Introduction
Chronic myeloid leukemia (CML) is a
myeloproliferative neoplasm in which granulocytes are the major
proliferative component (1). The
annual incidence of CML in Europe is 0.7-1 cases per 100,000
individuals, with a male predominance (male/female ratio: 1.2-1.7)
and an average age at presentation of 57-60 years (2).
CML is characterized by a reciprocal chromosomal
translocation between the chromosomes 9 and 22: t(9;22)(q34;q11),
which results in the formation of the Philadelphia (Ph) chromosome,
generating the fusion of BCR and ABL1 genes (3). BCR-ABL1 chimeric gene produces a
protein with increased tyrosine kinase activity, and this results
in constitutively activated cell-signaling pathways, causing
aberrant proliferation, apoptosis and cellular adhesion. Thus,
tyrosine kinase inhibitors (TKIs) are the treatment of choice for
these patients (3).
Nevertheless, BCR-ABL1 rearrangements are not
exclusive to CML, and they may also be found in acute lymphoblastic
leukemia and acute myeloid leukemia (4). At the time of diagnosis, 5-10% of
patients with CML possess a variant Ph chromosome translocation,
involving chromosome 22 and a chromosome other than chromosome 9
(simple variant translocation), or one or more chromosomes in
addition to chromosomes 9 and 22 (complex variant translocation)
(5). Their breakpoints are
frequently located in the G-light bands, in the CG-richest regions
(6). Different mechanisms have been
proposed to explain the variant Ph translocations (6). However, the specifics remain to be
determined. The prognosis of these patients has been analyzed in
case reports or small series, which have reported variable outcomes
(7,8).
Despite the fact that there are reports describing
the involvement of every chromosome in variant translocations with
different breakpoints, their frequency is highly variable (6,9). The X
chromosome(s) in particular, are very rarely affected.
In the present report, the case of a patient with
CML with a variant translocation involving the X chromosome is
described. Additionally, a review of the cases previously reported,
including their clinical and genetic features, the treatment they
received and the response to it was performed.
Materials and methods
Approval and consent
Human material and clinical information were
obtained in accordance with the Declaration of Helsinki (10). Informed consent was obtained from the
patient and the present study was approved by the Hospital Clínico
San Carlos Ethics Committee.
Literature review
A review of the literature was performed using the
PubMed database using the following search terms: ‘variant
Philadelphia translocation involving X chromosome’, ‘variant
Philadelphia translocation’ and ‘variant translocation in chronic
myeloid leukemia’. All the studies that were found describing CML
in chronic phase with variant Ph translocation involving the X
chromosome, and were written in English, were included. Cases that
were in the CML accelerated phase or blast crisis, and acute
leukemia with variant Ph translocations were excluded.
Karyotype
Based on the white blood cell counts, an appropriate
amount of heparinized bone marrow (~300 µl) was added to 5 ml
RPMI-1640 culture medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 2 mM L-glutamine (Biological Industries;
Sartorius AG), 30% FBS (Gibco; Thermo Fisher Scientific, Inc.), 15
mM HEPES (Gibco; Thermo Fisher Scientific, Inc.), and 75 U/ml
Penicillin-75 µg/ml Streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) per a culture tube. Bone marrow cells were cultured in this
mitogen-free media at 37˚C. After 24 or 48 h, 100 µl Colcemid was
added to the sample and incubated for 60 min at 37˚C. Then, the
cells were centrifuged for 10 min at room temperature at 500 x g,
the supernatant was removed, 5 ml KCl 0.075 M was added and the
sample was incubated for a further 30 min at 37˚C. Samples were
centrifuged again at room temperature centrifugation at 500 x g,
the supernatant was removed and 5 ml Carnoy's solution was added
gradually to resuspend the pellet. The previous steps were repeated
2-3 times, until the pellet was clear. Pellet smears were prepared
on glass slides. Each slide was incubated at 37˚C for 4 sec in
trypsin solution prepared with 270 ml DPBS (Gibco; Thermo Fisher
Scientific, Inc.; Thermo Fisher Scientific, Inc) and 30 ml Trypsin
(Lonza Group, Ltd.). Next, the samples were stained for 90 sec at
room temperature with Leishman stain prepared with 3 ml pH 6.8
buffer solution made with 1 Buffer tablet (Merck KGaA) in 1,000 ml
distilled water and 1 ml Leishman stain stock [1.5 g Leishman's
eosin methylene blue (Merck KGaA) in 1,000 ml methanol]. Finally,
G-banded cytogenetic analysis of 20 metaphases was performed.
