The polycomb group (PcG) is a class of highly
conserved transcriptional repressors. They are divided into two
core protein complexes: Polycomb repressive complex (PRC)1 and
PRC2. Both PRC1 and PRC2 serve an important role in the maintenance
of the inhibition state of chromatins by polycomb proteins. PRC2
binds to the target gene during the initial stage of transcription
and recruits the PRC1 complex to bind to the target gene, which
maintains the repressed state of the gene (1).
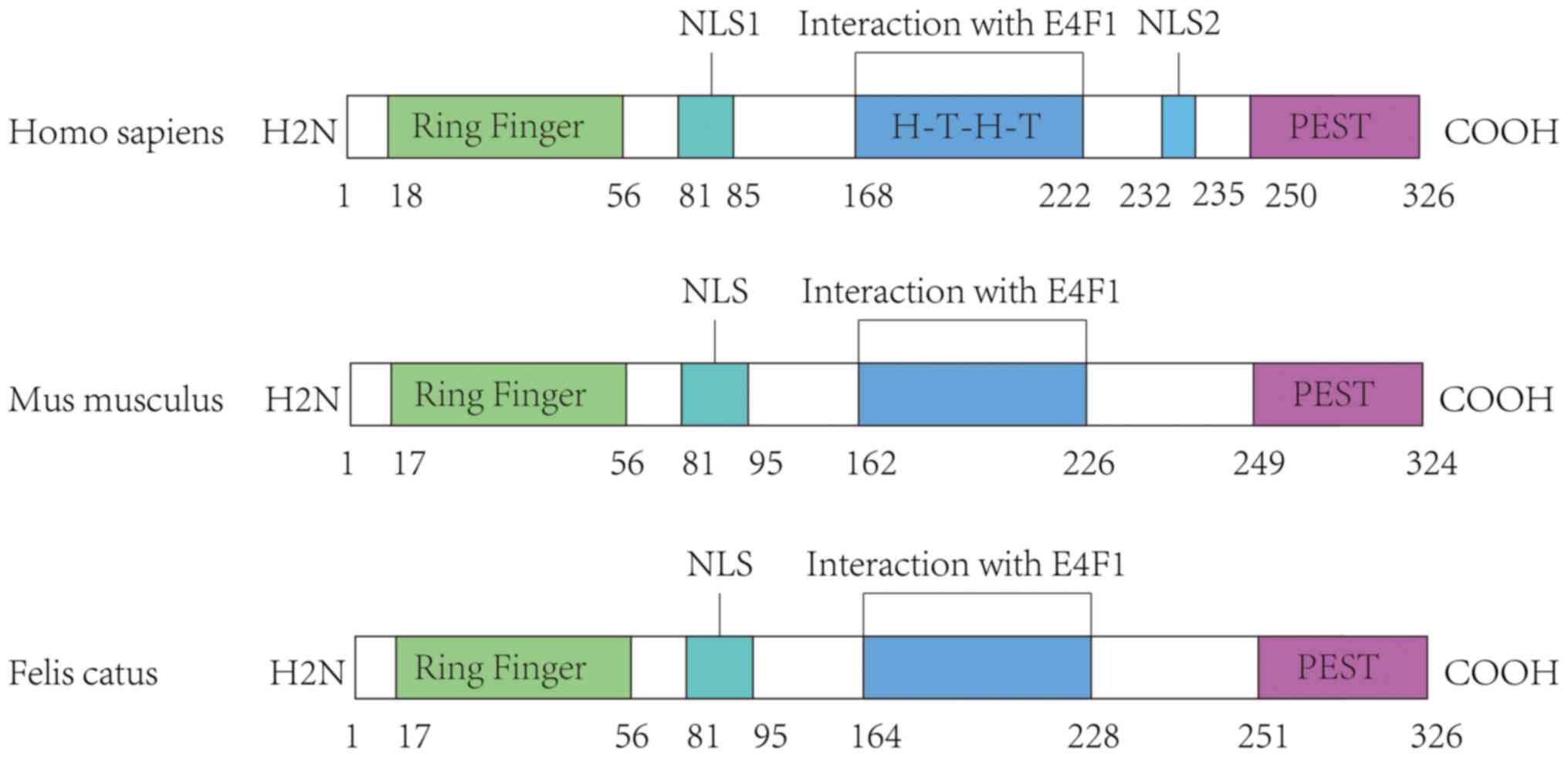
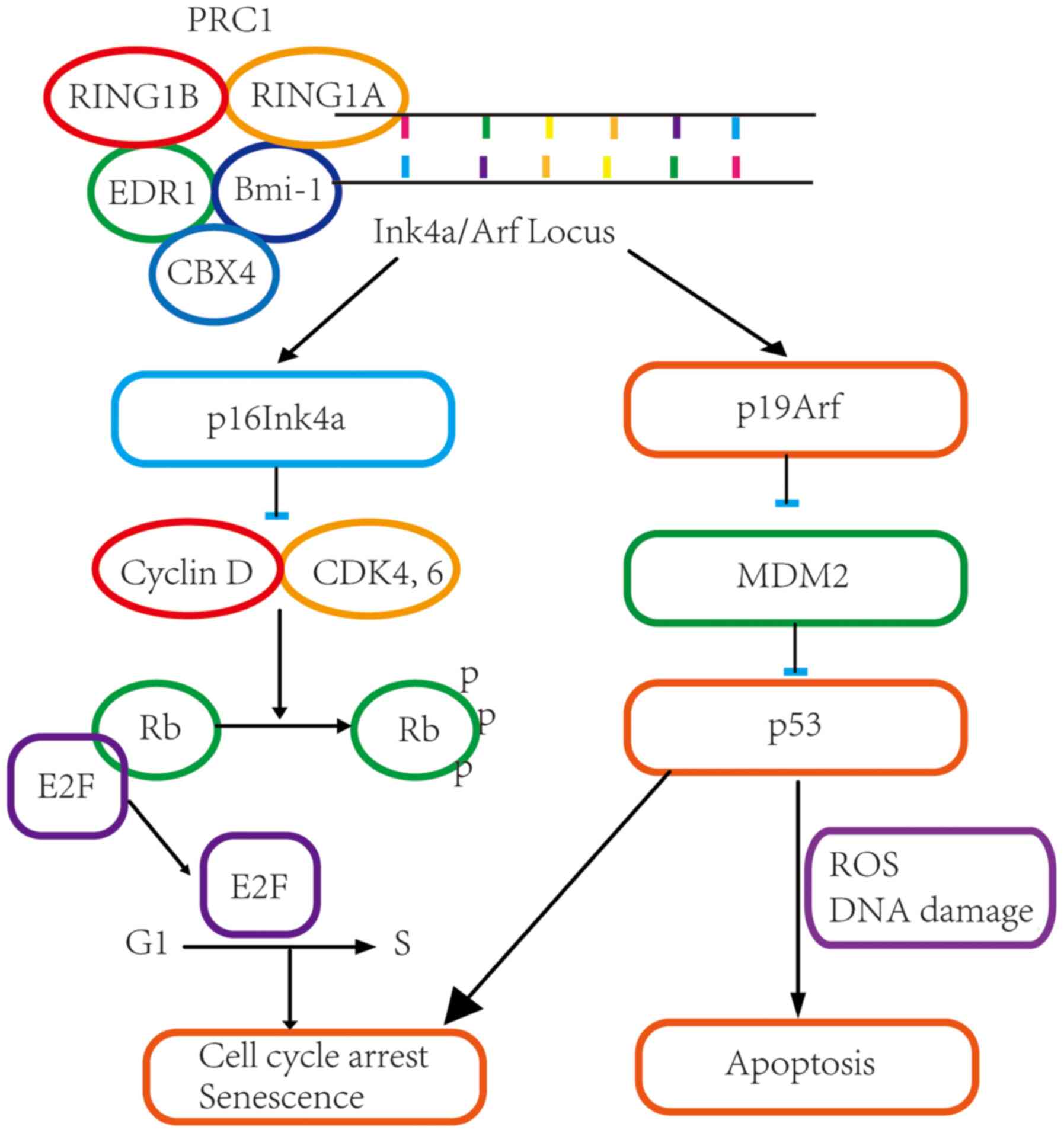
The PRC1 complex core proteins include RING1B (also
referred to as RNF2), RING1A (also referred to as RING1), B
lymphoma Mo-MLV insertion region 1 homolog (Bmi-1), EDR1 (also
referred to as HPH1) and CBX4 (also referred to as HPC2). Among
these components, Bmi-1 serves a pivotal role in the PRC1 complex
(1). The binding of Bmi-1 to
chromatin along with other PcG proteins of the PRC1 complex leads
to histone H3K27 methylation, which results in continued silencing
of the Ink4a/Arf locus. Decreased levels of p16Ink4a
and p19Arf lead to activation of nuclear E2F and
downregulation of p53(2), which in
turn promotes cell proliferation and self-renewal of cancer stem
cells (Fig. 1). In addition, Bmi-1
