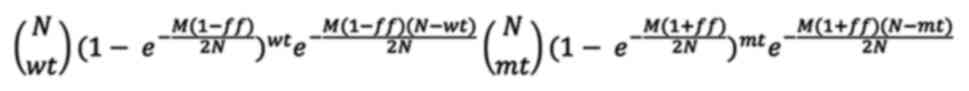Introduction
Thalassemia is an autosomal recessive disease that
is caused by the reduction in hemoglobin chain synthesis. It is
estimated that thalassemia affects 17% of over 330,000 newborns
worldwide each year (1). The two
major types of thalassemia are α- and β-thalassemia. α-thalassemia
is caused by the loss of one or both of the α-globin genes
(2). The Southeast Asian deletion
(SEA) variant is a large deletion of the α-globin gene region, and
is the most common cause of α-thalassemia in the Asian population.
Couples carrying the SEA deletion do not show symptoms, but will
have a 25% chance of conceiving a baby with Hb Bart's hydrops
fetalis and experiencing toxemia during pregnancy (3). β-thalassemia is caused by mutations in
β-globin genes, which lead to reduced β-globin chain synthesis
(4). Children born with
β-thalassemia may show symptoms of anemia and other complications
throughout their lives (5).
Thailand has a high (30-50%) prevalence of α- and β-thalassemia
(6), particularly in the northern
and northeastern regions of the country (7). As thalassemia treatments are costly
and time-consuming, the most effective means of limiting
thalassemia incidence is to screen for the mutant genes before
birth. Towards this goal, prenatal screening for thalassemia is
becoming a part of primary health services in the majority of
developed countries, particularly in Asia (8). Nonetheless, traditional prenatal
diagnoses, such as chorionic villi sampling and amniocentesis, are
invasive and may cause infection and miscarriage (9,10).
The discovery of cell-free fetal DNA (11), which is released from placental
tissue into the maternal bloodstream, has enabled the development
of non-invasive prenatal diagnosis (NIPD) for detecting
disease-causing mutations in the fetus (12,13).
In recent years, there has been substantial interest in developing
NIPD for thalassemia. Reverse transcription-quantitative PCR
(RT-qPCR) using the gap-PCR technique with primers designed to bind
at the deletion breakpoints have been used to diagnose common
deletions such as -a3.7, -a4.2,
-THAI, -MED and -SEA (14,15).
Similarly, the PCR-based techniques were also applied to diagnose
Hb Bart's hydrops fetalis (16) and
other forms of α-thalassemia (17-19).
Next-generation sequencing (NGS) is a powerful technique for NIPD,
as it has high sensitivity and enables the detection of low
abundance cell free-fetal DNA (20-23).
Several studies have shown high accuracy of NGS-based genome-wide
single-nucleotide polymorphism genotyping and targeted sequencing
for diagnosing β-thalassemia (24-26).
However, the high costs of NGS-based techniques make them
inappropriate in routine clinical practice.
Droplet digital PCR (ddPCR) is a sensitive and
quantitative PCR-based technique that can amplify a low initial
amount of target DNA molecules and enumerate different PCR products
using probe-specific fluorescent signals (27,28).
ddPCR has been used to detect circulating tumor DNA and cell-free
fetal DNA (29-31).
A study on SEA deletion α-(0) thalassemia showed a promising
prospect of ddPCR (32). However,
it is unclear whether the fetal DNA analyzed came directly from
fetal tissues or cell-free fetal DNA in the maternal bloodstream
(32). Furthermore, digital
PCR-based NIPD could be improved by utilizing a nucleic acid size
selection technique to enrich fetal DNA molecules (33).
The present study developed ddPCR-based assays for
detecting fetal α- and β-thalassemia mutations in cell-free DNA
from maternal plasma. The assays were evaluated on 22 pregnant
women carrying SEA deletions, 16 carrying HbE mutations, and 8
carrying 41/42 (-CTTT) mutations.
Materials and methods
Patient recruitment
In the present study 46 singleton pregnant women who
were thalassemia carriers [22 cases with SEA deletion, 16 cases
with HbE (G>A), and 8 cases with 41/42 (-CTTT)] were recruited
at Radjavithi Hospital, Bangkok, Thailand between March and July
2018 with approval from the Institutional Review Board of the
hospital. Peripheral blood samples (10 ml) were collected from each
participant for subsequent cell-free DNA extraction and ddPCR
analysis. The α- and β-thalassemia status of each parent and fetus
was determined by Hb typing, complete blood count tests and
genotyping by PCR from amniotic fluids. Samples were collected with
the approval from the Institutional Review Board of Radjavithi
Hospital, Bangkok (approval no. 59194; date of approval 17th
November 2016). Written informed consent was obtained from all
subjects prior to inclusion in the study. Written informed consent
from the parent or legal guardian of all subjects under the age of
18 was also obtained.
Cell-free DNA extraction from blood
samples
Maternal blood samples (10 ml) were placed in BCT
tubes (Streck™) and centrifuged at 1,600 x g for 10 min at 4˚C. The
supernatant was centrifuged again at 16,000 x g for 10 min at 4˚C.
The pellet was discarded and the supernatant was stored kept at
-20˚C until required for extraction. Cell-free DNA was extracted
from plasma (3 ml) using a QIAamp Circulating Nucleic acid kit
according to the manufacturer's protocol (Qiagen GmbH).
Amniotic fluid DNA extraction
Amniotic fluids (5 ml) were collected during weeks
15-18 of gestation and centrifuged at 16,000 x g for 10 min at 4˚C.
The supernatant was discarded, the pellet was washed twice with 1X
PBS (1 ml) and stored at -80˚C in a sterile tube. The extraction
step was performed according to the instruction manual of the
QIAamp Blood Mini kit (Qiagen GmbH).
ddPCR
A ddPCR assay was developed for determining the copy
number of the SEA allele. The HEX probe was designed to bind the
genomic region inside the SEA deletion
(NG_000006.1:g.26264_45564del19301) (34) whereas the FAM probe was designed to
bind the genomic region just outside of the SEA locus
(NG_000006.1:g.26140 to 26262), which is expected to be present in
both a wild type fetus and a fetus with an SEA deletion (Table SI). For detecting HbE (G>A) and
41/42 (-CTTT), a rare mutation assay with specific probes designed
to target the mutant and wild type amplicons was developed
(Table SI).
ddPCR experiments were performed according to the
manufacturer's protocol (Bio-Rad Laboratories, Inc.) with some
modifications as described here. The master mix solution was
prepared from 10 µl 2X ddPCR SuperMix for probes (Bio-Rad
Laboratories, Inc.), 1 µl 20X prime PCR assay (Bio-Rad
Laboratories, Inc.), and either 8 µl DNA (12 µg/ml) from plasma or
1 µl DNA (20 µg/ml) from amniotic fluid. Nuclease free water was
added to adjust the final volume to 20 µl. A total of 13,500-15,000
droplets per reaction were created in a DG8 cartridge with 70 µl
oil using a QX200 Droplet Generator (Bio-Rad Laboratories, Inc.).
Droplets (40 µl) were transferred to a 96-well PCR plate and sealed
with aluminium foil using PX1 (Bio-Rad Laboratories, Inc.) for 5
sec at 180˚C. Next the plate was placed in a T100 Thermal Cycler
(Bio-Rad Laboratories, Inc.). PCR reactions began with a cycle of
10 min at 95˚C, followed by 40 cycles of 30 sec at 94˚C and 1 min
at 54˚C, with an enzyme deactivation step for 10 min at 98˚C.