Fluorescence in situ hybridization
(FISH)
FISH was performed on cells from bone marrow samples
using a Vysis LSI BCR/ABL1/ASS1 Tri-Color Dual Fusion FISH Probe
kit and Vysis Whole Chromosome Painting for chromosome X (WCP X)
SpectrumGreen Probe (both from Abbott Laboratories) according to
the manufacturer's protocol. Briefly, the slides were immersed in
the denaturation solution (formamide) for 5 min at 73±1˚C. Then,
they were dehydrated for 1 min in 70% ethanol, followed by 1 min in
85% ethanol, and 1 min in 100% ethanol. A total of 10 µl probe
mixture (7 µl LSI/WCP hybridization buffer, 1 µl probe, 2 µl
purified water), pre-heated at 73±1˚C in a water bath for 5 min,
was applied to one target area and the coverslip was immediately
added. The slides were placed in a pre-warmed humidified box at
37˚C for 16 h. The coverslip was removed and the slide was washed
for 2 min in 0.4X saline-sodium citrate (SSC) buffer (0.3 M sodium
chloride and 30 mM trisodium citrate, adjusted to pH 7.0 with HCl)
at 73±1˚C, and then for 5-8 sec in 2X SSC at room temperature; 10
µl DAPI II counterstaining solution was applied at room temperature
and the coverslips were added. A minimum of 200 interphases were
observed for each Vysis probe.
Array-comparative genomic
hybridization (CGH)
The sample was hybridized against a female human
reference commercial DNA sample (Promega Corporation) using a 60k
KaryoNIM® array-CGH, designed by NIMGenetics®
for the detection of copy number variations (CNV) (Agilent
Technologies, Inc.). Data analysis was performed using hg19 genomic
build and the ADM-2 statistic (set as 6). At least five probes were
accepted as the threshold for detected CNVs. Analytical validation
of this platform confirms an informative range of 350 kb across the
whole genome. Additionally, the syndromic regions included in this
design were covered with an estimated average resolution of ~100
kb. The interpretation of the CNVs was performed as described by
Vermeesch et al (11).
RNA extraction and reverse
transcription
RNA was isolated from bone marrow mononuclear cells
or peripheral blood total leukocytes using TRIzol®
reagent (Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocol. RNA concentration was quantified using spectrophotometry
on a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific,
Inc.) and 2 µg RNA was reverse transcribed to cDNA using
SuperScript IV Reverse Transcriptase with random hexamers according
to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc.).
Reverse-transcription qualitative PCR
(RT-PCR)
RT-PCR analysis of bone marrow mononuclear cells,
obtained by Ficoll (Lymphoflot; Bio-Rad Laboratories, Inc.)
gradient centrifugation at 500 x g for 30 min at room temperature,
was performed in accordance with the BIOMED I protocols (12), using 50 ng cDNA for each assay and
ampliTaq Gold DNA polymerase for amplification (Applied Biosystems;
Thermo Fisher Scientific, Inc.). BCR/ABL1 MBCR fusion
transcripts were amplified with the primers BCR-b1-A,
5'-GAAGTGTTTCAGAAGCTTCTCC-3' and ABL-a3-B,
5'-GTTTGGGCTTCACACCATTCC-3' (12).
ABL1 control was amplified with ABL1 forward,
5'-CTTCTCGCTGGACCCAGTGA-3' and reverse,
5'-TGTGATTATAGCCTAAGACCCGGAG-3'. Amplified products were viewed
using 2% agarose gel electrophoresis using DNA Phi-X-174 Hae III
digested molecular weight marker (Sigma-Aldrich) and ethidium
bromide-staining.
Reverse transcription-quantitative PCR
(RT-qPCR)
A total of 9 ml peripheral blood was collected in
EDTA tubes. Total leukocytes were obtained by centrifugation at
2,000 x g for 15 min at 4˚C, and red blood cells were removed using
Erythrocyte Lysis Buffer-Buffer EL (Qiagen GmbH).
