Introduction
Rheumatoid arthritis (RA), a chronic inflammatory
disease, is primarily characterized by the symmetrical distribution
of invasive joint inflammation of the hands, feet, blood vessels or
related connective tissues (1,2). RA
occurs in patients of all ages, and is a serious health burden as
well as an economic and social burden (3).
In general, genetic and environmental factors are
the primary factors attributed to RA, and immune disorders are
considered to be the primary pathogenic factors (4-6).
Exogenous or endogenous antigens activate T cells, which release
inflammatory cytokines. T cells activate B cells to produce a
series of antibodies that induce immune disorders, which damage
various types of tissues (7).
However, the mechanism by which the immune disorder is induced, and
how the disease further worsens during the process of RA is not
well explained and requires in-depth research. The efficacy of
several drugs used to alleviate the pain of patients with RA is
limited, and the side effects are evident (8). Therefore, the development of novel and
effective drugs for RA treatment is required.
Expression profiling is increasingly used to analyze
disease-related genes or signaling pathways to determine the
pathogenesis of numerous diseases (9,10).
T-cell surface antigen CD2 (CD2) can interact with lymphocyte
function associated antigens CD58 and CD48/BCM1 to mediate adhesion
between T cells and other cell types. CD2 is also involved in T
cell induction, and the cytoplasmic domain is involved in signal
transduction. It also serves an important role in the inflammatory
response. LLDT-8 is a novel RA drug. Pharmacological analysis has
shown that LLDT-8 is a new diterpene compound of Tripterygium
wilfordii with low toxicity and high efficiency and strong
immunosuppressive activity in vitro and in vivo. In
addition, it has been reported that LLDT-8 inhibits
osteoclastogenesis via regulation of the RANKL/RANK/OPG signaling
pathway (11). LLDT-8 protects
against cerebral ischemia/reperfusion injury by suppressing
post-stroke inflammation (12).
LLDT-8 can protect against bleomycin-induced lung fibrosis in mice
(13). However, the effect of LLDT-8
as a treatment for RA remains to be further studied.
In the present study, differentially expressed genes
(DEGs) related to RA were screened through bioinformatics analysis
of the expression profile data of clinical RA and normal tissue
samples. A key node gene regulating RA was further selected using
topological analysis. Finally, traditional Chinese medicine (TCM)
libraries were molecularly screened on the basis of the key node
gene to identify potential therapeutic drugs.
Materials and methods
Microarray data information and data
preprocessing
GSE55235 and GSE84074 microarray data were
downloaded from the GEO database (ncbi.nlm.nih.gov/geo/) (14). The GSE55235 chip data was based on
GPL96, [hg-u133a] Affymetrix Human Genome U133A Array (15). The Affymetrix, calif. GSE84074 chip
was based on the GPL19640, agilent-062918 Human lncRNA array
version 4.0 (Probe name version) microarray data in Bioconductor
(version 1.46.1; bioconductor.org/) (16). The preprocessing stage included
background correction, normalization and expression calculation.
The Bioconductor Annotation data package was used to convert the
chip data probe into gene symbols. If multiple probes were mapped
to a gene symbol, the average was set for the final expression
value of the gene. The DEGs of each group were analyzed using the
limma package (17). The DEGs were
analyzed using the volcano map script in the R studio (version
1.2.5042) (18,19). P<0.05 and |log(fold change)
FC|>2 were used as the cutoff criteria. The Heatmap package was
used for hierarchical clustering analysis and visualization of DEGs
(20).
GO and pathway enrichment
analysis
The Clue-GO plugin in Cytoscape (21) was used to analyze the functions and
pathways of the DEGs. P<0.05 was used as the cutoff criteria.
Enrichment analysis was performed and visualized using ClueGO and
CluePedia with P<0.05 as the cutoff criterion (22-24).
The function and pathway enrichment analysis of DEGs following
LLDT-8 processing was performed using the clusterProfiler package
(25).
Cell culture
Synoviocytes were purchased from Shanghai Institute
of Biochemistry and Cell Biology. Synovial cells were grown in DMEM
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
in a humidified incubator at 37˚C with 5% CO2.
Cell transfection
Cells in the logarithmic growth phase were used for
transfection. A total of 4x105 cells/well were plated in
a 6-well plate and 2 ml supplemented media without antibiotics was
added. Transfection was performed when the confluence of cells
reached 70%. For transfection, the negative control small
interfering (si)RNA was used, and for the experimental group si-CD2
was added. Transfections were performed using Lipofectamine™ 2000
(Thermo Fisher Scientific, Inc.). The final siRNA concentration
used was 40 nmol/l. The sequences of the siRNAs were: si-NC,
5'-UUCUCCGAACGUGUCACG UTT-3' and 5'-ACGUGACACGUUCGGAGAATT-3'; and
for si-CD2, 5'-CCAAAGGUGCAGUCUCCAATT-3' and 5'-UUG
GAGACUGCACCUUUGGTT-3'. The siRNAs were purchased from Santa Cruz
Biotechnology, Inc. Follow-up experiments were performed 48 h after
transfection.
Cell proliferation assay
Cell proliferation was detected using a Cell
Counting Kit-8 (CCK-8) assay. A total of 1x103 cells in
200 µl were added per well to a 96-well plate. Following the
different treatments, 10 µl CCK-8 solution (Dojindo Molecular
Technologies, Inc.) was added to each well, and the plate was
further incubated for 2 h in a 5% CO2 incubator. The
absorbance of cells was measured using a microplate reader at 450
nm. All experiments were repeated 3 times.
Reverse transcription-quantitative
(RT-q)PCR
Cells were seeded in 6-well plates containing 2 ml
complete medium at a density of 1x106. The 6-well plates
were incubated at 37˚C, 5% CO2 and 100% humidity for 24
h. TNF-α (10 ng/ml; cat. no. 14-8329-62; Thermo Fisher Scientific,
Inc.) or IL-1β (10 mg/l; cat. no. SRP3083; Sigma-Aldrich; Merck
KGaA) was added to stimulate the cells for 24 h. Total RNA was
extracted using TRIzol® (cat. no. 15596026; Thermo
Fisher Scientific, Inc.). Isolated RNA was reverse transcribed into
cDNA using a reverse transcription kit according to the
manufacturer's protocol (cat. no. RR036A; Takara Bio, Inc.). The
sequences of the primers were: CD2 forward,
5'-CCCATGATTCCTTCATATTTGCA-3' and reverse, 5'-
GGTCCTTCTCCAGCCTAGT-3'; IL-6 forward, 5'-CCACTCACCTCTTCAGAACG-3'
and reverse, 5'-CATCTT TGGAAGGTTCAGGTTG-3'; LDH forward,
5'-GTTGGAACT GGTGCCGTAG-3' and reverse, 5'-GAAAAGACTGCCATG
CTGAAG-3'; IL-1β forward, 5'-CCTGTCCTGCGTGTTGAA AGA-3' and reverse,
5'-GGGAACTGGGCAGACTCAAA-3'; and β-actin forward,
5'-GATCTTGATCTTCATGGTGCTAG-3' and reverse,
5'-TTGTAACCACCTGGGACGATATGG-3'. β-actin was used as the internal
reference. For qPCR, 10 µl cDNA was used per sample, and the
thermocycling conditions were: 95˚C for 5 min; followed by 40
cycles of 95˚C for 10 sec and 60˚C for 30 sec. The
2-ΔΔCq method was used for relative quantitative
analysis (26).