Fluorescent signals from PCR products were measured using a QX200
Droplet Reader (Bio-Rad Laboratories, Inc.) and analyzed using
QuantaSoft version 1.7.4 (Bio-Rad Laboratories, Inc.). ddPCR
reactions for SEA cases were performed in duplicates. ddPCR
reactions for HbE and 41/42 (-CTTT) cases were performed in
triplicates.
Estimation of fetal fraction
Fetal fractions were estimated as previously
described (27,35). Briefly, RASSF1A and
ACTB alleles in plasma samples were digested with
BstU I (Vivantis Technologies) at a ratio of 1 µg DNA to 1 U
of enzyme for 1 h at 60˚C. The abundance of RASSF1A and
ACTB alleles before and after digestion were measured using
ddPCR analyses. ACTB locus, which is always unmethylated,
acts as a control of BstU I activity (36). Conversely, as only the fetal
RASSF1A locus is hypermethylated, the ratio of
RASSF1A positive droplets before and after BstU I
digestion approximate the fractional abundance of fetal DNA content
of the sample.
Copy number variation (CNV) analysis
to detect SEA deletion
QuantaSoft was used to determine the number of
positive and negative droplets and analyze the relative CNV of SEA
deletions. The average CNV value from triplicate wells was used to
classify the fetal genotype using the following equation:
CNV=(concentration of SEA allele/concentration of wild-type allele)
x copy number of wild type allele.
Rare mutation assay to detect HbE and
41/42 (-CTTT) mutations
QuantaSoft was used to determine the number of
positive droplets, negative droplets and double-positive droplets.
A previously described sequential probability ratio test (SPRT)
(33) was adopted to classify fetal
HbE and 41/42 (-CTTT) genotypes. Briefly, the total number of DNA
molecules in each sample was calculated under the assumption that
the number of positive droplets (droplets with DNA materials)
followed the Poisson distribution: The probability of observing
k positive droplets is (λke-λ)/k!, where λ
indicates the average number of DNA molecules per a droplet. Hence,
by setting k=0, the number of DNA molecules can be
calculated as: -ln(fraction of negative droplets) x number of
droplets.
Next, the likelihood that the fetus has a specific
genotype was calculated under the assumption that the numbers of
negative and positive droplets for each allele followed the
Binomial distribution. For example, if the fetal genotype is a
homozygous mutant, then the fraction of mutant allele would be
(1+fetal fraction)/2, whereas the fraction of wild type allele
would be (1-fetal fraction)/2. Then, letting N be the total
number of droplets, M be the total number of DNA molecules,
mt be the number of positive droplets for the mutant allele,
wt be the number of positive droplets for the wild type
allele, and ff be the estimated fetal fraction, the
likelihood of this observation can be expressed as:
Two likelihood ratio tests were performed for each
sample: One between the hypotheses that the fetal genotype is
homozygous mutant and heterozygous, and another between the
hypotheses that the fetal genotype is wild type and heterozygous.
The threshold for accepting each test was set at type I and type II
errors of 5% according to the SPRT criteria: ±log [(1-0.05)/0.05],
which is ~±1.27 (33,37).
Real time PCR for melting curve
analysis of SEA deletion from amniocentesis
The fetal genotypes were confirmed by
high-resolution melting curve analysis in 10 samples were randomly
selected from singleton parents for confirmation. Total reaction
volume of 20 µl contained 10 µl Precision melt supermix (Bio-Rad
Laboratories, Inc.), 1 µl of each primer and 5 µl of DNA sample.
The sequences of the primers were NaI forward,
5'AGAAGCTGAGTGATGGGTCCG-3' and reverse, 5'ACAAACGCCCGTCCGACTCAA-3';
and NaIII reverse 5'-TGGACTTAAGTGATCCTCCTGCCC-3',. The
amplification step began at 95˚C for 3 min followed by 40 PCR
cycles at 95˚C for 20 sec, 60˚C for 20 sec, and 72˚C for 20 sec.
Finally, a high-resolution melting cycle from 85-95˚C at a rate of
0.1˚C per 2 sec was performed (38). Dissociation curve analysis was
performed using CFX96 Manager version 1.3 (Bio-Rad Laboratories,
Inc.).
Statistical analyses
The sequential probability ratio test (SPRT)
technique was used to analyze ddPCR results to identify the most
likely fetal genotypes (39). For
analysis, 5% type I and type II error thresholds were set.
Numerical calculations were performed in Microsoft Excel according
to the Poisson and Binomial distribution models described
above.
Results
Patient characteristics
A total of 46 couples who were at high risk of
having children with thalassemia were recruited in the present
study. The median age (age range) of pregnant mothers was 27
(15-39)
years old. The median gestational age (range) was 18 (17-27)
weeks. The median BMI (range) of participants was 21.8 (15.7-32.8).
The cohort consisted of 22 (48%) couples with SEA deletion, 16
(35%) couples with an HbE (G>A) mutation, and 8 (17%) couples
with an 41/42 (-CTTT) mutation.
Classification performance of fetal
SEA genotype
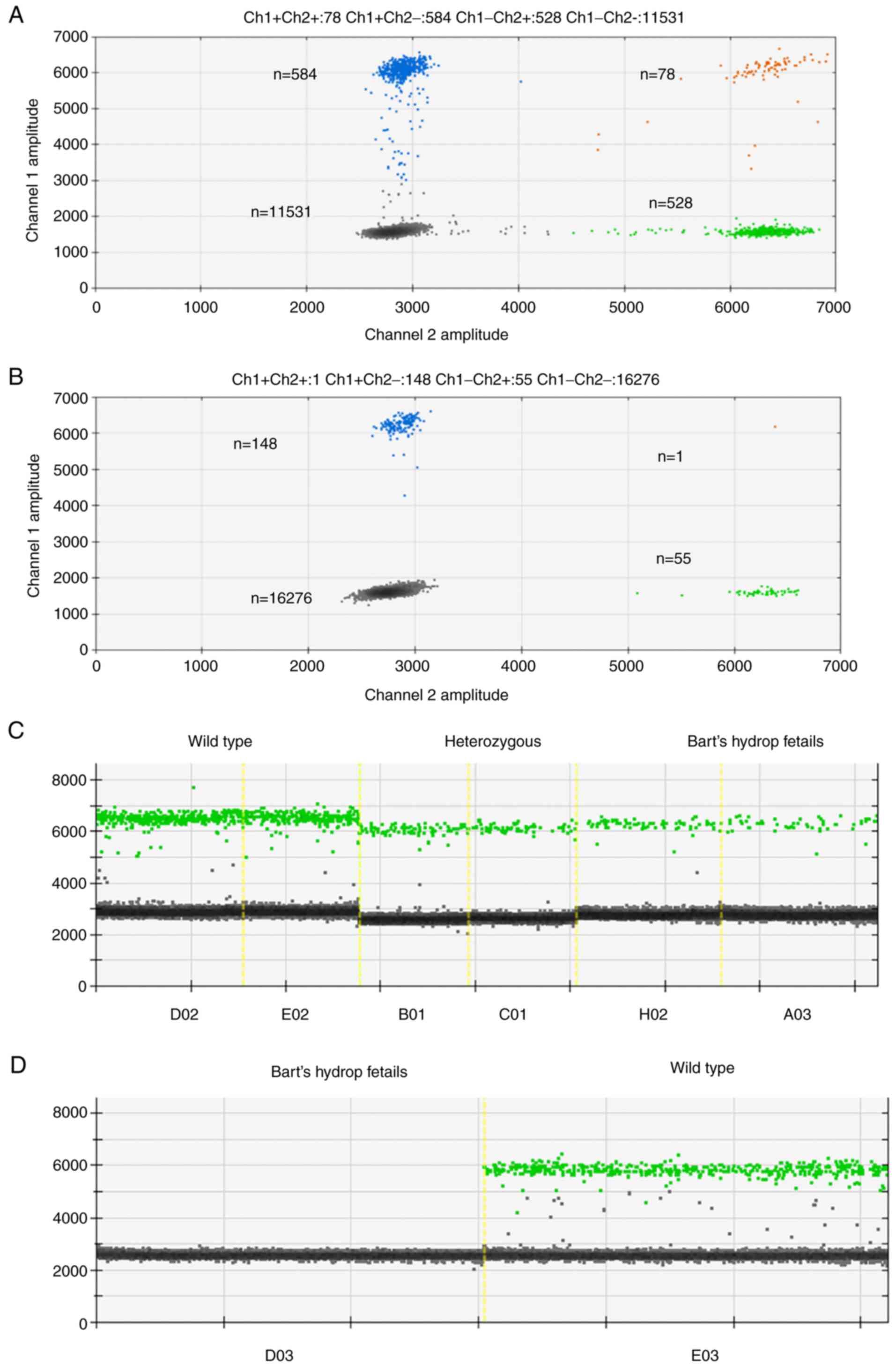
Analysis of ddPCR fluorescent signals showed clear
separations between negative and positive droplets as well as lower
abundances of the SEA locus in Bart's hydrops fetalis case as would
be expected (Fig. 1A and B). The complete absence of positive
droplets for the SEA locus in the amniocentesis sample of the
Bart's hydrops fetalis case confirmed that the designed probe was
highly specific to the SEA locus (Fig.