BCR-ABL1 expression was measured using
RT-qPCR as described previously by the Europe Against Cancer
Program (13). ABL1 was used
as a reference gene (14). The
reaction was performed in a 20 µl reaction mixture containing
TaqMan Universal PCR MasterMix or TaqMan Fast Universal PCR
MasterMix (Applied Biosystems; Thermo Fisher Scientific, Inc.) with
100 ng cDNA for each assay in duplicate, either in an ABI Prism
7700 Sequence Detection system or a 7500 Fast Real Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.).
Quantification was expressed on the International Scale (IS) as a
percentage of BCR-ABL1 copy number using ABL1 as a
reference gene (15), using a
BCR-ABL1 Mbcr IS-Major Molecular Response (MMR) kit (Qiagen
GmbH).
Case report
A 51-year-old female consulted her GP due to the
presence of abnormal bruises on her limbs. Laboratory results
revealed a leucocyte count of 90,300/µl, 12 g/dl hemoglobin and a
platelet count of 270,000/µl. Peripheral blood smears showed
neutrophilia with granulocytes at various stages of
differentiation: 49% neutrophils, 17% metamyelocytes, 13%
myelocytes, 1% promyelocytes, 9% lymphocytes, 5% eosinophils and 6%
basophils. Bone marrow aspiration morphology showed
hypercellularity with granulocytic proliferation without
maturational hiatus, a substantially increased myeloid: erythroid
ratio, sea-blue histiocytes, and augmented basophils and
eosinophils. The proportion of megakaryocytes was also increased;
several of which were small and exhibited hypolobulated nuclei. A
hematoma in the right upper limb was observed in the physical
examination. Abdominal ultrasound revealed a mild splenomegaly of
13.3 cm.
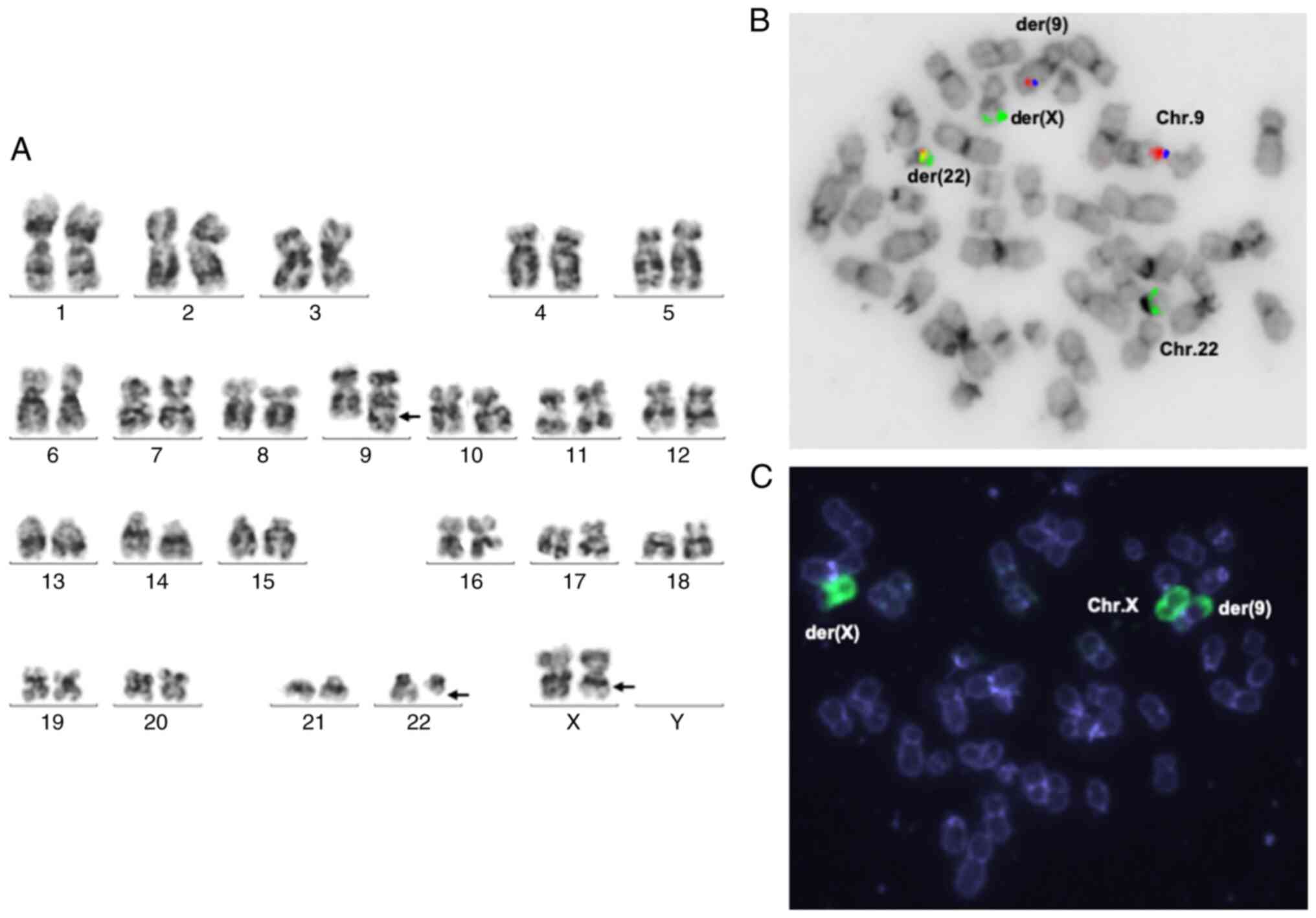
G-banded cytogenetic analysis of bone marrow cells
showed the following karyotype: 46,X,t(X;9;22)(q22?,q34;q11.2)
(Fig. 1A). FISH analysis of cells
from the bone marrow showed an atypical pattern of BCR-ABL1
rearrangement in >90% cells (Fig.