Detection of cellular inflammatory
factors
LDH and IL-6 ELISA kits (Beijing Solarbio Science
& Technology Co., Ltd.) were used to detect the levels of
inflammatory factors in cells following the different treatments,
according to the manufacturer's protocol.
Protein-protein interaction (PPI)
networks
The CentiScape plugin (27) was used to calculate the node degree,
and the molecular complex detection (MCODE) plugin (28) was employed to find clusters in the
entire PPI network. Nodal proteins may have important physiological
regulatory functions and may be key candidate genes. The genes in
the most significant modules were extracted for functional
enrichment analysis. P<0.05 was set as the cutoff value.
Integration of the PPI network
The online database STRING (string-db.org) was used to construct the PPI network
of DEGs (29). Cytoscape was used to
construct the protein interaction network and analyze the
interactive relationship of the candidate DEGs encoding proteins in
RA. Hub genes were determined on the basis of the degree
results.
Molecular docking
The protein structure of CD2 was downloaded from the
PDB database (rcsb.org; PDB ID, 1HNF) (30). Schrödinger (Maestro-2015-2) software
was used to optimize the structure of the protein, which included
assigning bond sequences, hydrogenating atoms, and removing
eclipsed conformations. Finally, the energy of CD2 was minimized.
The Schrodinger LigPred module was used to optimize the energy of
the small molecule structure (31).
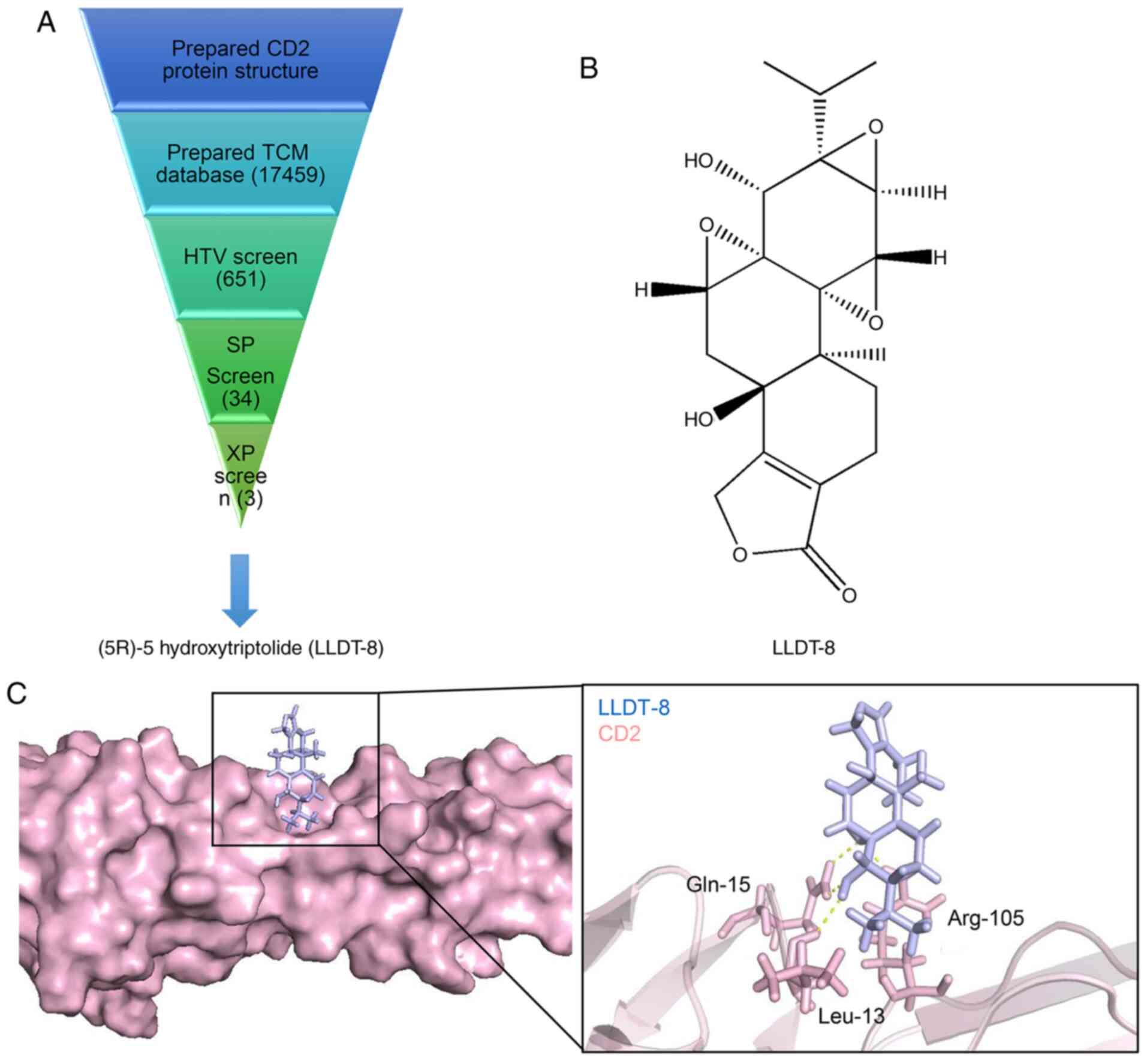
First, 651 Chinese medicine molecules with the highest CD2 docking
scores were screened using high throughput virtual (HTV) screening.
The standard precision (SP) screen was selected from 34 Chinese
medicine molecules. Finally, three candidate Chinese medicine
molecules (extra precision screen) were obtained via precise
docking. PyMOL (version 2.3) (32)
was used to visualize the structures of triptolide derivative
(5R)-5-hydroxytriptolide (LLDT-8) and CD2.
Gene set enrichment analysis
(GSEA)
Genes that may serve a key role in RA were
determined through the statistical calculation on the basis of GSEA
(version 3.0) (33), which was used
to compare the genes to be analyzed with those that were
pre-divided into gene sets. The input file in the GCT/CLS format
was created in accordance with the format requirements of the
software. The CLS file was the grouping description file of the
expression matrix of sequencing data, and the GCT file was the gene
expression matrix files in sequencing. For data analysis, the
parameters were set as: Number of permutations, 1,000; Collapse
dataset to gene symbols, true; Permutation type, gene_set; Max
size, exclude larger sets, 500; Min size, exclude smaller sets:
15.