1C and D). SEA locus copy
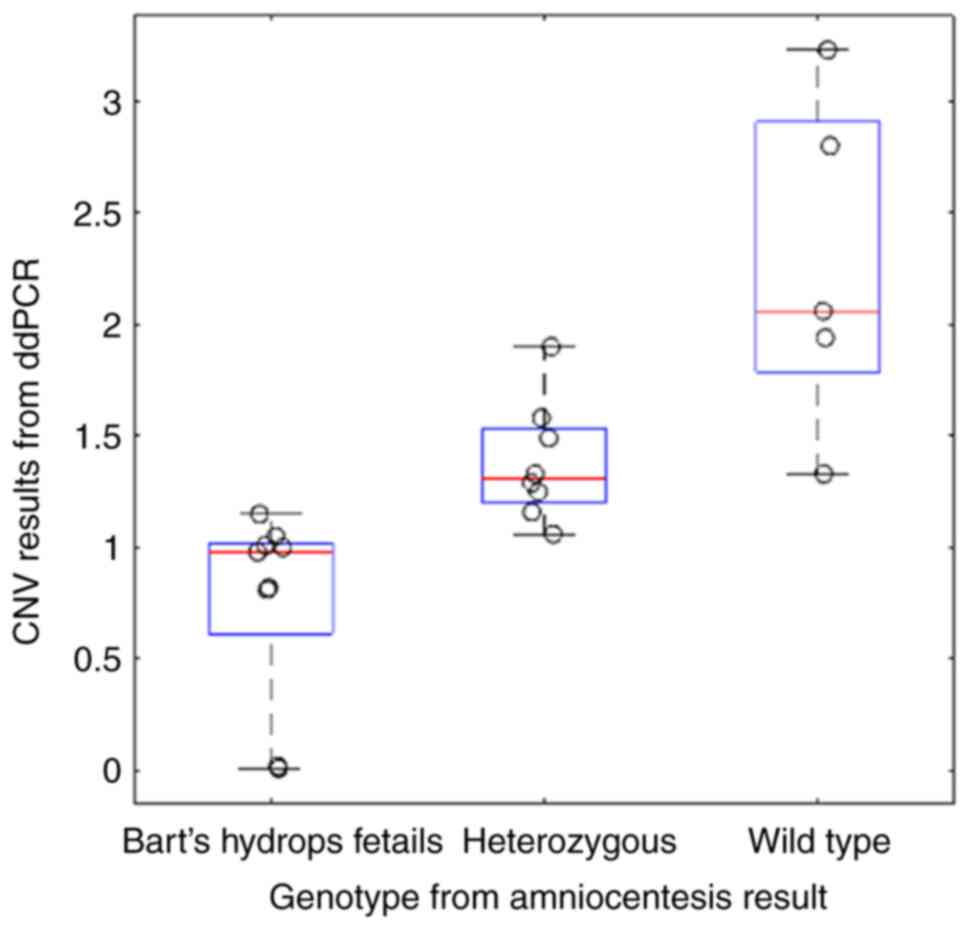
numbers estimated from cell-free DNA samples significantly differ
across wild type, heterozygous and Bart's hydrops fetalis samples
(Mann Whitney U test P=0.0124 for the comparison between the wild
type and heterozygous cases; P=0.000311 for the comparison between
heterozygous and Bart's hydrops fetalis cases). The average copy
numbers for the wild type, heterozygous and Bart's hydrops fetalis
groups were 2.27, 1.38 and 0.76, respectively (Fig. 2). To ensure the high quality of
ground truth fetal genotypes, high resolution melting curve
analyses on selected samples was also performed and 100%
concordance was obtained (Fig.
S1). Most samples exhibited the expected copy numbers of the
SEA locus, except for one wild type case that may have been
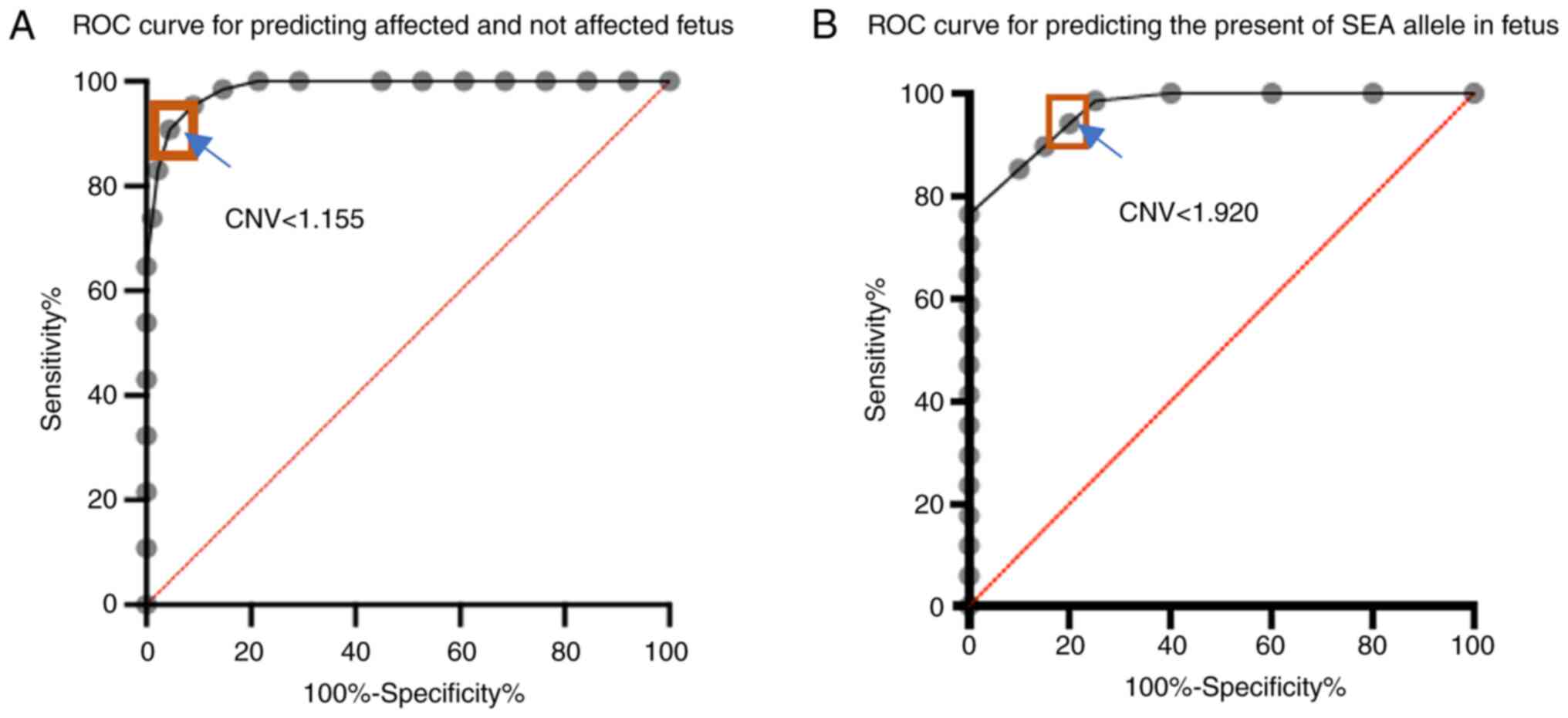
affected by a very low (3%) fetal fraction (Table I). The optimal copy number cut-off
for distinguishing affected (Bart's hydrops fetalis) from
unaffected (wild type and heterozygous) fetuses was ~1.155, which
yielded 95.38% sensitivity [95% confidence interval (CI):
87.29-98.74] and 91.01% specificity (95% CI: 83.25-95.37) (Fig. 3A). The area under the receiver
operating characteristic (AUROC) curve was 0.98. The optimal copy
number cut-off for distinguishing the presence of SEA deletion in
the fetus (Bart's hydrops fetalis and heterozygous) was 1.920,
which yielded 98.53% sensitivity (95% CI: 92.13-99.92) and 75%
specificity (95% CI: 53.13-88.81) (Fig.