1B and C). Analysis using
array-CGH 60k detected no deletions (Fig. S1). RT-PCR analysis of bone marrow
mononuclear cells demonstrated the expression of e13a2 (b2a2)
BCR-ABL1 RNA (Fig. 2A).
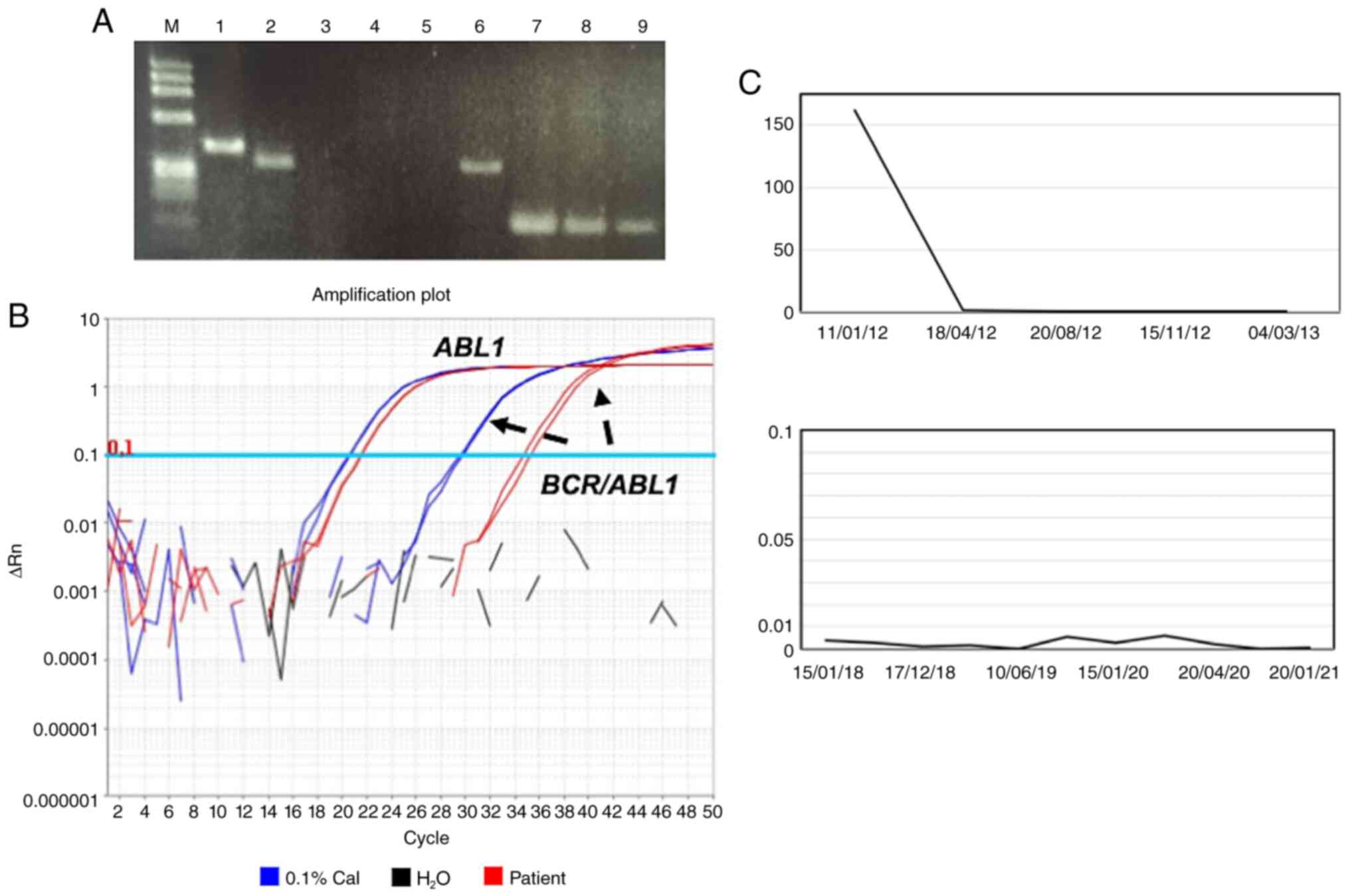 | Figure 2RT-qPCR follow-up shows a good
response to a second generation tyrosine kinase inhibitor in the
patient with variant Ph involving X chromosome. (A) Ethidium
bromide-stained agarose gel showing electrophoresis of
BCR-ABL1 RT-PCR (lanes 1-6) and ABL1 control (lanes
7-9). Lane descriptions: M, molecular weight marker DNA Phi-X-174
Hae III digested; 1, corresponds to a patient who expressed e14a2
(b3a2) BCR-ABL1 transcript; 2, shows the BCR/ABL1
amplification product of the case described in the present study
that possessed an e13a2 (b2a2) transcript; 3, negative case; 4 and
5, blank negative controls (water); 6, e13a2 (b2a2) transcript
positive control; 7-9, ABL1 control amplification of
patients 1-3, respectively. (B) Representative real time
amplification plot of the last year of follow-up showing the
patient results in red, water control in black and the 0.1% (major
molecular response) IS calibrator in blue. PCR duplicates are
shown, the curves on the left correspond to ABL1
amplification with similar results in the patient and the
calibrator. The curves on the right are BCR-ABL1 data
showing the difference between the patient and the calibrator. (C)
BCR-ABL1 RT-qPCR monitoring expressed on the IS as a
percentage of BCR-ABL1/ABL1 normalized copy number (Y axis),
only a selection of the dates are indicated at the X axis to
facilitate the reading. The first year follow-up is shown at the
top; the last 3 years, showing deep molecular response, can be seen