Statistical analysis
SPSS version 20.0 (IBM, Corp.) was used for
analysis, and measurement data are expressed as the mean ± standard
deviation. Comparisons between two groups were performed using a
Student's t-test, and comparisons between multiple groups were
performed using a one-way ANOVA followed by a post-hoc Tukey's
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of DEGs in RA, and GSEA
of the DEGs
Data were downloaded from NCBI-GEO, a public
functional genomics database that receives data from different
microarray platforms, and the gene expression profiles of RA and
normal synovial tissues were obtained from the GSE55235 dataset
(15). The data included synovial
tissues from 10 patients with RA and 10 individuals with healthy
joints. P<0.05 and |log(fold change) FC|>2 were used as the
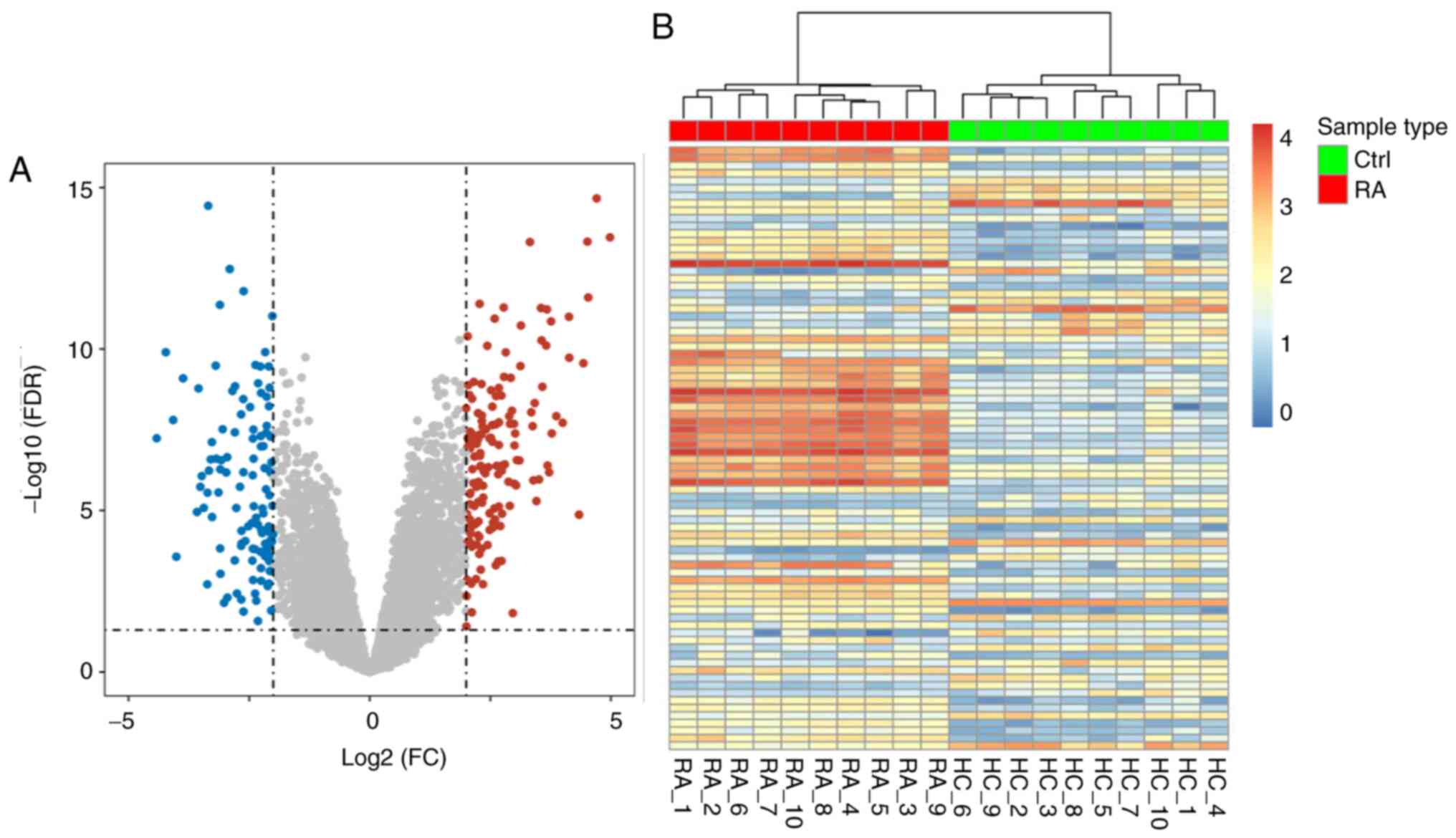
cutoff criteria. A total of 279 DEGs (166 upregulated and 113
downregulated genes) were extracted from the expression profile
dataset (Fig. 1A). The DEGs are
shown as a heatmap based on the |logFC| value in Fig. 1B. According to the results, the DEGs
identified could be used to distinguish RA from the normal group
and could be used for subsequent biological analysis. The GSEA
software package was used to analyze the different biological
processes of synovial tissues in patients with RA and healthy
individuals, and the truncated value was P<0.05. GSEA is a
computational method used to determine whether a predefined set of
genes can show significant consistent differences in two biological
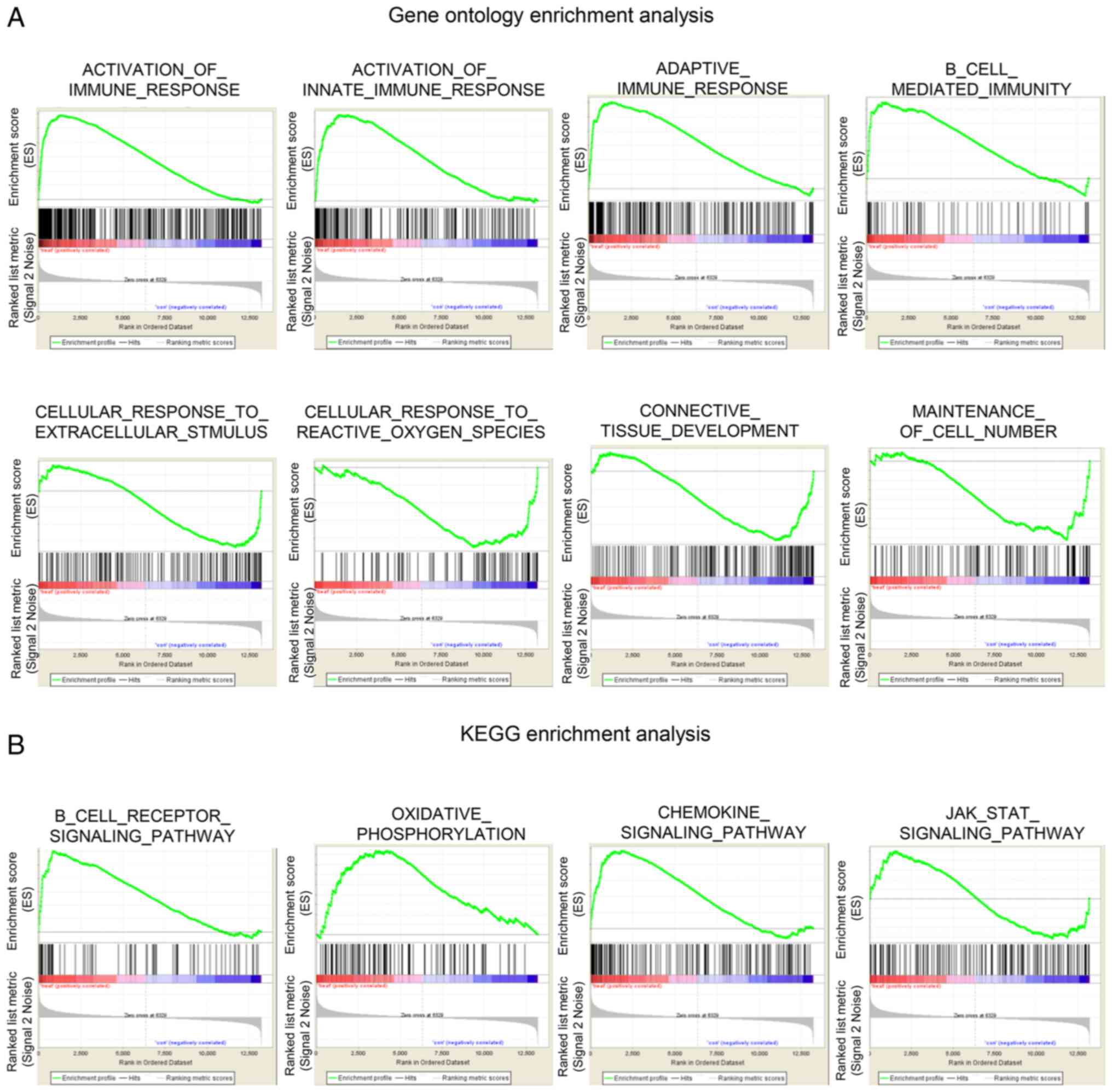
states. In the GO analysis, the upregulated functions associated
with the significantly enriched genes included the activation of
immune and adaptive immune responses. The connective tissue
development, cell number maintenance, and cellular response to
extracellular stimulus function decreased (Fig. 2A). KEGG enrichment analysis showed
that the B cell receptor, chemokine, the JAK-STAT signaling
pathways and oxidative phosphorylation were activated in RA lesions
(Fig. 2B). Identification of
significantly rich functions and pathways may assist in further
studying the role of DEGs in RA.