3B). The AUROC curve in this scenario was 0.96.
 | Table IDetailed parental genotypes and ddPCR
test results for α (0) thalassemia (SEA). |
Table I
Detailed parental genotypes and ddPCR
test results for α (0) thalassemia (SEA).
| Sample ID | Paternal
genotype | Maternal
genotype | Fetal
fraction,% | SEA deletion
conc. | Wild type
conc. | CNV by ddPCR | Interpretation | Fetal Genotype from
conventional amniocentesis |
|---|
| 1 | (αα/αα) |
(—SEA/αα) | 10 | 141.5 | 87.5 | 3.23 | Wild type | Wild type |
| 2 |
(—SEA/αα) |
(—SEA/αα) | 15 | 15.7 | 11.2 | 2.8 | Wild type | Wild type |
| 3 |
(—SEA/αα) |
(—SEA/αα) | 16 | 11.2 | 10.9 | 2.06 | Wild type | Wild type |
| 4 |
(—SEA/αα) |
(—SEA/αα) | 7 | 23.55 | 24.3 | 1.94 | Wild type | Wild type |
| 5 |
(—SEA/αα) |
(—SEA/αα) | 17 | 57.95 | 60.95 | 1.9 | Heterozygous | Heterozygous |
| 6 |
(—SEA/αα) |
(—SEA/αα) | 8 | 8.45 | 10.7 | 1.58 | Heterozygous | Heterozygous |
| 7 |
(—SEA/αα) |
(—SEA/αα) | 9 | 44.45 | 59.8 | 1.49 | Heterozygous | Heterozygous |
| 8 | (αα/αα) |
(—SEA/αα) | 10 | 53.3 | 80.3 | 1.33 | Heterozygous | Heterozygous |
| 9 |
(—SEA/αα) |
(—SEA/αα) | 3 | 3.15 | 4.8 | 1.33 |
Heterozygousa | Wild type |
| 10 |
(—SEA/αα) |
(—SEA/αα) | 11 | 7.05 | 10.9 | 1.29 | Heterozygous | Heterozygous |
| 11 |
(—SEA/αα) |
(—SEA/αα) | 9 | 11.5 | 18.45 | 1.25 | Heterozygous | Heterozygous |
| 12 |
(—SEA/αα) |
(—SEA/αα) | 8 | 11.8 | 20.35 | 1.16 | Heterozygous | Heterozygous |
| 13 |
(—SEA/αα) |
(—SEA/αα) | 10 | 8.05 | 14.05 | 1.15 |
Heterozygousa | Bart |
| 14 |
(—SEA/αα) |
(—SEA/αα) | 15 | 10.75 | 20.35 | 1.06 | Heterozygous | Heterozygous |
| 15 |
(—SEA/αα) |
(—SEA/αα) | 11 | 15.3 | 29.15 | 1.05 | Bart | Bart |
| 16 |
(—SEA/αα) |
(—SEA/αα) | 11 | 173.5 | 344 | 1.01 | Bart | Bart |
| 17 |
(—SEA/αα) |
(—SEA/αα) | 10 | 5.05 | 10.15 | 1 | Bart | Bart |
| 18 |
(—SEA/αα) |
(—SEA/αα) | 15 | 4.85 | 9.85 | 0.98 | Bart | Bart |
| 19 |
(—SEA/αα) |
(—SEA/αα) | 15 | 4.4 | 10.7 | 0.82 | Bart | Bart |
| 20 |
(—SEA/αα) |
(—SEA/αα) | 10 | 8.1 | 20.1 | 0.81 | Bart | Bart |
| 21 |
(—SEA/αα) |
(—SEA/αα) | 15 | 0.22 | 21.3 | 0.02 | Bart | Bart |
| 22 |
(—SEA/αα) |
(—SEA/αα) | 10 | 0.14 | 27.15 | 0.01 | Bart | Bart |
Amniocentesis results from high
resolution melting curve analysis
Amniocentesis DNA samples were collected from 10
randomly selected singleton parents to confirm the SEA genotypes.
Amongst the 10 samples, 4 were diagnosed as Bart's hydrops fetalis,
5 were from normal fetuses and 1 was from a heterozygous fetus. The
median maternal age was 27 (15-36)
years old. The median gestational age was 18 (17-19)
weeks. Dissociation curve analysis showed that the melting
temperature for the wild-type fetuses was 91.5±0.15˚C, whereas the
melting temperature for Bart's hydrops fetalis was 87.8±0.02˚C.
Heterozygous fetuses showed peaks at the melting temperatures of
both the wild-type and Bart's hydrops fetalis as expected.