inhibits the expression of E-cadherin by interacting with
epithelial to mesenchymal transition (EMT) regulatory molecules,
such as Twist1, Wnt, Snail and β-catenin to induce EMT, whereas
inhibition of Bmi-1 expression leads to EMT reversal and decreased
cell migratory ability (3).
BMI-1 is a highly conserved gene with rare mutations. It
serves as a central node of various oncogenes and plays an
important role in cell proliferation and tumorigenesis. Multiple
signaling pathways, including N-Myc (MYCN), c-Myc, (MYC)
(4), twist (3), Akt (5),
and MAPK upregulate BMI-1 expression. Under normal growth
conditions, BMI-1 expression is maintained within
physiological levels through a feed-back loop that involves the PcG
family members, PRC1 and PRC2(6).
However, BMI-1 expression is upregulated in malignant cells,
partly due to stimulation by oncogenes, such as E2F-1 and
c-MYC, and this allows for the maintenance of an
undifferentiated state of the cells. BMI-1 overexpression is
a biomarker of malignant tumors and is closely related to tumor
malignancy, invasion, metastasis and prognosis (7). Therefore, inhibition of BMI-1
expression, restoration of p16Ink4a and p19Arf
levels, and induction of cellular senescence are novel potential
therapeutic targets for anti-cancer targeted therapy (8).