PPI network analysis of DEGs in
RA
The study of protein interaction networks assist in
deepening our understanding of cell structure and function from a
systematic perspective and provide a theoretical basis for the
discovery of novel drug targets and drug design. Using
bioinformatics methods to study protein interaction network shows
great advantages, including protein interaction network mapping and
display, network topology analysis, network structure module
research and network comparison. Based on the obtained DEGs, the
interaction network of DEGs was further analyzed. The STRING
database was used to calculate the PPI network (Fig. S1). As observed from the results, the
PPI network can be divided into five major interaction subset
networks.
The key nodes in the PPI network were analyzed using
Cytoscape plugins, to further analyze the hub genes in RA and
determine the association of the protein interactions. The results
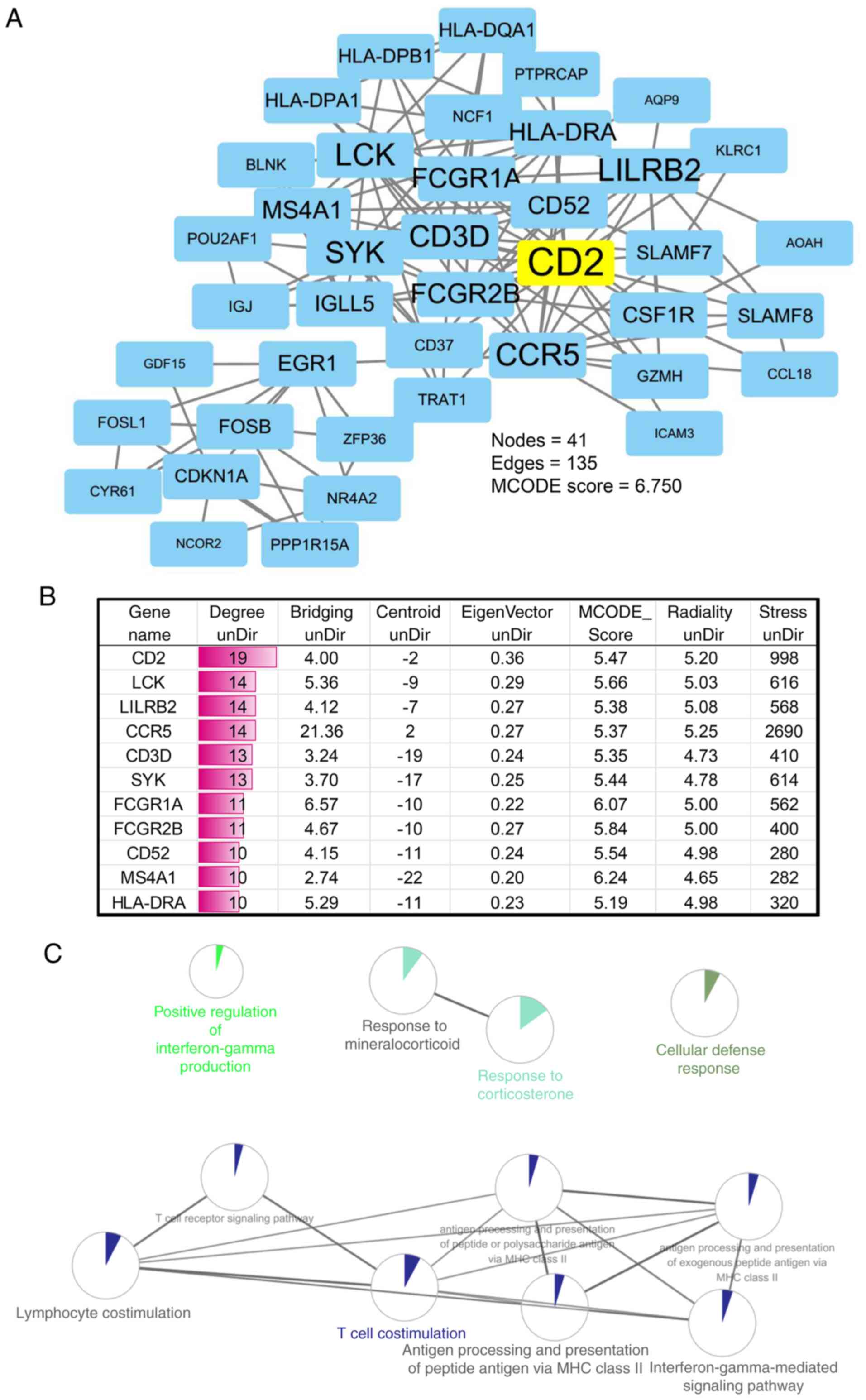
showed that the interactive network had 41 nodes, 135 edges and a
MCODE score of 6.750 (Fig. 3A). The
degree of each node gene was further calculated, and CD2 had the
highest topological connectivity (Fig.
3B). Furthermore, the Clue-GO plugin in Cytoscape software was
used to enrich and annotate the key PPI module genes. Results
showed that the key module genes were enriched primarily in
response to corticosterone, antigen processing, and presentation of
peptide or polysaccharide antigen via MHC class II, lymphocyte
co-stimulation, interferon-γ-mediated signaling pathway, T cell
co-stimulation and the T cell receptor signaling pathway (Fig. 3C).
