV, Buzaleh AM, Parera VE, Fukuda
H, Lombardo ME, Lavandera J, Gerez EN, Melito VA, Zuccoli JR,
Ruspini SV, et al: Metabolismo del Hemo: Las dos caras de los
efectos de la acumulación de precursores y porfirinas. Acta Bioquim
Clin Latinoamer Libro de Oro. 50:547–573. 2016.(In Spanish).
|
|
3
|
Méndez M, Rossetti MV, Gómez-Abecia S,
Morán-Jiménez MJ, Parera V, Batlle A and Enríquez de Salamanca R:
Molecular analysis of the UROD gene in 17 Argentinean patients with
familial porphyria cutanea tarda: Characterization of four novel
mutations. Mol Genet Metab. 105:629–633. 2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Phillips JD: Heme biosynthesis and the
porphyrias. Mol Genet Metab. 128:164–177. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Weiss Y, Chen B, Yasuda M, Nazarenko I,
Anderson KE and Desnick RJ: Porphyria cutanea tarda and
hepatoerythropoietic porphyria: Identification of 19 novel
uroporphyrinogen III decarboxylase mutations. Mol Genet Metab.
128:282–287. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Jalil S, Grady JJ, Lee C and Anderson KE:
Associations among behavior-related susceptibility factors in
porphyria cutanea tarda. Clin Gastroenterol Hepatol. 8:297–302.e1.
2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Singal AK, Venkata KVR, Jampana S, Islam
FU and Anderson KE: Hepatitis C treatment in patients with
porphyria Cutanea Tarda. Am J Med Sci. 353:523–528. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
To-Figueras J: Association between
hepatitis C virus and porphyria Cutanea Tarda. Mol Genet Metab.
128:363–366. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ryan Caballes F, Sendi H and Bonkovsky HL:
Hepatitis C, porphyria cutanea tarda, and liver iron: An update.
Liver Int. 32:880–893. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Usta Atmaca H and Akbas F: Porphyria
cutanea tarda: A case report. J Med Case Rep. 13(17)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Franzon VA, Mikilita ES, Camelo FH and
Camargo R: Porphyria cutanea tarda in a HIV-positive patient. An
Bras Dermatol. 91:520–523. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Melito VA, Parera VE, Rossetti MV and
Batlle A: Manifestación de porfiria cutánea tardía en pacientes
infectados con el virus de la inmunodeficiencia humana. Acta
Bioquím Clín Latinoamer. 40:29–34. 2006.
|
|
13
|
Jalil SJ, Grady JJ, Lee C and Anderson KE:
Associations among behavior-related susceptibility factors in
porphyria cutanea tarda. Clin Gastroenterol Hepatol. 8(3):297–302.
2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Aguilera P, Laguno M and To-Figueras J:
Human immunodeficiency virus and risk of porphyria Cutanea Tarda: A
possible association examined in a large hospital. Photodermatol
Photoimmunol Photomed. 32:93–97. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hoffmeyer S, Burk O, von Richter O, Arnold
HP, Brockmöller J, Johne A, Cascorbi I, Gerloff T, Roots I,
Eichelbaum M and Brinkmann U: Functional polymorphisms of the human
multidrug-resistance gene: Multiple sequence variations and
correlation of one allele with P-glycoprotein expression and
activity in vivo. Proc Natl Acad Sci USA. 97:3473–3478.
2000.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fung K and Gottesman M: A synonymous
polymorphism in a common MDR1 (ABCB1) haplotype shapes protein
function. Biochim Biophys Acta. 1794:860–871. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Brambila-Tapia AJ: MDR1 (ABCB1)
polymorphisms: Functional effects and clinical implications. Rev
Invest Clin. 65:445–454. 2013.PubMed/NCBI
|
|
18
|
Yano K, Tomono T and Ogihara T: Advances
in studies of P-Glycoprotein and its expression regulators. Biol
Pharm Bull. 41:11–19. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Arana MR and Altenberg GA: ATP-binding
cassette exporters: Structure and mechanism with a focus on
P-glycoprotein and MRP1. Curr Med Chem. 26:1062–1078.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wielandt AM, Vollrath V and Chianale J:
Polymorphisms of the multiple drug resistance gene (MDR1) in
Mapuche, Mestizo and Maori populations in Chile. Rev Med Chil.