Classification performance of fetal
HbE (G>A) and 41/42 (-CTTT) genotype
Cell-free DNA from maternal plasma of 16 HbE
carriers and 8 41/42(-CTTT) carriers was next investigated. ddPCR
was performed using probes targeting either the wild type or mutant
allele. The ground truth genotypes for several samples were
confirmed with high-resolution melting curve analyses (Figs. S1 and 2). ddPCR results were then analyzed based
on a published sequential probability ratio test method (33). At type I and type II errors of 5%,
out of 16 HbE cases, the results for 3 patients were inconclusive,
and 10 out of the remaining 13 cases were correctly classified. For
41/42 (-CTTT), the results for 4 out of 8 cases were inconclusive,
and 2 out of the remaining 4 cases were correctly classified
(Tables II and III). It should be noted that all
inconclusive results still retain the correct genotypes and may be
correctly classified if more ddPCR reactions are performed.
 | Table IIDetailed parental genotypes and
droplet digital PCR test results for HbE β-thalassemia. |
Table II
Detailed parental genotypes and
droplet digital PCR test results for HbE β-thalassemia.
| Sample ID | Total droplet | Wild type
droplet | Mutant droplet | Double- positive
droplet | Average
concentration | Fetal fraction,
% | Mutant/mutant to
mutant/wild type LLR | P<0.05 | Wild type/wild type
to mutant/wild type LLR | P<0.05 | Interpretation | Fetal genotype from
conventional amniocentesis |
|---|
| 1 | 38,941 | 396 | 234 | 205 | 10.9 | 10 | -8.5 | Yes | 5.7 | Yes | Wild type | Wild type |
| 2 | 41,227 | 315 | 321 | 117 | 9.1 | 24.5 | -7.9 | Yes | -9.2 | Yes |
Heterozygousa | Wild type |
| 3 | 44,109 | 243 | 141 | 56 | 14.6 | 26.1 | -17.8 | Yes | 6 | Yes | Wild type | Wild type |
| 4 | 38,124 | 130 | 89 | 143 | 7.3 | 12.7 | -3 | Yes | 1.5 | Yes | Wild type | Wild type |
| 5 | 37,237 | 510 | 389 | 134 | 10.6 | 11.1 | -8.3 | Yes | 3.5 | Yes | Wild type | Wild type |
| 6 | 42,706 | 548 | 102 | 84 | 9.23 | 15.3 | -33.4 | Yes | 26.7 | Yes | Wild type | Wild type |
| 7 | 43,713 | 75 | 66 | 61 | 12.3 | 11.5 | -0.9 | No | 0 | No | Inconclusive | Heterozygous |
| 8 | 42,939 | 407 | 405 | 401 | 11.6 | 26.6 | -13.2 | Yes | -12.7 | Yes | Heterozygous | Heterozygous |
| 9 | 37,535 | 332 | 348 | 338 | 11 | 26 | -8.5 | Yes | -12.2 | Yes | Heterozygous | Heterozygous |
| 10 | 37,001 | 175 | 181 | 178 | 6.46 | 12.6 | -0.9 | No | -1.6 | Yes | Mutant or
Heterozygous | Heterozygous |
| 11 | 37,322 | 234 | 243 | 225 | 8.64 | 27.6 | -7.1 | Yes | -9.3 | Yes | Heterozygous | Heterozygous |
| 12 | 42,845 | 863 | 191 | 186 | 14.62 | 19.2 | -65.8 | Yes | 48.6 | Yes | Wild
typea | Heterozygous |
| 13 | 37,355 | 145 | 150 | 144 | 5.1 | 24.2 | -3.3 | Yes | -4.4 | Yes | Heterozygous | Heterozygous |
| 14 | 39,795 | 713 | 761 | 713 | 18.4 | 17.6 | -6.3 | Yes | -13.8 | Yes | Heterozygous | Heterozygous |
| 15 | 37,309 | 94 | 15 | 91 | 11.32 | 18.8 | -7.4 | Yes | 5.7 | Yes | Wild
typea | Heterozygous |
| 16 | 38,696 | 113 | 91 | 76 | 5.97 | 13.5 | -2.1 | Yes | 0.5 | No | Wild type or
heterozygous | Heterozygous |
 | Table IIIDetailed parental genotypes and
droplet digital PCR test results for 41/42 (-CTTT) β
thalassemia. |
Table III
Detailed parental genotypes and
droplet digital PCR test results for 41/42 (-CTTT) β
thalassemia.
| Sample ID | Total droplet | Wild type
droplet | Mutant droplet | Double- positive
droplet | Average
concentration | Fetal fraction,
% | Mutant/mutant to
mutant/wild type LLR | P<0.05 | Wild type/wild type
to mutant/wild type LLR | P<0.05 | Interpretation | Fetal genotype from
conventional amniocentesis |
|---|
| 1 | 38,196 | 139 | 205 | 137 | 16.6 | 24 | 2.6 | Yes | -11.5 | Yes | Mutanta | Wild type |
| 2 | 33,835 | 357 | 17 | 15 | 8.5 | 14 | -22.5 | Yes | 19.3 | Yes | Wild type | Wild type |
| 3 | 29,144 | 505 | 440 | 108 | 22.1 | 13.5 | -7.7 | Yes | 0.1 | No | Wild type or
heterozygous | Heterozygous |
| 4 | 31,830 | 197 | 159 | 7 | 9.5 | 13.9 | -3.8 | Yes | 0.8 | No | Wild type or
heterozygous | Heterozygous |
| 5 | 33,596 | 549 | 631 | 37 | 25.2 | 12.7 | 0.4 | No | -8.8 | Yes | Mutant or
heterozygous | Heterozygous |
| 6 | 32,369 | 254 | 277 | 0 | 12.2 | 22.8 | -3.8 | Yes | -8.5 | Yes | Heterozygous | Heterozygous |
| 7 | 33,437 | 157 | 123 | 12 | 6.02 | 10 | -2.1 | Yes | 0.9 | No | Wild type or
heterozygous | Heterozygous |
| 8 | 37,846 | 571 | 35 | 23 | 12.2 | 11.5 | -28.8 | Yes | 25.3 | Yes | Wild
typea | Heterozygous |
Typical causes of poor ddPCR assay performances
include a low fetal fraction, low amount of amplifiable DNA, and
poor binding between ddPCR probes and target alleles, all of which
would hinder confident determination of the fetal genotype.