CD2 is a key protein involved in the
development of RA
According to the results of bioinformatics analysis,
CD2 was determined to be a key protein involved in the development
of RA. In order to verify the role of CD2 in RA, IL-1 and TNF-α
were used to induce synovial cells for establishment of an in
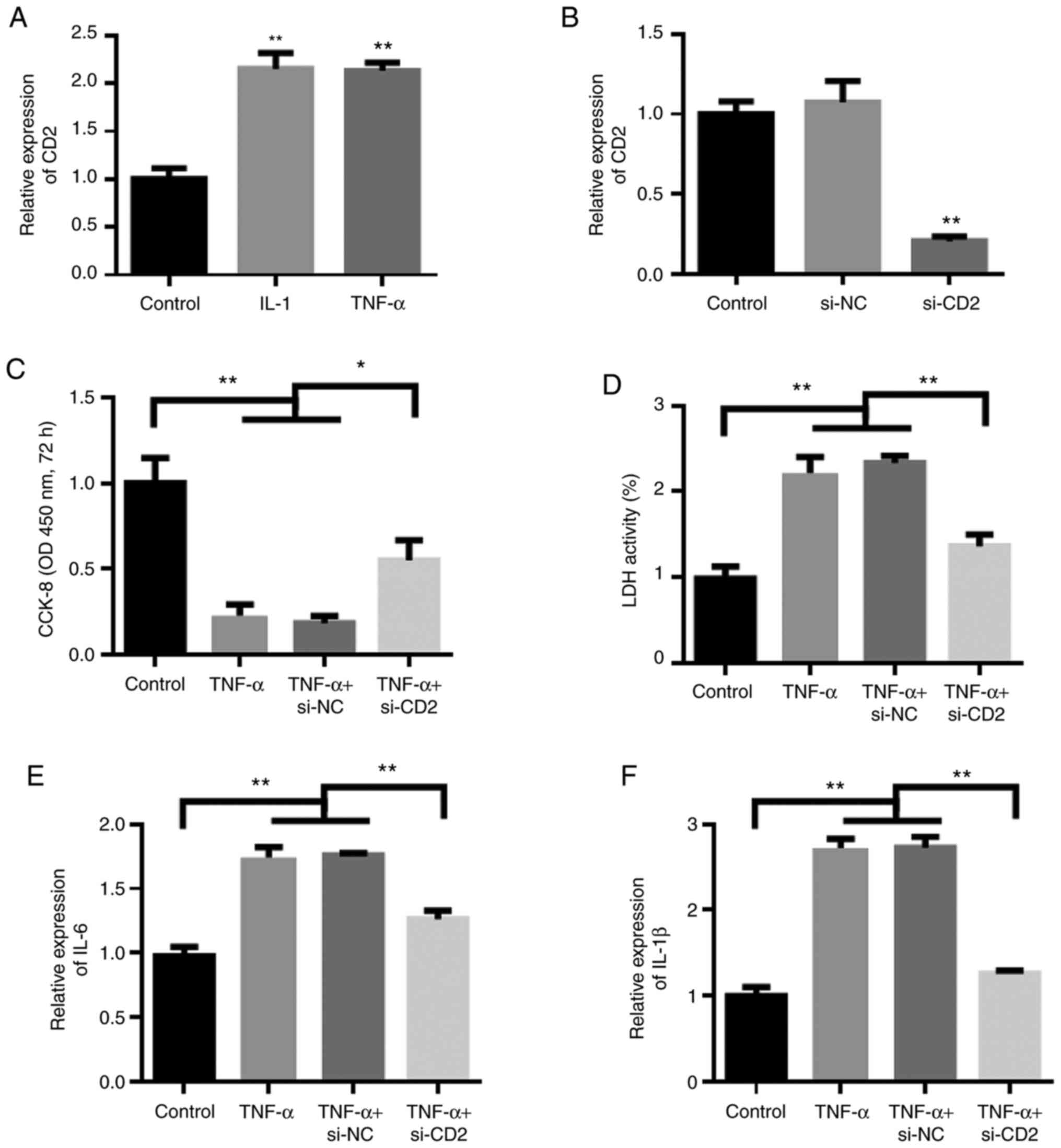
vitro RA cell model. The experimental results showed that IL-1
and TNF-α could induce the upregulation of CD2, consistent with the
results of the bioinformatics analysis (Fig. 4A). Next, siRNAs were used to
knockdown the expression of CD2. The experimental results show that
the siRNAs effectively reduced the expression of CD2 (Fig. 4B). The results of cell proliferation
experiments showed that compared with the control group, CD2
knockdown reduced the inhibitory effect of TNF-α on proliferation
in synovial cells (Fig. 4C).
Furthermore, changes in the levels of cellular inflammatory factors
were assessed. The results showed that compared with the control
group, TNF-α stimulation increased the levels of inflammatory
factors, whereas knockdown of CD2 reduced the effects of TNF-α
(Fig. 4D-F).
Screening of TCMs that may inhibit key
genes based on the structure of CD2
By analyzing the PPI network, CD2 was found to be a
key node gene that serves an important role in the pathogenesis of
RA. CD2 was significantly upregulated in RA (logFC=2.731) and
therefore was chosen as the basis for the selection of candidate
compounds. A set of molecular screening strategies for TCM was
designed on the basis of high-throughput virtual screening method.
The schematic diagram of the screening process is shown in Fig. 5A. First, 651 Chinese medicine
molecules with the highest CD2 docking scores were screened using a
HTV screening. The SP screen was selected from 34 Chinese medicine
molecules. Finally, three candidate Chinese medicine molecules
(extra precision screen) were obtained via precise docking. Amongst
these molecules, LLDT-8 and CD2 were found to have the highest
docking score and the best binding mode (Fig. 5B). The binding between LLDT-8 and the
target protein CD2 is shown in Fig.
5C. The key amino acid residues (Gln-15, Leu-13 and Arg-105)
interacted with LLDT-8 to form a hydrogen bond interaction. The 3D
conformation of LLDT-8 interacting with CD2 was used to further
clarify their interaction. LLDT-8 may inhibit the function of CD2
by inhibiting its biological activity center, and this inhibition
may be beneficial for the management of RA.
LLDT-8 may exert its beneficial
effects on RA through regulation of immunity and suppression of key
pathways
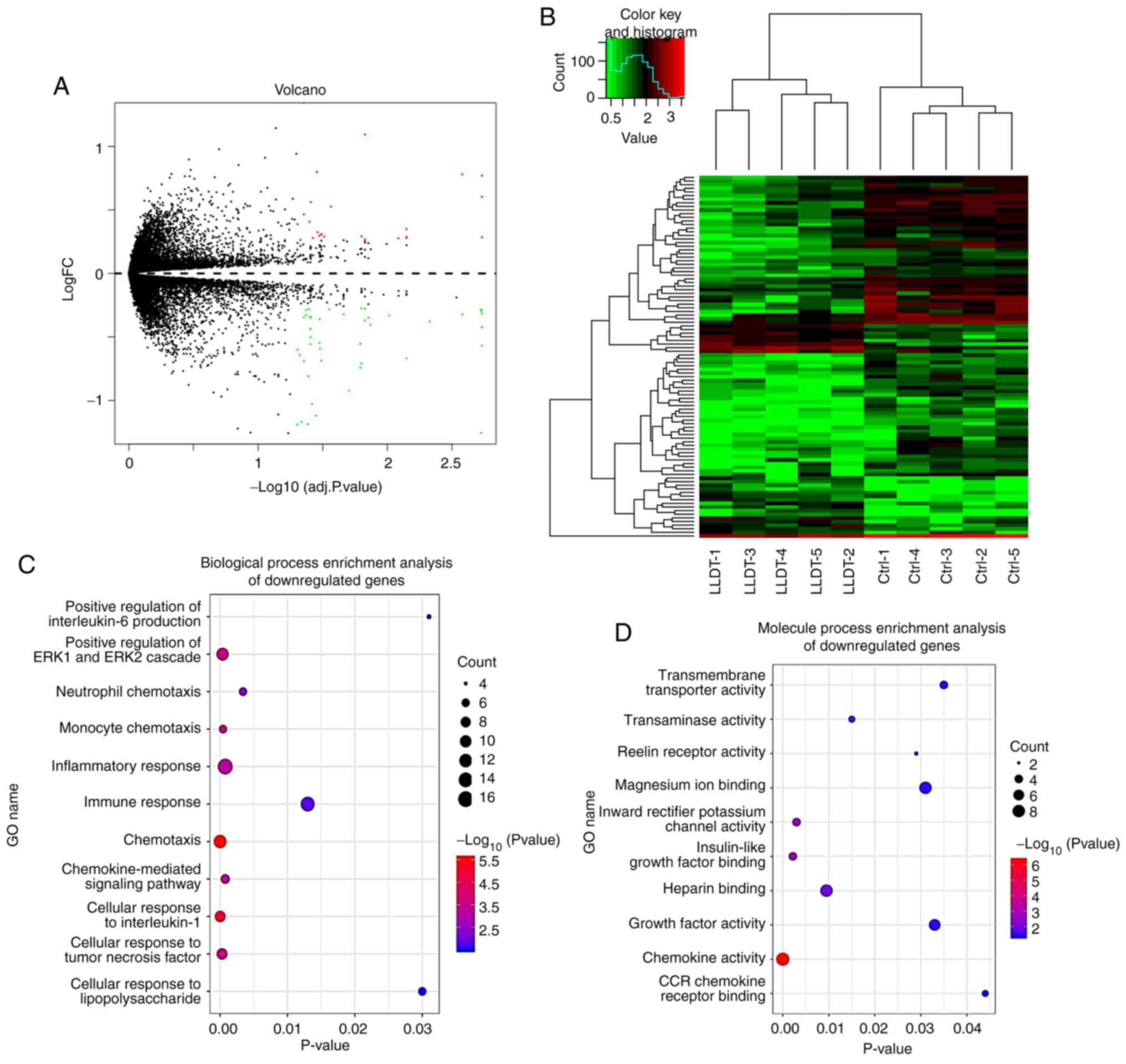
A total of 324 DEGs were identified following
pretreatment and batch elimination of DEGs in the GSE84074 dataset
using the limma software package (16). Amongst these genes, 139 upregulated
and 311 downregulated genes were identified in the LLDT-8 treatment
group compared with the control group (Fig. 6A). DEGs are shown in the volcano
plot, and the first 100 DEGs shown based on the value of |logFC|
are also illustrated in the heatmap (Fig. 6B).