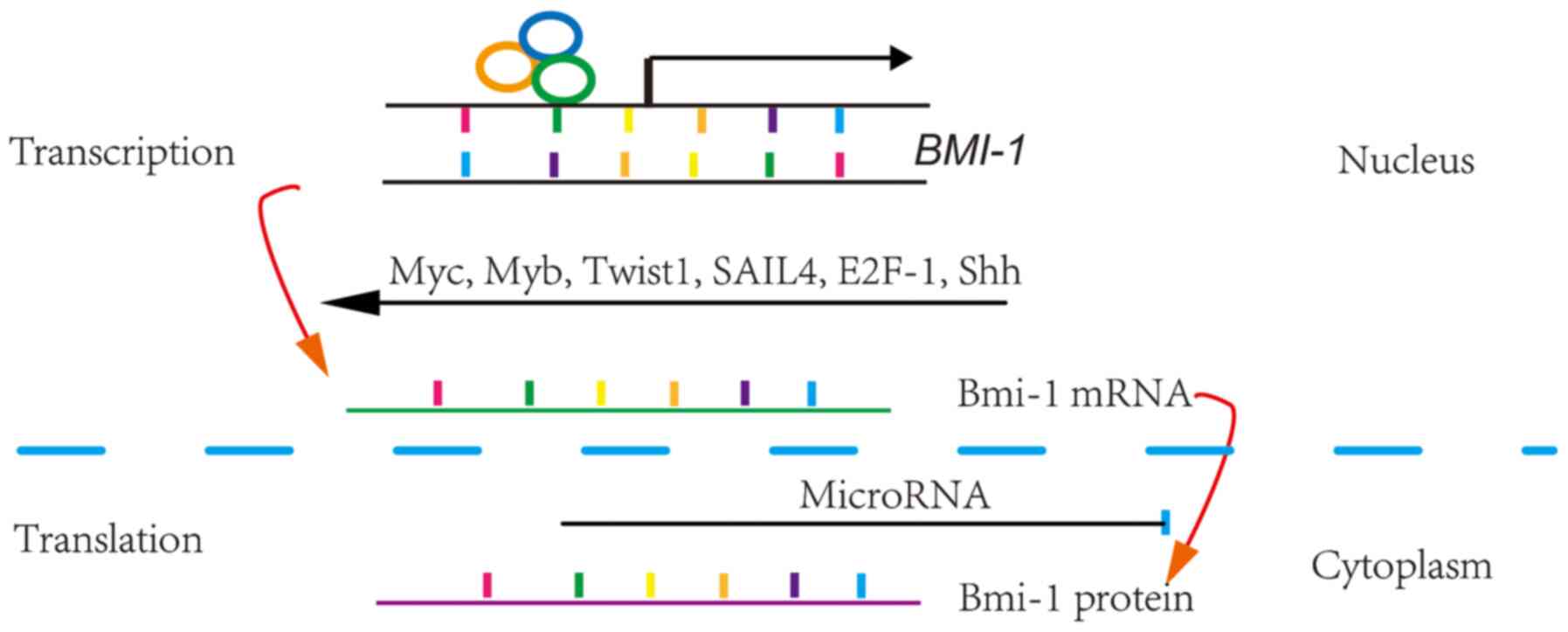
Bmi-1 is a short-lived protein, and its expression
levels are controlled by various mechanisms. Bmi-1 expression is
primarily controlled by transcriptional and post-transcriptional
regulation (1,8). Transcriptional regulation of eukaryotic
genes involves DNA methylation, histone modification, chromatin
remodeling and transcription factors. Post-transcriptional
regulation is predominantly achieved through regulation of RNA,
which includes RNA processing and maturation, RNA transport and
subcellular localization, mRNA translation and mRNA degradation
(9). In this review, the recent
advances in the understanding of transcriptional (Table I) and post-transcriptional regulation
(Table II) of BMI-1
expression are summarized.
The Myc family is a group of important oncogenes,
including MYC, MYCN, L-Myc, S-Myc and B-Myc. Of these, MYC and
MYCN are involved in BMI-1 transcription (4). MYC plays an important role in cell
proliferation, differentiation and apoptosis, and is abnormally
expressed in several types of cancer (17). In murine lymphoma and human malignant
glioma, both MYC and BMI-1 are highly expressed, and
exhibit a positive correlation with each other (18,19). The
MYC protein is a transcription factor of the basic helix-loop-helix
leucine zipper family. MYC forms a functional DNA-binding complex
with Max, another member of the same family, and this complex
specifically recognizes the E-box sequence (CACGTG) of the
BMI-1 gene promoter, thereby increasing the expression of
BMI-1 at the transcriptional level (4,19,20).
Twist1 belongs to the family of basic
helix-loop-helix transcription factors. It promotes EMT by
inhibiting E-cadherin expression (32). Mechanistically, Twist1 binds directly
to the E-cadherin promoter to inhibit its expression, and it also
directly binds to the E-box site of the -732 to -727 nucleotide
sequence in intron 1 of the BMI-1 promoter to initiate the
transcriptional upregulation of the BMI-1 gene (5). Twist1-mediated suppression of
E-cadherin and upregulation of Bmi-1 leads to disruption of the
tight junction between cells, thereby increasing tumor cell
metastasis (33,35).