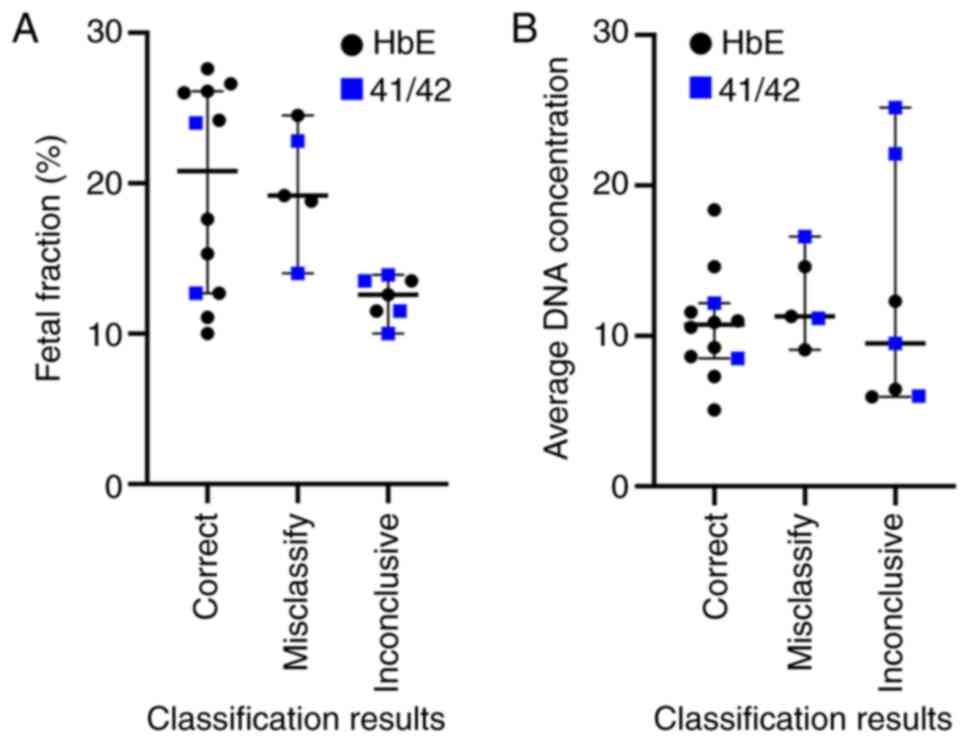
Although a low fetal fraction is linked to inconclusive results
(Fig. 4A, lower fetal fractions
among inconclusive cases; Mann-Whitney U test P=0.0173), it cannot
explain misclassifications. The 12-25% fetal fractions amongst
misclassified samples are high and should be adequate for NIPD
applications. There is also no apparent difference in the amount of
amplified DNA between correctly classified and misclassified cases
(Fig. 4B). Finally, to test whether
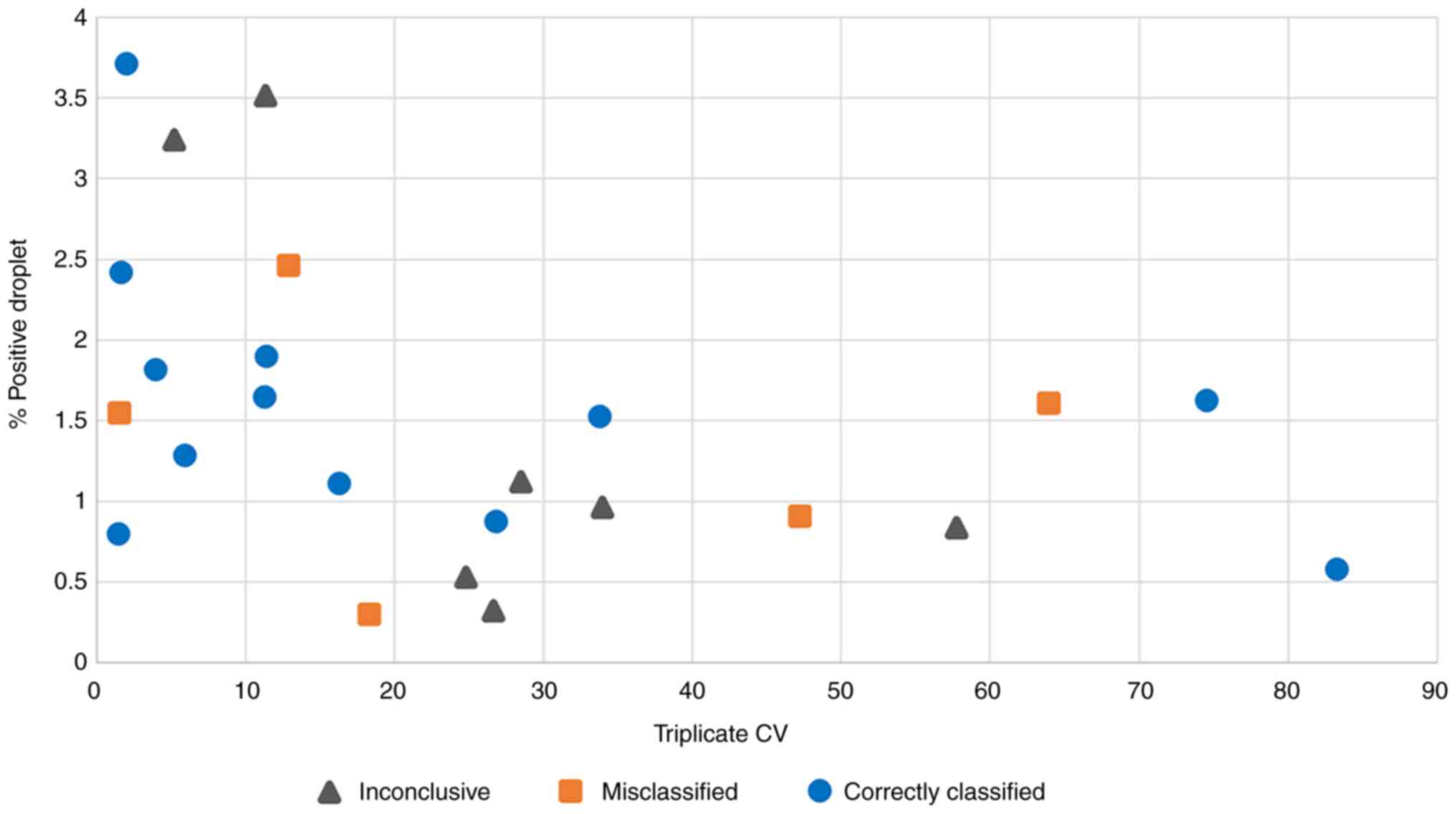
poor probe binding could explain the observed results, the
coefficient of variation (CV) of the mutant-to-wild type droplet
ratios from the triplicate ddPCR experiments and the percentage of
positive droplets of each patient sample to the classification
result were compared. The rationale here was that poor probe
binding should result in a low percentage of positive droplets and
a high variability across replicate experiments. This revealed a
clear association between the variability of triplicate ddPCR
experiments and the genotype classification accuracy (Fig. 5). Amongst the 11 samples with
<15% CV, seven were correctly classified, two were
misclassified, and two were inconclusive. Conversely, amongst the
remaining 13 samples with 16-83% CV, only five were correctly
classified, three were misclassified and five were inconclusive.
The analysis also showed a borderline significant correlation
between the CV of triplicate ddPCR results and the fraction of
total positive droplets (Spearman Rank correlation=-0.4417 with
permutation test P=0.0306).
Discussion
The growing interest in non-invasive prenatal
testing has driven the development of various assays for diagnosing
monogenic diseases from cell-free fetal DNA (24-26,30,31,33).
One of the main challenges is the interference from high maternal
DNA background that can overwhelm the signal from circulating fetal
DNA fragments (40). Prior studies
have focused on autosomal dominant monogenic diseases, such as
achondroplasia, myotonic dystrophy and some forms of β-thalassemia,
where the mutated fetal allele could be clearly distinguished from
wild type maternal DNA (41-44).
However, this consideration does not apply to autosomal recessive
diseases and several other situations that involve maternal
inherited alleles (44). Notably,
the digital relative mutation dosage and SPRT methods have improved
the confidence of assessment of the fetal genotype in the presence
of the background maternal DNA (33,45).
These techniques enabled interpretation of fetal status and
detection of small allelic imbalances when the mother carries the
pathologic allele.
ddPCR is a high-sensitivity and high-specificity
technique that improves upon conventional PCR and has been utilized
in prenatal diagnoses of thalassemia (29,46).
Early studies of SEA deletion α-(0) thalassemia using RT-qPCR
demonstrated high sensitivities for Bart's hydrops fetalis
detection. However, these methods could not clearly distinguish
between heterozygous and wild type fetuses (37,46,47). A
study using the ddPCR technique on fetal genomic DNA samples
provided the first accurate classification of fetal SEA genotype,
suggesting that ddPCR is suitable for non-invasive prenatal
diagnosis of SEA deletion (32).
Nonetheless, no prior study had applied ddPCR to cell-free DNA
without sample preprocessing, such as enrichment of fetal DNA by
size selection, to the best of our knowledge.
The present study is the first to showcase the
application of ddPCR to identify the copy number of SEA deletion in
unprocessed cell-free DNA obtained from maternal plasma, thereby
eliminating the need for invasive fetal DNA collection and sample
preprocessing. The SEA CNV assay accurately classified 20 out of 22
samples, and detected Hb Bart's hydrops fetalis with 95%
sensitivity and 91% specificity. A possible explanation for the
misclassified cases may be a low fetal fraction (3% in one case)
and instability of the cell-free fetal DNA in the SEA region
(16). These findings demonstrate
the readiness of ddPCR-based assay for NIPD of SEA deletion α-(0)
thalassemia.