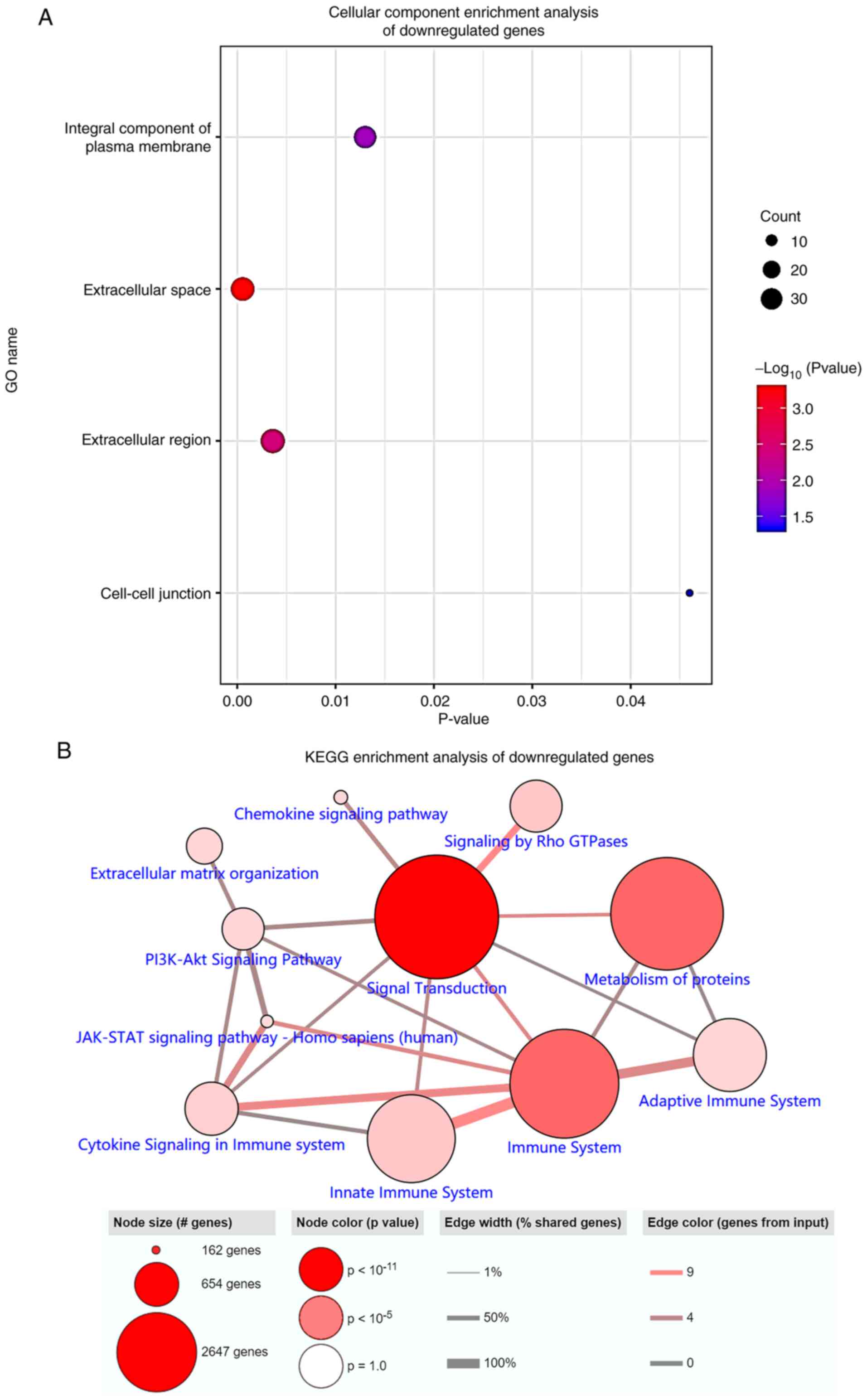
The function and pathway enrichment analysis of DEGs
after LLDT-8 processing was performed using the clusterProfiler
package (25). DEGs were divided
into three functional groups, namely, biological process, molecular
function and cellular component (Figs.
6C-D and 7A). LLDT-8 primarily
inhibited the cellular response to IL-1, the positive regulation of
ERK1 and ERK2 cascades, the chemokine-mediated signaling pathway,
inflammatory and immune responses, the positive regulation of IL-6
production function and chemokine signaling pathway, innate and
adaptive immune systems, and the JAK-STAT signaling pathway in the
RA process (Fig. 7B).