at the bottom. RT-qPCR, Reverse transcription-quantitative PCR;
RT-PCR, Reverse transcription-qualitative PCR; IS, International
Scale; ∆Rn, Δ normalized reporter value, where Rn is the reporter
fluorescence signal normalized to the signal of the passive
reference dye: ΔRn=Rn - baseline. |
Given the above findings, the patient was diagnosed
with chronic phase CML with a variant Ph translocation. The risk
group for this patient was low, according to Sokal and European
Treatment and Outcome Study for CML (EUTOS) scores, and
intermediate using the Hasford Score (16-18).
The patient was treated with nilotinib 300 mg twice a day, and
exhibited a complete hematologic response after 2 weeks of
treatment, and a complete cytogenetic response after 3 months. The
patient was monitored by RT-qPCR following the Europe Against
Cancer Program (13), using
ABL1 as a reference gene (14) and expressed on the IS as a percentage
of BCR-ABL1 (15). The
patient had an optimal molecular response according to the ELN
criteria (15): BCR-ABL1 ≤10%
after 3 months of treatment (1.12%) and a MMR after 12 months
(0.02%), that was subsequently maintained. A deep molecular
response was assessed following EUTOS laboratory recommendations
(19), and this was achieved after
32 months of starting treatment, and has been maintained even after
9 years of follow-up (Fig. 2B and
C).
Discussion
CML is characterized by the presence of a
t(9;22)(q34;q11.2) translocation, which results in the Ph
chromosome. Of CML cases, >85% of patients possess a classic
translocation, 5-10% have a variant translocation and <5%
possess a masked Ph translocation that is not detected by
conventional cytogenetic analysis; however, the BCR-ABL1
fusion gene is identified by FISH and/or PCR (20).