Although the current performances of ddPCR for HbE
and 41/42 (-CTTT) point mutations still do not meet the
requirements for clinical use, the results for HbE are promising.
Out of 16 samples, 10 were correctly classified, and three may be
correctly classified with additional ddPCR experiments, whilst
three misclassifications were made. The association between
inaccurate classifications and high variability amongst triplicate
ddPCR experiments and a low percentage of positive droplets
suggests that these errors were likely caused by poor binding
between ddPCR probes and target alleles. Some remedies for this
issue include designing additional probes for targeting mutated DNA
molecules, pre-amplifying the sample, and performing size selection
to enrich fetal DNA fragments (48,49).
The present study developed ddPCR-based assays for
identifying the fetal thalassemia status from unprocessed cell-free
DNA extracted from maternal plasma. Although the 20 kb deletions of
the SEA region can be reliably detected, the detection accuracies
for HbE and 41/42 (-CTTT) point mutations were too low for
recommendation of this technique for clinical use. The critical
limitation of this work is the small number of HbE and 41/42
(-CTTT) samples, especially the complete lack of cases with a
homozygous mutant allele. Further studies with a larger sample size
and additional ddPCR probe designs are required to improve and
validate the utility of these assays.
Supplementary Material
Dissociation curve analysis of SEA
deletion. (A) The wild type fetus melting temperature peak was
91.5±0.15˚C. (B) The Bart's hydrops fetalis melting temperature
peak was 87.8±0.02˚C. (C) Heterozygous fetus shows the melting
temperature of both types.
Real time PCR graph showing the
decrease in fluorescence signal of the different genotypes Graph of
the fluorescence signal for the (A) wild type, (B) Bart's hydrops
fetalis and (C) Heterozygous genotypes. RFU, relative fluorescence
unit.
Oligonucleotide sequences of the
digital droplet PCR probes and the PCR conditions used.
Acknowledgements
Not applicable.
Funding
This study was supported by funding from Radjavithi Hospital
(grant no. 59194) and the Research Fund of Chulabhorn International
College of Medicine, Thammasat University.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SPO, SJ managed the project. SPO, WQ, SC and KS
conceived and designed the study. SJ recruited patients and
collected data. WN, SPR and KT collected samples and extracted
cell-free DNA. KS performed the experiments. SS and KS analyzed the
data and interpreted the results. KS, SPO and SS wrote and revised
the manuscript. SS, SJ and SPO supervised the project. All authors
provided critical comments and contributed to the revision of the
manuscript. KS and SPO confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Samples were collected with the approval from the
Institutional Review Board of Radjavithi Hospital, Bangkok
(approval no. 59194 date of approval, 17th November 2016). Written
informed consent was obtained from all subjects prior to inclusion
in the study. Written informed consent from the parents or legal
guardians of all subjects under the age of 18 was also
obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Modell B and Darlison M: Global
epidemiology of haemoglobin disorders and derived service
indicators. Bull World Health Organ. 86:480–487. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Galanello R and Cao A: Gene test review.
Alpha-thalassemia. Genet Med. 13:83–88. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Fucharoen S, Winichagoon P, Siritanaratkul
N, Chowthaworn J and Pootrakul P: Alpha- and beta-thalassemia in
Thailand. Ann N Y Acad Sci. 850:412–414. 1998.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cao A and Galanello R: Beta-thalassemia.
Genet Med. 12:61–76. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Origa R: β-thalassemia. Genet Med.
19:609–619. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chaibunruang A, Sornkayasit K,
Chewasateanchai M, Sanugul P, Fucharoen G and Fucharoen S:
Prevalence of thalassemia among newborns: A re-visited after 20
years of a prevention and control program in Northeast Thailand.
Mediterr J Hematol Infect Dis. 10(e2018054)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Fucharoen S and Winichagoon P: Thalassemia
in SouthEast Asia: Problems and strategy for prevention and
control. Southeast Asian J Trop Med Public Health. 23:647–655.
1992.PubMed/NCBI
|
|
8
|
Cao A and Kan YW: The prevention of
thalassemia. Cold Spring Harb Perspect Med.
3(a011775)2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Old J: Chapter 71 - Hemoglobinopathies and
thalassemias. In: Emery and rimoin's principles and practice of
medical genetics. 6th edition. Academic Press, Cambridge, MA,
pp1-44, 2013.
|
|
10
|
Evans MI and Wapner RJ: Invasive prenatal
diagnostic procedures 2005. Semin Perinatol. 29:215–218.
2005.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lo YM, Corbetta N, Chamberlain PF, Rai V,
Sargent IL, Redman CW and Wainscoat JS: Presence of fetal DNA in
maternal plasma and serum. Lancet. 350:485–487. 1997.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jenkins LA, Deans ZC, Lewis C and Allen S:
Delivering an accredited non-invasive prenatal diagnosis service
for monogenic disorders and recommendations for best practice.
Prenat Diagn. 38:44–51. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
Rabinowitz T and Shomron N: Genome-wide
noninvasive prenatal diagnosis of monogenic disorders: Current and
future trends. Comput Struct Biotechnol J. 18:2463–2470.
2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Baysal E and Huisman TH: Detection of
common deletional alpha-thalassemia-2 determinants by PCR. Am J
Hematol. 46:208–213. 1994.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bowden DK, Vickers MA and Higgs DR: A
PCR-based strategy to detect the common severe determinants of
alpha thalassaemia. Br J Haematol. 81:104–108. 1992.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sirichotiyakul S, Charoenkwan P and
Sanguansermsri T: Prenatal diagnosis of homozygous
alpha-thalassemia-1 by cell-free fetal DNA in maternal plasma.
Prenat Diagn. 32:45–49. 2012.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Tungwiwat W, Fucharoen S, Fucharoen G,
Ratanasiri T and Sanchaisuriya K: Development and application of a
real-time quantitative PCR for prenatal detection of fetal
alpha(0)-thalassemia from maternal plasma. Ann N Y Acad Sci.
1075:103–107. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yan TZ, Mo QH, Cai R, Chen X, Zhang CM,
Liu YH, Chen YJ, Zhou WJ, Xiong F and Xu XM: Reliable detection of
paternal SNPs within deletion breakpoints for non-invasive prenatal
exclusion of homozygous α-thalassemia in maternal plasma. PLoS One.
6(e24779)2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pornprasert S, Sukunthamala K, Kunyanone
N, Sittiprasert S, Thungkham K, Junorse S, Pongsawatkul K,
Pattanaporn W, Jitwong C and Sanguansermsri T: Analysis of
real-time PCR cycle threshold of alpha-thalassemia-1 Southeast
Asian type deletion using fetal cell-free DNA in maternal plasma
for noninvasive prenatal diagnosis of bart's hydrops fetalis. J Med
Assoc Thai. 93:1243–1248. 2010.PubMed/NCBI
|
|
20
|
Norton ME, Brar H, Weiss J, Karimi A,
Laurent LC, Caughey AB, Rodriguez MH, Williams J III, Mitchell ME,
Adair CD, et al: Non-invasive chromosomal evaluation (NICE) study:
Results of a multicenter prospective cohort study for detection of
fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol.
207(137)2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bianchi DW, Platt LD, Goldberg JD,
Abuhamad AZ, Sehnert AJ and Rava RP: MatErnal BLood IS Source to
Accurately diagnose fetal aneuploidy (MELISSA) Study Group.