SALL4 is a more recently identified zinc finger
transcription factor that plays an important role in the
maintenance of pluripotency of embryonic stem cells and the
self-renewal capacity of hematopoietic stem cells (36). Significant upregulation of SALL4 and
Bmi-1 expression has been reported in patients with myeloid
leukemia (37). Additionally, high
expression levels of these two genes were shown to be associated
with the expansion of hematopoietic progenitor cells. This suggests
that the expression of SALL4 and Bmi-1 is a prognostic biomarker of
acute leukemia. Results of luciferase reporter assays by Yang et
al (36) showed that the
BMI-1 gene is a target of SALL4, and, increased expression
of SALL4 was found to upregulate the activity of the BMI-1
promoter. Further analysis of the binding sites revealed that the
SALL4 protein binds to a functional site in the -450 to -1
nucleotide sequence of the BMI-1 promoter to initiate
transcription of the BMI-1 gene.
E2F-1 is a member of the E2F transcription factor
family. E2F-1 is involved in cell cycle progression and regulates
cell viability via both p53-independent and p53-dependent pathways
(38). E2F-1 initiates transcription
of the BMI-1 gene and upregulates BMI-1 expression by
directly binding to the E2F binding site in the BMI-1 gene
promoter; interestingly, when the cell is in the cell cycle or
differentiation phase, BMI-1 is not regulated by
E2F-1(39). In androgen-deficient
prostate cancer cells, the IκB kinase α (IKKα)-E2F1-Bmi-1 cascade
is activated. In these cells, activated IKKα phosphorylates E2F-1
to promote E2F-1 nuclear localization, whereby E2F-1 binds to the
co-activator CREB binding protein (histone H3 acetyltransferase)
and recruits the target genes, including BMI-1, thereby
resulting in activation of BMI-1 (40).
Hedgehog signaling is a major regulator of
vertebrate embryonic development, as it is involved in stem cell
maintenance and cell differentiation and proliferation. Abnormal
activation of the Hedgehog signaling pathway was shown to be
associated with the development of lung, prostate and breast cancer
(41). The primary components of the
Hedgehog signaling pathway include patched, Smoothened and
glioma-associated oncogene (GLI) (42). In a study by Liu et al
(43), addition of sonic Hedgehog to
activate the Hedgehog signaling pathway increased the expression of
Bmi-1, whereas inhibition of the Hedgehog signaling pathway using
cyclopamine resulted in downregulated expression of Bmi-1. Wang
et al (44) found that GLI1
binds to the promoter of the BMI-1 gene, and that the
BMI-1 transcription level changes in accordance with the
increase or decrease in GLI1 expression.
In addition to the above transcription factors,
several other factors may regulate BMI-1. Estrogen receptor
α activates the transcriptional activity of the BMI-1 gene
by interacting with the -327 to -172 bp nucleotide sequence
upstream of the BMI-1 promoter, thereby increasing BMI-1
gene expression (45). The
transcription factor Sp1 binds to the +181 to +214 region of the
BMI-1 promoter and increases BMI-1 gene expression (46). Krüppel-like factor 4 (KLF4) is a zinc
finger protein that is normally expressed in the intestines and
skin, and plays an important role in the regulation of stem cells.
Yu et al (47) found that
KLF4 binds directly to the -233 to 0 sequence of the BMI-1
gene promoter and inhibits the transcriptional activity of
BMI-1, thereby reducing the expression of Bmi-1. The binding
site of KLF4 to the BMI-1 gene is different from the binding
site of c-Myc to the BMI-1 promoter. Redox sensing factor
Nrf2 was shown to increase the expression of BMI-1 at the
transcriptional level, thereby promoting the proliferation of
cancer stem cells and inhibiting cancer cell apoptosis (48). The helix-loop-helix inhibitor of
differentiation and DNA binding facilitates tumorigenesis by
increasing the expression of Bmi-1 via c-Myc (49). Bommi et al (50) found that histone deacetylase
inhibitors (HDACi) inhibit BMI-1 gene transcription in
breast cancer cells via an indirect mechanism. In certain cancer
cell lines, c-Myc is the target gene of HDACi, whereas in
breast cancer cells, the inhibitory effect of HDACi on BMI-1
gene expression is not dependent on downregulation of c-Myc;
however, the precise mechanism is not clear. Thus, there are
various transcription factors that bind the promoter of
BMI-1 and regulate BMI-1 gene expression at the
transcriptional level.
Post-transcriptional regulation primary involves the
regulation of RNA and is divided into the following steps in
chronological order: RNA processing and maturation, RNA transport
and subcellular localization, mRNA translation and mRNA
degradation. MicroRNAs (miRNAs) block gene expression primarily by
preventing mRNA translation and/or promoting mRNA degradation
(51). miRNAs are non-coding,
short-stranded RNAs, which typically consist of 18-22 nucleotides.
An miRNA complements the 3'-untranslated region (UTR) of its target
mRNA and directs RNA-induced silencing complex to a specific region
of the mRNA, thereby inhibiting mRNA translation or promoting mRNA
degradation (8).
Bmi-1 expression is inhibited by several miRNAs,
including miR-135a, miR-141, miR-183, miR-15a, miR-194, miR-203,
miR-200b, miR-320a, miR-200c, miR-16, miR-495, miR-221, miR-30d,
miR-128a, miR-34a, miR-452, miR-302 and miR-30e (51-67).
For example, the expression of miR-218 in cancer tissues is lower
than that in the paratumoral normal tissues, whereas the expression
of Bmi-1 in cancer tissues is higher than that in the paratumoral
normal tissues. An inverse correlation between Bmi-1 expression and
miR-128 has been demonstrated in glioma and rectal cancer cells.