The involvement of the X chromosome in the variant
Ph has seldom been described. It has been detected in case series;
however, in the majority of these, the description of clinical
features, the treatment administered, the response to treatment and
other data are not individually available (reviewed in Table I).
 | Table IDescription of previously published
cases of a variant translocation involving chromosome X focusing on
chronic phase CML. |
Table I
Description of previously published
cases of a variant translocation involving chromosome X focusing on
chronic phase CML.
| Patient no. | Sex/age | Diagnosis | Description of the
studies | Variant
karyotype | Treatment and
response | Survival | Refs |
|---|
| 1 | NA | CML CP (0.29%). No
deletion on der (9). | 336 CML cases: 25
cases in CP with vPh (7.4%), 1 of them involving X | 46,XY,t(X;9;22)
(p22;q34;q11.2)[20] | NA | NA | (21) |
| 2 | M/33 | CML | 1 ALL Ph+ case, 1
AML Ph+ case and 5 CML cases (2 cases with masked Ph and 3 cases
with vPh), 1 of them with vPh involving X. | 46,Y,t(X;9;22)
(p11.2;q34;q11.2)[3]/ 47,idem,+der(9)t(X;9;22)
(p11.2;q34;q11.2)[17] | NA | NA | (20) |
| 3 | NA | CML | 22 CML cases with
vPh, 1 of them involving X. |
46,X,t(X;9;22)(q24;q34;q11.2)[20] | HU-IFNα; mCgR at 1
year. | NA | (22) |
| 4 | F/13 | CML CP | 93 CML CP cases
treated with imatinib: 8 with vPh, 1 of them involving X. Hb, 11
g/dl; WBC, 134,000/µl; platelet count, 222,000/µl. Sokal score,
low. |
46,X,t(X;9;22)(q28;q34;q11.2)[20] | Imatinib; CCgR
after 6, 12 and 18 months. | NA | (23) |
| 5 | M/54 | CML | 2 CML cases with
vPh: 1 involving X. Hb, 12 g/dl; WBC, 76,500/µl; platelet count,
134,000/µl. |
47,Y,t(X;22)(p22;q11), +19 | Busulfan | NA | (24) |
| 6 | NA | CML | 721 CML cases
treated with imatinib: 44 cases with vPh (6.1%), 1 of them
involving X (0.13%). |
46,X,t(9;22;X)(q34;q11.2;p11.2)[20] | Imatinib | NA | (25) |
| 7 | NA | CML | 94 cases with vPh:
3 involving X; 1 simplea, 1 complex
three-wayb and 1 complex four-way. | t(X;9;17;22) | NA | NA | (26,27) |
| 8 | F/27 | CML from ET | 5 CML cases with
vPh: 1 case involving X. Hb, 13 g/dl; WBC, 12,200/µl; platelets,
2,030,000/µl. |
46,X,t(X;9;22)(q11;q34;q11)[14] | Aspirin and
pipobroman, no response. Thrombocytapheresis, nitrogen mustard and
busulfan. | NA | (28,29) |
| 9 | NA | CML CP | 615 CML CP cases:
72 with vPh (11.7%), 1 of them involving X (0.16%). |
46,Y,t(X;9;22)(p11;q34;q11) | Imatinib | NA | (30) |
| 10 | NA | CML CP | 452 CML CP cases:
43 with vPh (9.5%), 1 of them involving X (0.22%), presented del
(9)(q33q34). |
t(9;X;22)(q34;q13.1;q11) | NA | NA | (31) |
| 11 and | M/54 | CML CP | 7 CML CP cases with
vPh: 2 of them involving X, | 46,Y,t(X;9;11;22)
(p11;q34;q12;q11)[20] | NA | NA | (32) |
| 12 | M/25 | | 1 complex and 1
simple. |
46,Y,t(X;22)(q28;q11)[12] | NA | NA | (32) |
| 13 and 14 | NA | CML | 3,361 newly
diagnosed CML: 109 vPh (3.2%), 2 of them involving X (0.06%). | NA | NA | NA | (33) |
| 15 | F/48 | CML CP | 4 CML cases with
vPh: 1 case involving X. Hb, 9 g/dl; WBC, 4,800/µl; platelets,
9,150/µl. |
46,XX,t(X;9;22)(q13;q34;q11.2)[10] | HU and Imatinib,
CHR | 4 months | (34) |
| 16 | NA | CML | 507 CML: 28 vPh
(5.5%); 1 of them involving X (0.19%). | Xq24 | NA | NA | (35) |
In the present report, the case of a new patient
with X chromosome-variant Ph CML is described, and a review of
cases reported in the literature is provided. A total of 16 cases
were identified (Table I) (20-35).