132:1061–1068. 2004.PubMed/NCBI View Article : Google Scholar : (In Spanish).
|
|
21
|
Thuerauf N and Fromm MF: The role of the
transporter P-glycoprotein for disposition and effects of centrally
acting drugs and for the pathogenesis of CNS diseases. Eur Arch
Psychiatry Clin Neurosci. 256:281–286. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fathy M, Kamal M, Mohy A and Nabil A:
Impact of CYP3A5 and MDR-1 gene polymorphisms on the dose and level
of tacrolimus among living-donor liver transplanted patients:
Single center experience. Biomarkers. 21:335–341. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Marzolini C, Paus E, Buclin T and Kim RB:
Polymorphisms in human MDR1 (Pglycoprotein): Recent advances and
clinical relevance. Clin Pharmacol Ther. 75:13–33. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sharom FJ: ABC multidrug transporters:
Structure, function and role in chemoresistance. Pharmacogenomics.
9:105–127. 2008.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bellusci CP, Rocco C, Aulicino P,
Mecikovsky D, Curras V, Hegoburu S, Bramuglia GF, Bologna R, Sen L
and Mangano A: Influence of MDR1 C1236T polymorphism on lopinavir
plasma concentration and virological response in HIV-1-infected
children. Gene. 522:96–101. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yan Y, Liang H, Xie L, He Y, Li M, Li R,
Li S and Qin X: Association of MDR1 G2677T polymorphism and
leukemia risk: Evidence from a meta-analysis. Tumour Biol.
35:2191–2197. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Strange RC, Spiteri MA, Ramachandran S and
Fryer AA: Glutathione-S-transferase family of enzymes. Mutat Res.
482:21–26. 2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hayes JD, Flanagan JU and Jowsey IR:
Glutathione transferases. Ann Rev Pharmacol Toxicol. 45:51–88.
2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Schnekenburger M, Karius T and Diederich
M: Regulation of epigenetic traits of the glutathione S-transferase
P1 gene: From detoxification toward cancer prevention and
diagnosis. Front Pharmacol. 16(170)2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Weich N, Ferri C, Moiraghi B, Bengió R,
Giere I, Pavlovsky C, Larripa IB and Fundia AF: GSTM1 and GSTP1,
but not GSTT1 genetic polymorphisms are associated with chronic
myeloid leukemia risk and treatment response. Cancer Epidemiol.
44:16–21. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Brind AM, Hurlstone A, Edrisinghe D,
Gilmore I, Fisher N, Pirmohamed M and Fryer AA: The role of
polymorphisms of glutathione S-transferases GSTM1, M3, P1, T1 and
A1 in susceptibility to alcoholic liver disease. Alcohol Alcohol.
39:478–483. 2004.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bowatte G, Lodge CJ, Perret JL, Matheson
MC and Dharmage SC: Interactions of GST polymorphisms in air
pollution exposure and respiratory diseases and allergies. Curr
Allergy Asthma Rep. 16(85)2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Singh HO, Lata S, Angadi M, Bapat S, Pawar
J, Nema V, Ghate MV, Sahay S and Gangakhedkar RR: Impact of GSTM1,
GSTT1 and GSTP1 gene polymorphism and risk of ARV-associated
hepatotoxicity in HIV-infected individuals and its modulation.
Pharmacogenomics J. 17:53–60. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
WMA Declaration of Helsinki-Ethical
Principles for Medical Research Involving Human Subjects. World
Medical Association: July 9, 2018 (urihttps://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjectssimplehttps://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects).
|
|
35
|
Kim HJ, Hwang SY, Kim JH, Park HJ, Lee SG,
Lee SW, Joo JC and Kim YK: Association between genetic polymorphism
of multidrug resistance 1 gene and Sasang constitutions. Evid Based
Complement Alternat Med. 6 (Suppl 1):S73–S80. 2009.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Taheri M, Mahjoubi F and Omranipour R:
Effect of MDR1 polymorphism on multidrug resistance expression in
breast cancer patients. Gen Mol Res. 9:34–40. 2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zuccoli J, Melito V, Ruspini S, Lavandera
J, Abelleyro M, Parera V, Rossetti MV, Batlle A and Buzaleh AM:
Análisis de polimorfismos del exón 21 del gen MDR1 en la asociación
Porfiria Cutánea Tardia-VIH. J Basic Appl Genetics. 25(278)2014.(In
Spanish).
|
|
38
|
Solé X, Guinó E, Valls J, Iniesta R and
Moreno V: SNPStats: A web tool for the analysis of association
studies. Bioinformatics. 22:1928–1929. 2006.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Moher D, Liberati A, Tetzlaff J and Altman
DG: Preferred reporting items for systematic reviews and
meta-analyses: The PRISMA Statement. PLoS Med.
6(e1000097)2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Conde Almeida AC, Tadeu Villa R and Bedin
V: Porfiria cutánea tarda no paciente infectado pelo vírus da
imunodeficiência adquirida. Med Cutan Iber Lat Am. 38:91–93.