LLDT-8 treats RA by inhibiting
CD2
Based on high-throughput molecular screening
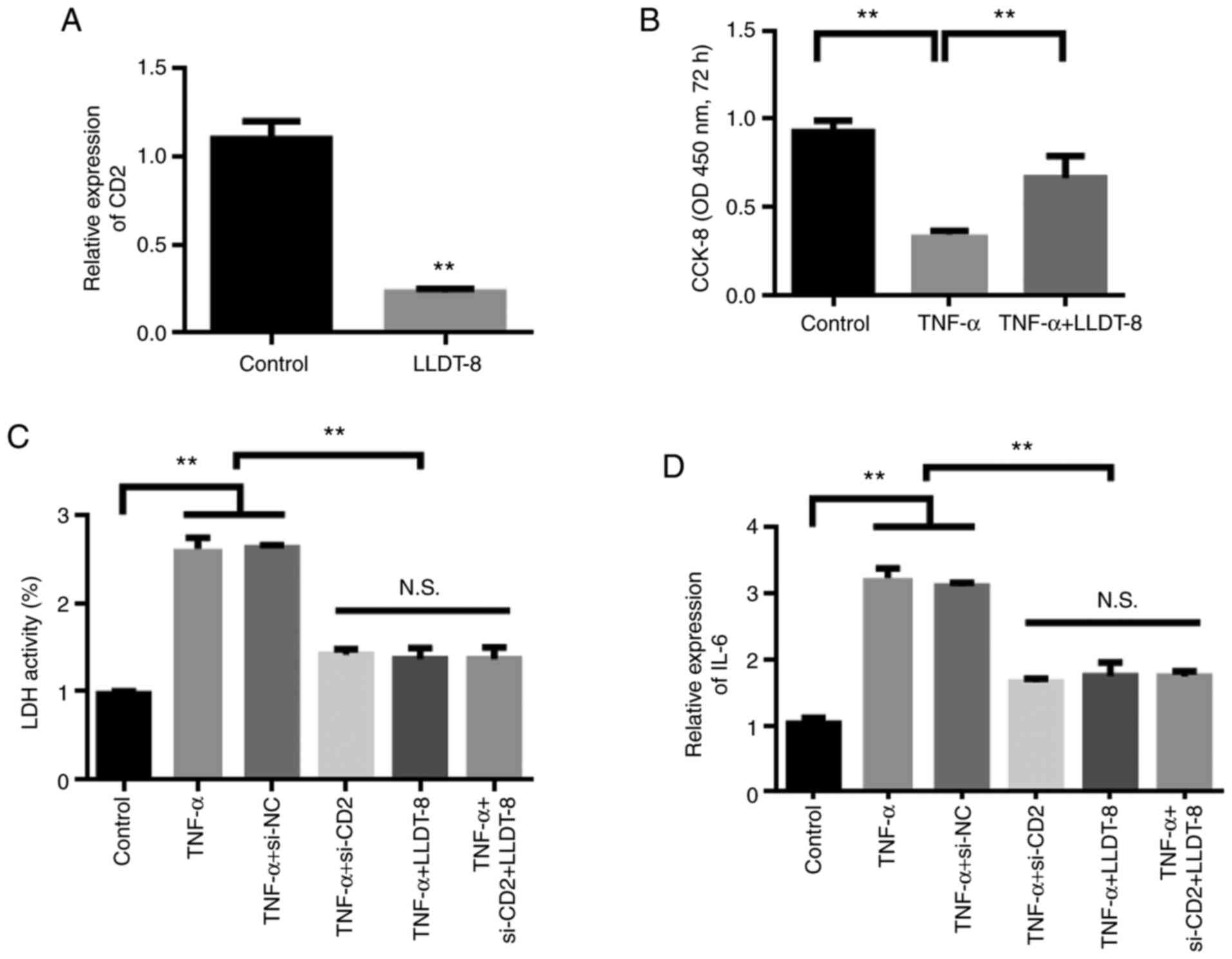
results, it was found that LLDT-8 inhibited CD2. Therefore, the
changes in the levels of CD2 were detected in the LLDT-8 treated
synovial cells. The experimental results showed that compared with
the control group, LLDT-8 reduced the expression of CD2 (Fig. 8A). The results of cell proliferation
experiments showed that TNF-α could inhibit the proliferation of
synovial cells, whereas LLDT-8 treatment restored the proliferation
of synovial cells (Fig. 8B).
Further, changes in cellular inflammatory factors
were detected. The experimental results showed that, compared with
the control group, TNF-α stimulation increased the levels of
inflammatory factors, whereas knockdown of CD2 reduced the
stimulation of TNF-α. After knocking down CD2, the administration
of LLDT-8 did not further reduce the levels of inflammatory
factors, suggesting that CD2 was the target of LLDT-8 (Fig. 8C and D).
Discussion
RA is a chronic systemic disease with an unclear
etiology, although genetic and environmental factors may affect its
pathogenesis (1). Several mechanisms
regarding the immune disorders in RA have been proposed (34), but none can fully explain its exact
pathogenesis. Therefore, screening key node genes that regulate
immune disorders combined with drugs targeting these key genes may
have important potential value for the clinical treatment of
RA.
Key signaling pathways or node genes can be screened
from the cluttered omics data through joint bioinformatics
analysis, and this method is widely used to improve our
understanding of the pathogenesis of various diseases, such as
cancer and Alzheimer's disease (35,36). In
the present study, the DEGs between normal and RA tissues, based on
data obtained from GEO, were identified. Topological analysis
revealed that CD2, a special marker protein distributed on the
surface of T and NK cells (37), was
a key node gene regulating immune disorders during the pathogenesis
of RA. As a receptor, CD2 transmitted signals into the cells to
activate T and NK cell activities (38,39).
During the progression of RA, disordered immune system function
primarily manifests as the activation of T and NK cells. Thus, the
CD2 protein may serve a key role in the immune disorder of RA, and
CD2 may serve as a potential target for treating RA. Synovial cell
level studies have shown that knockdown of CD2 can reduce the
progression of RA.
Through molecular simulation docking, the potential
drug LLDT-8 was screened to obtain the molecule with the optimal
targeting activity of CD2. LLDT-8, one of the primary active
ingredients in Tripterygium wilfordii plants (40), is an epoxy diterpene lactone
compound. Tripterygium wilfordii has therapeutic value for
the management of RA due to the presence of a triptolide; however,
its pharmacological properties remain unclear (41). The present study further discussed
the specific mechanism of Tripterygium wilfordii-related
compounds with regard to their anti-inflammatory and analgesic
properties. According to the omics data analysis, LLDT-8
effectively inhibited the immune- and inflammation-related
signaling pathways, showing the potential therapeutic value of
LLDT-8 in RA. In vitro functional and phenotypic experiments
further explained the role of CD2 in RA progression and the
potential therapeutic activity of LLDT-8.
The present study has some limitations. This study
only verified the effect of LLDT-8 at the cellular level, and
subsequent studies should also verify the efficacy of LLDT-8 in
vivo. In addition, the downstream molecular mechanism of LLDT-8
targeting CD2 inhibition also needs to be further studied.
In conclusion, the results of the present study may
improve our understanding of the developmental process of RA and
highlight LLDT-8 as potential novel and effective drug for the
treatment of RA.
Supplementary Material
Protein-protein interaction analysis
of the differentially expressed genes associated with rheumatoid
arthritis for major interaction subset small networks.
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science Foundation of
Tianjin (grant no. 16ZXMJSY00220).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YL conceived and designed the study. WQ and LY
performed the bioinformatics analysis. YL and MW performed the in
situ analysis. YL and LZ wrote the manuscript. All authors read and
approved the final manuscript. YL and LZ confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Falconer J, Murphy AN, Young SP, Clark AR,
Tiziani S, Guma M and Buckley CD: Review: Synovial Cell Metabolism
and Chronic Inflammation in Rheumatoid Arthritis. Arthritis
Rheumatol. 70:984–999. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Firestein GS and McInnes IB:
Immunopathogenesis of Rheumatoid Arthritis. Immunity. 46:183–196.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kim SY, Schneeweiss S, Liu J, Daniel GW,
Chang CL, Garneau K and Solomon DH: Risk of osteoporotic fracture
in a large population-based cohort of patients with rheumatoid
arthritis. Arthritis Res Ther. 12(R154)2010.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Källberg H, Ding B, Padyukov L, Bengtsson
C, Rönnelid J, Klareskog L and Alfredsson L: EIRA Study Group.