Results of luciferase reporter assays showed that miR-128 inhibits
Bmi-1 protein expression by binding to the 476 to 488 region of
BMI-1 3'-UTR (54). several
transcription factors and cytokines affect the expression levels of
Bmi-1 by altering the expression of miRNAs. For example, Zeb1 was
shown to downregulate Bmi-1 expression by inducing upregulation of
the expressions of miR-203 and miR-183(32).
The polycomb family protein member Bmi-1 acts as an
oncogene and maintains the undifferentiated state of malignant
tumor cells. Bmi-1 expression levels are closely related to the
degree of malignancy, invasion and metastasis, and is a biomarker
of adverse prognosis in cancer patients. As a pivotal node of
multiple signaling pathways, Bmi-1 regulates the function of
several downstream transcription factors and cytokines. Therefore,
inhibition of Bmi-1 expression is a promising strategy for
anticancer drug development. It has been shown that NVP-LDE-225
(Erismodegib) inhibits Bmi-1 expression by inducing upregulation of
miR-128(68). In addition, PTC-209
is a small molecule drug that specifically inhibits Bmi-1
expression at the post-transcriptional level by binding to the 5'
and 3' non-coding regions of BMI-1 mRNA (69). Transcriptional and
post-transcriptional regulation are the primary means of regulation
of Bmi-1 expression. Therefore, regulatory factors are potential
therapeutic targets to reduce Bmi-1 expression in cancer cells.
Not applicable.
Funding: This study was supported by the National Nature Science
Foundation of China (grant no. 82060450), Nature Science Foundation
of Jiangxi province of China (grant nos. 20192BAB205072 and
20181BAB205050), and The Education Department of Jiangxi Province
of China (grant no. 160032).
Not applicable.
MZ and QX searched the literature, reviewed the
articles and collected the relevant data from the selected papers.
DH and LL wrote the manuscript. All authors have read and approved
the final manuscript.
Not applicable.
Not applicable.
All authors declare no competing interests.
|
1
|
Park IK, Qian D, Kiel M, Becker MW,
Pihalja M, Weissman IL, Morrison SJ and Clarke MF: Bmi-1 is
required for maintenance of adult self-renewing haematopoietic stem
cells. Nature. 423:302–305. 2003.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Park IK, Morrison SJ and Clarke MF: Bmi1,
stem cells, and senescence regulation. J Clin Invest. 113:175–179.
2004.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY,
Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, et al: Bmi1 is
essential in Twist1-induced epithelial-mesenchymal transition. Nat
Cell Biol. 12:982–992. 2010.PubMed/NCBI View
Article : Google Scholar : Erratum in: Nat Cell
Biol 21, 533, 2019.
|
|
4
|
Adhikary S and Eilers M: Transcriptional
regulation and transformation by Myc proteins. Nat Rev Mol Cell
Biol. 6:635–645. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Kim J, Hwangbo J and Wong PK: p38
MAPK-Mediated Bmi-1 down-regulation and defective proliferation in
ATM-deficient neural stem cells can be restored by Akt activation.
PLoS One. 6(e16615)2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Cao R, Tsukada Y and Zhang Y: Role of
Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol
Cell. 20:845–854. 2005.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bhattacharyya J, Mihara K, Ohtsubo M,
Yasunaga S, Takei Y, Yanagihara K, Sakai A, Hoshi M, Takihara Y and
Kimura A: Overexpression of BMI-1 correlates with drug resistance
in B-cell lymphoma cells through the stabilization of survivin
expression. Cancer Sci. 103:34–41. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cao L, Bombard J, Cintron K, Sheedy J,
Weetall ML and Davis TW: BMI1 as a novel target for drug discovery
in cancer. J Cell Biochem. 112:2729–2741. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Venkatesh S and Workman JL: Histone
exchange, chromatin structure and the regulation of transcription.
Nat Rev Mol Cell Biol. 16:178–189. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Siddique HR, Parray A, Tarapore RS, Wang
L, Mukhtar H, Karnes RJ, Deng Y, Konety BR and Saleem M: BMI1
polycomb group protein acts as a master switch for growth and death
of tumor cells: Regulates TCF4-transcriptional factor-induced BCL2
signaling. PLoS One. 8(e60664)2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Alkema MJ, Wiegant J, Raap AK, Berns A and
van Lohuizen M: Characterization and chromosomal localization of
the human proto-oncogene BMI-1. Hum Mol Genet. 2:1597–1603.
1993.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Freemont PS, Hanson IM and Trowsdale J: A
novel cysteine-rich sequence motif. Cell. 64:483–484.
1991.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Dimri GP, Martinez JL, Jacobs JJL,
Keblusek P, Itahana K, Van Lohuizen M, Campisi J, Wazer DE and Band
V: The Bmi-1 oncogene induces telomerase activity and
immortalizes human mammary epithelial cells. Cancer Res.