2010.
|
|
41
|
Quansah R, Cooper CJ, Said S, Bizet J,
Paez D and Hernandez GT: Hepatitis C- and HIV-induced porphyria
Cutanea Tarda. Am J Case Rep. 15:35–40. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Farabaugh PJ and Björk G: How
translational accuracy influences reading frame maintenance. EMBO
J. 18:1427–1434. 1999.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kimchi-Sarfaty V, Oh J, Kim I, Sauna Z,
Calcagno A, Ambudkar S and Gottesman MA: ‘Silent’ polymorphism in
the MDR1 gene changes substrate specificity. Science. 315:525–528.
2007.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wen J and Brogna S: Nonsense-mediated mRNA
decay. Biochem Soc Trans. 36:514–516. 2008.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kong LL, Zhuang XM, Yang HY, Yuan M, Xu L
and Li H: Inhibition of P-glycoprotein gene expression and function
enhances triptolide-induced hepatotoxicity in mice. Sci Rep.
5(11747)2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Sakurai A, Onishi Y, Hirano H, Seigneuret
M, Obanayama K, Kim G, Liew E, Sakaeda T, Yoshiura K, Niikawa N, et
al: Quantitative Structure-activity relationship analysis and
molecular dynamics simulation to functionally validate
nonsynonymous polymorphisms of human ABC Transporter ABCB1
(P-Glycoprotein/MDR1). Biochemistry. 46:7678–7693. 2007.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ozdemir S, Uludag A, Silan F, Atik SY,
Turgut B and Ozdemir O: Possible roles of the xenobiotic
transporter P-glycoproteins encoded by the MDR1 3435 C>T gene
polymorphism in differentiated thyroid cancers. Asian Pac J Cancer
Prev. 14:3213–7321. 2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Muralidharan N, Antony PT, Jain VK,
Mariaselvam CM and Negi VS: Multidrug resistance 1 (MDR1)
3435C>T gene polymorphism influences the clinical phenotype and
methotrexate-induced adverse events in South Indian Tamil
rheumatoid arthritis. Eur J Clin Pharmacol. 71:959–965.
2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Puoti M, Moioli MC, Travi G and Rossotti
R: The burden of liver disease in human immunodeficiency
virus-infected patients. Semin Liver Dis. 32:103–113.
2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Debes JD, Bohjanen PR and Boonstra A:
Mechanisms of accelerated liver fibrosis progression during HIV
infection. J Clin Transl Hepatol. 4:328–335. 2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ganesan M, Poluektova LY, Kharbanda KK and
Osna NA: Liver as a target of human immunodeficiency virus
infection. World J Gastroenterol. 24:4728–4737. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ganesan M, New-Aaron M, Dagur RS, Makarov
E, Wang W, Kharbanda KK, Kidambi S, Poluektova LY and Osna NA:
Alcohol Metabolism potentiates HIV-induced hepatotoxicity:
Contribution to End-stage liver disease. Biomolecules.
9(851)2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Bhattacharjee P, Paul S, Banerjee M, Patra
D, Banerjee P, Ghoshal N, Bandyopadhyay A and Giri AK: Functional
compensation of glutathione S-transferase M1 (GSTM1) null by
another GST superfamily member, GSTM2. Sci Rep.
3(2704)2013.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Doukali H, Ben Salah G, Hamdaoui L,
Hajjaji M, Tabebi M, Ammar-Keskes L, Masmoudi ME and Kamoun H:
Oxidative stress and glutathione S-transferase genetic
polymorphisms in medical staff professionally exposed to ionizing
radiation. Int J Radiat Biol. 93:697–704. 2017.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Datta SK, Kumar V, Pathak R, Tripathi AK,
Ahmed RS, Kalra OP and Banerjee BD: Association of glutathione
S-transferase M1 and T1 gene polymorphism with oxidative stress in
diabetic and nondiabetic chronic kidney disease. Ren Fail.
32:1189–1195. 2010.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Suvakov S, Damjanovic T, Stefanovic A,
Pekmezovic T, Savic-Radojevic A, Pljesa-Ercegovac M, Matic M,
Djukic T, Coric V, Jakovljevic J, et al: Glutathione S-transferase
A1, M1, P1 and T1 null or low-activity genotypes are associated
with enhanced oxidative damage among haemodialysis patients.
Nephrol Dial Transplant. 28:202–212. 2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Mhaidat N, Alshogran O, Khabour O, Alzoubi
K, Matalka I, Haddadin W, Mahasneh I and Aldaher A: Multi-drug
resistance 1 genetic polymorphism and prediction of chemotherapy
response in Hodgkin's Lymphoma. J Exp Clin Cancer Res.