Smoking is a major preventable risk factor for rheumatoid
arthritis: Estimations of risks after various exposures to
cigarette smoke. Ann Rheum Dis. 70:508–511. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Klareskog L, Gregersen PK and Huizinga TW:
Prevention of autoimmune rheumatic disease: State of the art and
future perspectives. Ann Rheum Dis. 69:2062–2066. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Viatte S, Plant D, Bowes J, Lunt M, Eyre
S, Barton A and Worthington J: Genetic markers of rheumatoid
arthritis susceptibility in anti-citrullinated peptide antibody
negative patients. Ann Rheum Dis. 71:1984–1990. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hogan DB: Did Osler suffer from ‘paranoia
antitherapeuticum baltimorensis’? A comparative content analysis of
The Principles and Practice of Medicine and Harrison's Principles
of Internal Medicine, 11th edition. CMAJ. 161:842–845.
1999.PubMed/NCBI
|
|
8
|
McInnes IB and Schett G: Pathogenetic
insights from the treatment of rheumatoid arthritis. Lancet.
389:2328–2337. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Reis-Filho JS and Pusztai L: Gene
expression profiling in breast cancer: Classification,
prognostication, and prediction. Lancet. 378:1812–1823.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tan L, Yu JT, Tan MS, Liu QY, Wang HF,
Zhang W, Jiang T and Tan L: Genome-wide serum microRNA expression
profiling identifies serum biomarkers for Alzheimer's disease. J
Alzheimers Dis. 40:1017–1027. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shen Y, Jiang T, Wang R, He S, Guo M, Zuo
J and He D: (5R)-5-Hydroxytriptolide (LLDT-8) inhibits
osteoclastogenesis via RANKL/RANK/OPG signaling pathway. BMC
Complement Altern Med. 15(77)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen Y, Zhang L, Ni J, Wang X, Cheng J, Li
Y, Zhen X, Cao T and Jia J: LLDT-8 protects against cerebral
ischemia/reperfusion injury by suppressing post-stroke
inflammation. J Pharmacol Sci. 131:131–137. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ren YX, Zhou R, Tang W, Wang WH, Li YC,
Yang YF and Zuo JP: (5R)-5-hydroxytriptolide (LLDT-8) protects
against bleomycin-induced lung fibrosis in mice. Acta Pharmacol
Sin. 28:518–525. 2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xie ZC, Dang YW, Wei DM, Chen P, Tang RX,
Huang Q, Liu JH and Luo DZ: Clinical significance and prospective
molecular mechanism of MALAT1 in pancreatic cancer exploration: A
comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA
databases. OncoTargets Ther. 10:3991–4005. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Woetzel D, Huber R, Kupfer P, Pohlers D,
Pfaff M, Driesch D, Häupl T, Koczan D, Stiehl P, Guthke R, et al:
Identification of rheumatoid arthritis and osteoarthritis patients
by transcriptome-based rule set generation. Arthritis Res Ther.
16(R84)2014.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Guo S, Liu J, Jiang T, Lee D, Wang R, Zhou
X, Jin Y, Shen Y, Wang Y, Bai F, et al: (5R)-5-Hydroxytriptolide
(LLDT-8) induces substantial epigenetic mediated immune response
network changes in fibroblast-like synoviocytes from rheumatoid
arthritis patients. Sci Rep. 9(11155)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e47. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
R Core Team: A Language and Environment
for Statistical Computing. R Foundation for Statistical Computing,
Vienna, 2013. Available from https://www.R-project.org/.
|
|
19
|
Team R: RStudio: Integrated Development
for R. RStudio, Inc., Boston MA, 2015. Available from URL
http://www.rstudio.com.
|
|
20
|
Xi X, Liu N, Wang Q, Chu Y, Yin Z, Ding Y
and Lu Y: ACT001, a novel PAI-1 inhibitor, exerts synergistic
effects in combination with cisplatin by inhibiting PI3K/AKT
pathway in glioma. Cell Death Dis. 10(757)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Xi X, Chu Y, Liu N, Wang Q, Yin Z, Lu Y
and Chen Y: Joint bioinformatics analysis of underlying potential
functions of hsa-let-7b-5p and core genes in human glioma. J Transl
Med. 17(129)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhong W, Sun B, Gao W, Qin Y, Zhang H,
Huai L, Tang Y, Liang Y, He L, Zhang X, et al: Salvianolic acid A
targeting the transgelin-actin complex to enhance vasoconstriction.
EBioMedicine. 37:246–258. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhong W, Yang W, Qin Y, Gu W, Xue Y, Tang
Y, Xu H, Wang H, Zhang C, Wang C, et al: 6-Gingerol stabilized the
p-VEGFR2/VE-cadherin/β-catenin/actin complex promotes microvessel
normalization and suppresses tumor progression. J Exp Clin Cancer
Res. 38(285)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Scardoni G, Petterlini M and Laudanna C:
Analyzing biological network parameters with CentiScaPe.
Bioinformatics. 25:2857–2859. 2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4(2)2003.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Bodian DL, Jones EY, Harlos K, Stuart DI
and Davis SJ: Crystal structure of the extracellular region of the
human cell adhesion molecule CD2 at 2.5 A resolution. Structure.
2:755–766. 1994.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhong W, Liu P, Zhang Q, Li D and Lin J:
Structure-based QSAR, molecule design and bioassays of
protease-activated receptor 1 inhibitors. J Biomol Struct Dyn.
35:2853–2867. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
DeLano WL: Pymol: An open-source molecular
graphics tool. CCP4 Newsletter on protein crystallography.
40:82–92. 2002.
|
|
33
|
Reimand J, Isserlin R, Voisin V, Kucera M,
Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, et
al: Pathway enrichment analysis and visualization of omics data
using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc.
14:482–517. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Catrina AI, Joshua V, Klareskog L and
Malmström V: Mechanisms involved in triggering rheumatoid
arthritis. Immunol Rev. 269:162–174. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Glaab E, Baudot A, Krasnogor N and
Valencia A: TopoGSA: Network topological gene set analysis.
Bioinformatics. 26:1271–1272. 2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bao R, Huang L, Andrade J, Tan W, Kibbe
WA, Jiang H and Feng G: Review of current methods, applications,
and data management for the bioinformatics analysis of whole exome
sequencing. Cancer Inform. 13 (Suppl 2):67–82. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yang JJ, Ye Y, Carroll A, Yang W and Lee
HW: Structural biology of the cell adhesion protein CD2:
Alternatively folded states and structure-function relation. Curr
Protein Pept Sci. 2:1–17. 2001.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Watzl C and Long EO: Signal transduction
during activation and inhibition of natural killer cells. Curr
Protoc Immunol. 11(17)2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
James JR and Vale RD: Biophysical
mechanism of T-cell receptor triggering in a reconstituted system.
Nature. 487:64–69. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wong KF, Yuan Y and Luk JM:
Tripterygium wilfordii bioactive compounds as anticancer and
anti-inflammatory agents. Clin Exp Pharmacol Physiol. 39:311–320.
2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Bao J and Dai SM: A Chinese herb
Tripterygium wilfordii Hook F in the treatment of rheumatoid
arthritis: Mechanism, efficacy, and safety. Rheumatol Int.
31:1123–1129. 2011.PubMed/NCBI View Article : Google Scholar
|