62:4736–4745. 2002.PubMed/NCBI
|
|
14
|
Cohen KJ, Hanna JS, Prescott JE and Dang
CV: Transformation by the Bmi-1 oncoprotein correlates with its
subnuclear localization but not its transcriptional suppression
activity. Mol Cell Biol. 16:5527–5535. 1996.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Buchwald G, van der Stoop P, Weichenrieder
O, Perrakis A, van Lohuizen M and Sixma TK: Structure and E3-ligase
activity of the Ring-Ring complex of polycomb proteins Bmi1 and
Ring1b. EMBO J. 25:2465–2474. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rogers S, Wells R and Rechsteiner M: Amino
acid sequences common to rapidly degraded proteins: The PEST
hypothesis. Science. 234:364–368. 1986.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Hoffman B and Liebermann DA: Apoptotic
signaling by c-MYC. Oncogene. 27:6462–6472. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jacobs JJL, Scheijen B, Voncken JW,
Kieboom K, Berns A and van Lohuizen M: Bmi-1 collaborates with
c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via
INK4a/ARF. Genes Dev. 13:2678–2690. 1999.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Cenci T, Martini M, Montano N,
D'Alessandris QG, Falchetti ML, Annibali D, Savino M, Bianchi F,
Pierconti F, Nasi S, et al: Prognostic relevance of c-Myc and BMI1
expression in patients with glioblastoma. Am J Clin Pathol.
138:390–396. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guo WJ, Datta S, Band V and Dimri GP:
Mel-18, a polycomb group protein, regulates cell proliferation and
senescence via transcriptional repression of Bmi-1 and c-Myc
oncoproteins. Mol Biol Cell. 18:536–546. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Weiss WA, Aldape K, Mohapatra G,
Feuerstein BG and Bishop JM: Targeted expression of MYCN causes
medulloblastoma in transgenic mice. EMBO J. 16:2985–2995.
1997.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ochiai H, Takenobu H, Nakagawa A,
Yamaguchi Y, Kimura M, Ohira M, Okimoto Y, Fujimura Y, Koseki H,
Kohno Y, et al: Bmi1 is a MYCN target gene that regulates
tumorigenesis through repression of KIF1Bβ and TSLC1
in neuroblastoma. Oncogene. 29:2681–2690. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang R, Cheung NKV, Vider J, Cheung IY,
Gerald WL, Tickoo SK, Holland EC and Blasberg RG: MYCN and MYC
regulate tumor proliferation and tumorigenesis directly through
BMI1 in human neuroblastomas. FASEB J. 25:4138–4149.
2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ishida A, Asano H, Hasegawa M, Koseki H,
Ono T, Yoshida MC, Taniguchi M and Kanno M: Cloning and chromosome
mapping of the human Mel-18 gene which encodes a DNA-binding
protein with a new ‘RING-finger’ motif. Gene. 129:249–255.
1993.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tetsu O, Ishihara H, Kanno R, Kamiyasu M,
Inoue H, Tokuhisa T, Taniguchi M and Kanno M: mel-18 negatively
regulates cell cycle progression upon B cell antigen receptor
stimulation through a cascade leading to c-myc/cdc25. Immunity.
9:439–448. 1998.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Guo WJ, Zeng MS, Yadav A, Song LB, Guo BH,
Band V and Dimri GP: Mel-18 acts as a tumor suppressor by
repressing Bmi-1 expression and down-regulating Akt activity in
breast cancer cells. Cancer Res. 67:5083–5089. 2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu-Bryan R and Terkeltaub R: Chondrocyte
innate immune myeloid differentiation factor 88-dependent signaling
drives procatabolic effects of the endogenous Toll-like receptor
2/Toll-like receptor 4 ligands low molecular weight hyaluronan and
high mobility group box chromosomal protein 1 in mice. Arthritis
Rheum. 62:2004–2012. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li SKM, Smith DK, Leung WY, Cheung AM, Lam
EW, Dimri GP and Yao KM: FoxM1c counteracts oxidative
stress-induced senescence and stimulates Bmi-1 expression. J Biol
Chem. 283:16545–16553. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cheasley D, Pereira L, Lightowler S,
Vincan E, Malaterre J and Ramsay RG: Myb controls intestinal stem
cell genes and self-renewal. Stem Cells. 29:2042–2050.
2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Vargova K, Curik N, Burda P, Basova P,
Kulvait V, Pospisil V, Savvulidi F, Kokavec J, Necas E, Berkova A,
et al: MYB transcriptionally regulates the miR-155 host gene in
chronic lymphocytic leukemia. Blood. 117:3816–3825. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Waldron T, De Dominici M, Soliera AR,
Audia A, Iacobucci I, Lonetti A, Martinelli G, Zhang Y, Martinez R,
Hyslop T, et al: c-Myb and its target Bmi1 are required for
p190BCR/ABL leukemogenesis in mouse and human cells. Leukemia.
26:644–653. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Martin A and Cano A: Tumorigenesis: Twist1
links EMT to self-renewal. Nat Cell Biol. 12:924–925.
2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang MH, Hsu DSS, Wang HW, Wang HJ, Lan
HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, et al: Bmi1 is
essential in Twist1-induced epithelial-mesenchymal transition. Nat
Cell Biol. 12:982–992. 2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu KJ: Direct activation of Bmi1 by
Twist1: Implications in cancer stemness, epithelial-mesenchymal
transition, and clinical significance. Chang Gung Med J.
34:229–238. 2011.PubMed/NCBI
|
|
35
|
Wu CY, Hung JJ and Wu KJ: Linkage between
Twist1 and Bmi1: Molecular mechanism of cancer metastasis/stemness
and clinical implications. Clin Exp Pharmacol Physiol. 39:668–673.