30(68)2011.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Milojkovic M, Stojnev S, Jovanovic I,
Ljubisavljevic S, Stefanovic V and Sunder-Plassman R: Frequency of
the C1236T, G2677T/A and C3435T MDR1 gene polymorphisms in the
Serbian population. Pharmacol Rep. 63:808–814. 2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Komoto C, Nakamura T, Sakaeda T, Kroetz
DL, Yamada T, Omatsu H, Koyama T, Okamura N, Miki I, Tamura T, et
al: MDR1 haplotype frequencies in Japanese and Caucasian, and in
Japanese patients with colorectal cancer and esophageal cancer.
Drug Metab Pharmacokinet. 21:126–132. 2006.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Weich N, Roisman A, Cerliani B, Aráoz HV,
Chertkoff L, Richard SM, Slavutsky I, Larripa IB and Fundia AF:
Gene polymorphism profiles of drug-metabolising enzymes GSTM1,
GSTT1 and GSTP1 in an Argentinian population. Ann Hum Biol.
44:379–383. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Rossini A, Rapozo D, Amorim L, Macedo J,
Medina R, Neto J, Gallo C and Pinto L: Frequencies of GSTM1, GSTT1,
and GSTP1polymorphisms in a Brazilian population. Genet Mol Res.
1:233–240. 2002.PubMed/NCBI
|
|
62
|
Pinheiro D, Santos R, Brito R, Cruz A,
Ghedini P and Reis A: GSTM1/GSTT1 double-null genotype increases
risk of treatment-resistant schizophrenia: A genetic association
study in Brazilian patients. PLoS One. 12(e0183812)2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Klusek J, Nasierowska-Guttmejer A, Kowalik
A, Wawrzycka I, Lewitowicz P, Chrapek M and Głuszek S: GSTM1,
GSTT1, and GSTP1 polymorphisms and colorectal cancer risk in Polish
nonsmokers. Oncotarget. 9:21224–21230. 2018.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Srivastava DSL, Jain VK, Verma P and Yadav
JP: Polymorphism of glutathione S-transferase M1 and T1 genes and
susceptibility to psoriasis disease: A study from North India.
Indian J Dermatol Venereol Leprol. 84:39–44. 2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Stamenkovic M, Lukic V, Suvakov S, Simic
T, Sencanic I, Pljesa-Ercegovac M, Jaksic V, Babovic S, Matic M,
Radosavljevic A, et al: GSTM1-null and GSTT1-active genotypes as
risk determinants of primary open angle glaucoma among smokers. Int
J Ophthalmol. 11:1514–1520. 2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
ThekkePurakkal AS, Nicolau B, Burk RD,
Franco EL and Schlecht NF: Genetic variants in CYP and GST genes,
smoking and risk for head and neck cancers: A gene-environment
interaction hospital-based case-control study among Canadian
Caucasians. Carcinogenesis: Apr 2, 2019 doi: 10.1093/carcin/bgz051
(Epub ahead of print).
|
|
67
|
Musavi Z, Moasser E, Zareei N, Azarpira N
and Shamsaeefar A: Glutathione S-transferase gene polymorphisms and
the development of new-onset diabetes after liver transplant. Exp
Clin Transplant. 17:375–380. 2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zehra A, Zehra S, Ismail M and Azhar A:
Glutathione S-transferase M1 and T1 gene deletions and
susceptibility to acute lymphoblastic leukemia (ALL) in adults. Pak
J Med Sci. 34:666–670. 2018.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Saravani S, Miri-Moghaddam M, Bazi A and
Miri-Moghaddam E: Association of glutathione-S-transferases M1 and
T1 deletional variants with development of oral squamous cell
carcinoma: A study in the South-East of Iran. Asian Pac J Cancer
Prev. 20:1921–1926. 2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Farmohammadi A, Arab-Yarmohammadi V and
Ramzanpour R: Association analysis of rs1695 and rs1138272
variations in GSTP1 gene and breast cancer susceptibility. Asian
Pac J Cancer Prev. 21:1167–1172. 2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Oshodi Y, Ojewunmi O, Oshodi TA, Ijarogbe
GT, Ogun OC, Aina OF and Lesi F: Oxidative stress markers and
genetic polymorphisms of glutathione S-transferase T1, M1, and P1
in a subset of children with autism spectrum disorder in Lagos,
Nigeria. Niger J Clin Pract. 20:1161–1167. 2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Idris HM, Elderdery AY, Khalil HB and
Mills J: Genetic Polymorphism of GSTP1, GSTM1 and GSTT1 genes and
susceptibility to chronic myeloid leukaemia. Asian Pac J Cancer
Prev. 21:499–503. 2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Rebai A, Chbili C, Ben Amor S, Hassine A,
Ben Ammou S and Saguem S: Effects of glutathione S-transferase M1
and T1 deletions on Parkinson's disease risk among a North African
population. Rev Neurol (Paris): Apr 29, 2020 (Epub ahead of
print).
|