Genome-wide fetal aneuploidy detection by maternal plasma DNA
sequencing. Obstet Gynecol. 119:890–901. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Rava RP, Srinivasan A, Sehnert AJ and
Bianchi DW: Circulating fetal cell-free DNA fractions differ in
autosomal aneuploidies and monosomy X. Clin Chem. 60:243–250.
2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W,
Leung TY, Foo CH, Xie B, Tsui NB, Lun FM, et al: Noninvasive
prenatal diagnosis of fetal chromosomal aneuploidy by massively
parallel genomic sequencing of DNA in maternal plasma. Proc Natl
Acad Sci USA. 105:20458–20463. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xiong L, Barrett AN, Hua R, Ho S, Jun L,
Chan K, Mei Z and Choolani M: Non-invasive prenatal testing for
fetal inheritance of maternal β-thalassaemia mutations using
targeted sequencing and relative mutation dosage: A feasibility
study. BJOG. 125:461–468. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lam KW, Jiang P, Liao GJ, Chan KC, Leung
TY, Chiu RW and Lo YM: Noninvasive prenatal diagnosis of monogenic
diseases by targeted massively parallel sequencing of maternal
plasma: Application to β-thalassemia. Clin Chem. 58:1467–1475.
2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lo YM, Chan KC, Sun H, Chen EZ, Jiang P,
Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR and Chiu RWK:
Maternal plasma DNA sequencing reveals the genome-wide genetic and
mutational profile of the fetus. Sci Transl Med.
2(61ra91)2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Orhant L, Anselem O, Fradin M, Becker PH,
Beugnet C, Deburgrave N, Tafuri G, Letourneur F, Goffinet F, El
Khattabi LA, et al: Droplet digital PCR combined with
minisequencing, a new approach to analyze fetal DNA from maternal
blood: Application to the non-invasive prenatal diagnosis of
achondroplasia. Prenat Diagn. 36:397–406. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Hudecova I: Digital PCR analysis of
circulating nucleic acids. Clin Biochem. 48:948–956.
2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
van Ginkel JH, Huibers MMH, van Es RJJ, de
Bree R and Willems SM: Droplet digital PCR for detection and
quantification of circulating tumor DNA in plasma of head and neck
cancer patients. BMC Cancer. 17(428)2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gruber A, Pacault M, El Khattabi LA,
Vaucouleur N, Orhant L, Bienvenu T, Girodon E, Vidaud D, Leturcq F,
Costa C, et al: Non-invasive prenatal diagnosis of paternally
inherited disorders from maternal plasma: Detection of NF1 and CFTR
mutations using droplet digital PCR. Clin Chem Lab Med. 56:728–738.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hudecova I and Chiu RWK: Non-invasive
prenatal diagnosis of thalassemias using maternal plasma cell free
DNA. Best Pract Res Clin Obstet Gynaecol. 39:63–73. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Pornprasert S and Prasing W: Detection of
alpha(0)-thalassemia South-East Asian-type deletion by droplet
digital PCR. Eur J Haematol. 92:244–248. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lun FM, Tsui NB, Chan KC, Leung TY, Lau
TK, Charoenkwan P, Chow KCK, Lo WYW, Wanapirak C, Sanguansermsri T,
et al: Noninvasive prenatal diagnosis of monogenic diseases by
digital size selection and relative mutation dosage on DNA in
maternal plasma. Proc Natl Acad Sci USA. 105:19920–19925.
2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kho SL, Chua KH, George E and Tan JA: A
novel gap-PCR with high resolution melting analysis for the
detection of alpha-thalassaemia Southeast Asian and Filipino
β˚-thalassaemia deletion. Sci Rep. 5(13937)2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chan KC, Ding C, Gerovassili A, Yeung SW,
Chiu RW, Leung TN, Lau TK, Chim SS, Chung GT, Nicolaides KH and Lo
YM: Hypermethylated RASSF1A in maternal plasma: A universal fetal
DNA marker that improves the reliability of noninvasive prenatal
diagnosis. Clin Chem. 52:2211–2218. 2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Manokhina I, Singh TK, Peñaherrera MS and
Robinson WP: Quantification of cell-free DNA in normal and
complicated pregnancies: Overcoming biological and technical
issues. PLoS One. 9(e101500)2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
El Karoui N, Zhou W and Whittemore AS:
Getting more from digital SNP data. Stat Med. 25:3124–3133.
2006.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Pornprasert S, Phusua A, Suanta S, Saetung
R and Sanguansermsri T: Detection of alpha-thalassemia-1 Southeast
Asian type using real-time gap-PCR with SYBR Green1 and high
resolution melting analysis. Eur J Haematol. 80:510–514.
2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tsui NB, Kadir RA, Chan KC, Chi C, Mellars
G, Tuddenham EG, Leung TY, Lau TK, Chiu RW and Lo YM: Noninvasive
prenatal diagnosis of hemophilia by microfluidics digital PCR
analysis of maternal plasma DNA. Blood. 117:3684–3691.
2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lun FM, Chiu RW, Chan KC, Leung TY, Lau TK
and Lo YM: Microfluidics digital PCR reveals a higher than expected
fraction of fetal DNA in maternal plasma. Clin Chem. 54:1664–1672.
2008.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Amicucci P, Gennarelli M, Novelli G and
Dallapiccola B: Prenatal diagnosis of myotonic dystrophy using
fetal DNA obtained from maternal plasma. Clin Chem. 46:301–302.
2000.PubMed/NCBI
|
|
42
|
Saito H, Sekizawa A, Morimoto T, Suzuki M
and Yanaihara T: Prenatal DNA diagnosis of a single-gene disorder
from maternal plasma. Lancet. 356(1170)2000.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chiu RW, Lau TK, Leung TN, Chow KC, Chui
DH and Lo YM: Prenatal exclusion of beta thalassaemia major by
examination of maternal plasma. Lancet. 360:998–1000.
2002.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Ding C, Chiu RW, Lau TK, Leung TN, Chan
LC, Chan AY, Charoenkwan P, Ng IS, Law HY, Ma ES, et al: MS
analysis of single-nucleotide differences in circulating nucleic
acids: Application to noninvasive prenatal diagnosis. Proc Natl
Acad Sci USA. 101:10762–10767. 2004.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hudecova I, Jiang P, Davies J, Lo YMD,
Kadir RA and Chiu RWK: Noninvasive detection of F8 int22h-related
inversions and sequence variants in maternal plasma of hemophilia
carriers. Blood. 130:340–347. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Taylor SC, Carbonneau J, Shelton DN and
Boivin G: Optimization of droplet digital PCR from RNA and DNA
extracts with direct comparison to RT-qPCR: Clinical implications
for quantification of Oseltamivir-resistant subpopulations. J Virol
Methods. 224:58–66. 2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Sirichotiyakul S, Charoenkwan P and
Sanguansermsri T: Prenatal diagnosis of homozygous
alpha-thalassemia-1 by cell-free fetal DNA in maternal plasma.
Prenat Diagn. 32:45–49. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Whale AS, Cowen S, Foy CA and Huggett JF:
Methods for applying accurate digital PCR analysis on low copy DNA
samples. PLoS One. 8(e58177)2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Vermeulen J, Derveaux S, Lefever S, De
Smet E, De Preter K, Yigit N, De Paepe A, Pattyn F, Speleman F and
Vandesompele J: RNA pre-amplification enables large-scale RT-qPCR
gene-expression studies on limiting sample amounts. BMC Res Notes.
2(235)2009.PubMed/NCBI View Article : Google Scholar
|