2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yang J, Chai L, Liu F, Fink LM, Lin P,
Silberstein LE, Amin HM, Ward DC and Ma Y: Bmi-1 is a target
gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad
Sci USA. 104:10494–10499. 2007.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Vander Griend DJ, D'Antonio J, Gurel B,
Antony L, Demarzo AM and Isaacs JT: Cell-autonomous intracellular
androgen receptor signaling drives the growth of human prostate
cancer initiating cells. Prostate. 70:90–99. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Irwin M, Marin MC, Phillips AC, Seelan RS,
Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH, et al:
Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature.
407:645–648. 2000.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Nowak K, Kerl K, Fehr D, Kramps C, Gessner
C, Killmer K, Samans B, Berwanger B, Christiansen H and Lutz W:
BMI1 is a target gene of E2F-1 and is strongly expressed in
primary neuroblastomas. Nucleic Acids Res. 34:1745–1754.
2006.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ammirante M, Kuraishy AI, Shalapour S,
Strasner A, Ramirez-Sanchez C, Zhang W, Shabaik A and Karin M: An
IKKα-E2F1-BMI1 cascade activated by infiltrating B cells controls
prostate regeneration and tumor recurrence. Genes Dev.
27:1435–1440. 2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Gupta S, Takebe N and Lorusso P: Targeting
the Hedgehog pathway in cancer. Ther Adv Med Oncol. 2:237–250.
2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Briscoe J and Thérond PP: The mechanisms
of Hedgehog signalling and its roles in development and disease.
Nat Rev Mol Cell Biol. 14:416–429. 2013.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Liu S, Dontu G, Mantle ID, Patel S, Ahn
NS, Jackson KW, Suri P and Wicha MS: Hedgehog signaling and Bmi-1
regulate self-renewal of normal and malignant human mammary stem
cells. Cancer Res. 66:6063–6071. 2006.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang X, Venugopal C, Manoranjan B,
McFarlane N, O'Farrell E, Nolte S, Gunnarsson T, Hollenberg R,
Kwiecien J, Northcott P, et al: Sonic hedgehog regulates Bmi1 in
human medulloblastoma brain tumor-initiating cells. Oncogene.
31:187–199. 2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang H, Liu H, Li X, Zhao J, Zhang H, Mao
J, Zou Y, Zhang H, Zhang S, Hou W, et al: Estrogen receptor
α-coupled Bmi1 regulation pathway in breast cancer and its clinical
implications. BMC Cancer. 14(122)2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wang HB, Liu GH, Zhang H, Xing S, Hu LJ,
Zhao WF, Xie B, Li MZ, Zeng BH, Li Y, et al: Sp1 and c-Myc regulate
transcription of BMI1 in nasopharyngeal carcinoma. FEBS J.
280:2929–2944. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yu T, Chen X, Zhang W, Colon D, Shi J,
Napier D, Rychahou P, Lu W, Lee EY, Weiss HL, et al: Regulation of
the potential marker for intestinal cells, Bmi1, by β-catenin and
the zinc finger protein KLF4: Implications for colon cancer. J Biol
Chem. 287:3760–3768. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Jia Y, Chen J, Zhu H, Jia ZH and Cui MH:
Aberrantly elevated redox sensing factor Nrf2 promotes cancer stem
cell survival via enhanced transcriptional regulation of ABCG2 and
Bcl-2/Bmi-1 genes. Oncol Rep. 34:2296–2304. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Qian T, Lee JY, Park JH, Kim HJ and Kong
G: Id1 enhances RING1b E3 ubiquitin ligase activity through the
Mel-18/Bmi-1 polycomb group complex. Oncogene. 29:5818–5827.
2010.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Bommi PV, Dimri M, Sahasrabuddhe AA,
Khandekar J and Dimri GP: The polycomb group protein BMI1 is a
transcriptional target of HDAC inhibitors. Cell Cycle. 9:2663–2673.
2010.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Slaby O, Svoboda M, Michalek J and Vyzula
R: MicroRNAs in colorectal cancer: Translation of molecular biology
into clinical application. Mol Cancer. 8(102)2009.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Bhattacharyya J, Mihara K, Yasunaga S,
Tanaka H, Hoshi M, Takihara Y and Kimura A: BMI-1 expression is
enhanced through transcriptional and posttranscriptional regulation
during the progression of chronic myeloid leukemia. Ann Hematol.
88:333–340. 2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca
EA and Lawler S: Targeting of the Bmi-1 oncogene/stem cell renewal
factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008.PubMed/NCBI View Article : Google Scholar
|
|
55
|
He X, Dong Y, Wu CW, Zhao Z, Ng SS, Chan
FK, Sung JJ and Yu J: MicroRNA-218 inhibits cell cycle progression
and promotes apoptosis in colon cancer by downregulating BMI1
polycomb ring finger oncogene. Mol Med. 18:1491–1498.
2013.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Dang Z, Xu WH, Lu P, Wu N, Liu J, Ruan B,
Zhou L, Song WJ and Dou KF: MicroRNA-135a inhibits cell
proliferation by targeting Bmi1 in pancreatic ductal
adenocarcinoma. Int J Biol Sci. 10:733–745. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Xu L, Li Y, Yan D, He J and Liu D:
MicroRNA-183 inhibits gastric cancer proliferation and invasion via
directly targeting Bmi-1. Oncol Lett. 8:2345–2351. 2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wu C, Zheng X, Li X, Fesler A, Hu W, Chen
L, Xu B, Wang Q, Tong A, Burke S, et al: Reduction of gastric
cancer proliferation and invasion by miR-15a mediated suppression
of Bmi-1 translation. Oncotarget. 7:14522–14536. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Dong P, Kaneuchi M, Watari H, Hamada J,
Sudo S, Ju J and Sakuragi N: MicroRNA-194 inhibits epithelial to
mesenchymal transition of endometrial cancer cells by targeting
oncogene BMI-1. Mol Cancer. 10(99)2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Dimri M, Carroll JD, Cho JH and Dimri GP:
microRNA-141 regulates BMI1 expression and induces senescence in
human diploid fibroblasts. Cell Cycle. 12:3537–3546.
2013.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Sugihara H, Ishimoto T, Watanabe M,
Sawayama H, Iwatsuki M, Baba Y, Komohara Y, Takeya M and Baba H:
Identification of miR-30e* regulation of Bmi1 expression
mediated by tumor-associated macrophages in gastrointestinal
cancer. PLoS One. 8(e81839)2013.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Yu J, Lu Y, Cui D, Li E, Zhu Y, Zhao Y,
Zhao F and Xia S: miR-200b suppresses cell proliferation, migration
and enhances chemosensitivity in prostate cancer by regulating
Bmi-1. Oncol Rep. 31:910–918. 2014.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhang Y, Zhou SY, Yan HZ, Xu DD, Chen HX,
Wang XY, Wang X, Liu YT, Zhang L, Wang S, et al: miR-203 inhibits
proliferation and self-renewal of leukemia stem cells by targeting
survivin and Bmi-1. Sci Rep. 6(19995)2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Qi X, Li J, Zhou C, Lv C and Tian M:
MicroRNA-320a inhibits cell proliferation, migration and invasion
by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett.
588:3732–3738. 2014.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Liu S, Tetzlaff MT, Cui R and Xu X:
miR-200c inhibits melanoma progression and drug resistance through
down-regulation of BMI-1. Am J Pathol. 181:1823–1835.
2012.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Bhattacharya R, Nicoloso M, Arvizo R, Wang
E, Cortez A, Rossi S, Calin GA and Mukherjee P: miR-15a and miR-16
control Bmi-1 expression in ovarian cancer. Cancer Res.
69:9090–9095. 2009.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Venkataraman S, Alimova I, Fan R, Harris
P, Foreman N and Vibhakar R: MicroRNA 128a increases intracellular
ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer
cell growth by promoting senescence. PLoS One.
5(e10748)2010.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Nanta R, Kumar D, Meeker D, Rodova M, Van
Veldhuizen PJ, Shankar S and Srivastava RK: NVP-LDE-225
(Erismodegib) inhibits epithelial-mesenchymal transition and human
prostate cancer stem cell growth in NOD/SCID IL2Rγ null mice by
regulating Bmi-1 and microRNA-128. Oncogenesis.
2(e42)2013.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Kreso A, van Galen P, Pedley NM,
Lima-Fernandes E, Frelin C, Davis T, Cao L, Baiazitov R, Du W,
Sydorenko N, et al: Self-renewal as a therapeutic target in human
colorectal cancer. Nat Med. 20:29–36. 2014.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Li Y, Hu J, Guan F, Song L, Fan R, Zhu H,
Hu X, Shen E and Yang B: Copper induces cellular senescence in
human glioblastoma multiforme cells through downregulation of
Bmi-1. Oncol Rep. 29:1805–1810. 2013.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Xuan HQ, Xue W, Pan JH, Sha JJ, Dong BJ
and Huang YR: Downregulation of miR-221,-30d, and-15a contributes
to pathogenesis of prostate cancer by targeting Bmi-1. Biochemistry
(Mosc). 80:276–283. 2015.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Guo JL, Li WP, Shi HL, Xie XH, Li LS, Tang
HL, Wu MQ, Kong Y, Yang L, Gao J, et al: Synergistic effects of
curcumin with emodin against the proliferation and invasion of
breast cancer cells through upregulation of miR-34a. Mol Cell
Biochem. 382:103–111. 2013.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Lin SL, Chang DC, Ying SY, Leu D and Wu
DTS: MicroRNA miR-302 inhibits the tumorigenecity of human
pluripotent stem cells by coordinate suppression of the CDK2 and
CDK4/6 cell cycle pathways. Cancer Res. 70:9473–9482.
2010.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Li TT, Jian XY, He H, Lai QH, Li XZ, Deng
DL, Liu TF, Zhu JH, Jiao HL, Ye YP, et al: MiR-452 promotes an
aggressive colorectal cancer phenotype by regulating a
Wnt/β-catenin positive feedback loop. J Exp Clin Cancer Res.
37(238)2018.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Liu LP, Chen K, Wu JH, Shi L, Hu B, Cheng
SY, Li MF and Song LB: Downregulation of miR-452 promotes stem-like
traits and tumorigenicity of gliomas. Clin Cancer Res.
19:3429–3438. 2013.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Li X, Song Y, Liu D, Zhao J, Xu J, Ren J,
Hu Y, Wang Z, Hou Y and Zhao G: MiR-495 promotes senescence of
mesenchymal stem cells by targeting Bmi-1. Cell Physiol Biochem.
42:780–796. 2017.PubMed/NCBI View Article : Google Scholar
|