Using this data, it was estimated that the frequency of variant Ph
CML involving the X chromosome is <1% of all CML cases.
Bonifacio et al (33), in the
most recent and largest study identified on this topic, described
only 2 patients with complex variant translocations involving the X
chromosome amongst 3,361 newly diagnosed CML cases; thus, it only
accounted for 0.06% of cases (Table
I). The mean age of patients with CML with the variant Ph
translocation involving the X chromosome, including just the cases
for which data are available (7 cases out of 16), was 36 years
(range 13-54 years; Table I).
The reported breakpoints were Xp22, Xp11.2, Xq24,
Xq28, Xp22, Xp11.2, Xq11, Xp11, Xq13.1, Xp11, Xq28, Xq13 and Xq24,
with Xp11 being the most frequent breakpoint (four cases). In three
cases, the breakpoint was unknown. Most cases were complex
three-way variant Ph; cases 5 and 12 were simple variant Ph and
cases 7 and 11 were four-way variant Ph (Table I).
The treatment administered is detailed in a few
cases (7 out of 16 cases): Hydroxyurea-interferon α for case 3,
busulfan for case 5, aspirin and pipobroman, thrombocytapheresis,
nitrogen mustard and busulfan for case 8, imatinib for cases 4, 6
and 9, and hydroxyurea and imatinib for case 15. The response to
treatment was specified in 3 cases (3, 4 and 15). In case 3, a
minor cytogenetic response was described 1 year after starting
therapy, whereas case 4 presented a complete cytogenetic response
after 6, 12 and 18 months, and case 15 achieved complete a
hematologic response, although follow-up cytogenetic studies were
not possible as the patient passed away 4 months after diagnosis.
Survival was only reported in case 15 (Table I).
Data concerning prognosis of these patients is
controversial. Certain studies have shown that patients with a
variant Ph chromosome translocation have a worse response to
treatment compared with those with the classic Ph chromosome
(8,22), whereas several other studies have
reported no differences with regard to treatment outcomes (7,25,36).
Certain studies have described a higher frequency of deletions on
the derivative chromosome 9 in patients with variant
translocations, which may be responsible for the worse prognosis
reported (21,22). Case 10 presented a deletion on
derivative chromosome 9, but the treatment administrated, the
response to it and the survival information is not available.
In summary, the literature review revealed that the
studied translocation is a very rare rearrangement, as only 16
cases involving chromosome X have been described thus far.
Additionally, it does not offer enough data to allow drawing
definitive conclusions on the prognosis of patients with variant
translocations involving the X chromosome.
The mechanism of production of the variant
translocation may represent a clonal evolution of the malignant
cells, a phenomenon which is known to precede or to coincide with
the progression of the disease (22), and therefore may be associated with
the worse prognosis of some of these patients. However, the origin
of the variant translocations is not well understood, and it may be
heterogeneous in nature. The patient described here did not present
a deletion of derivative chromosome 9, nor progression of the
disease to accelerated phase or blast crisis in a long follow-up
period. In fact, the patient achieved ELN optimal molecular
response (MR) after 3 and 12 months and has remained in deep MR,
BCR-ABL1 expression on the IS ≥4 log10 reduction
from IRIS baseline -MR4 or more (19), 110 months after starting treatment.
This shows that some patients with variant translocations can be
successfully treated with second generation TKIs.
The 2020 ELN recommendations have recently
established that generic imatinib is a cost effective first line
treatment for chronic phase CML with low toxicity, when compared
with second generation TKIs. This guide also gives clear rules as
to when the therapy should be discontinued (37). However, when the patient in the
present report was diagnosed 9 years ago, the treatment selection
was based on the quicker and deeper MR achieved with second
generation TKIs, which was considered a potential advantage with
more options for treatment discontinuation, that requires the
maintained deep MR (38,39). Although this option is particularly
recommended for women of fertile age, this patient had several
decades of life expectancy, and the best opportunity to cease
treatment was prioritized. This means of clinical reasoning is
still under consideration as a possible exception in first line
treatment choice (40).
The case in the present report had a good response
to second generation TKIs. However, clinical behavior of these
variant translocations requires further characterization, as well
as additional investigation on their prognostic impacts,
particularly with regard to sensitivity to imatinib and second
generation TKIs.
Supplementary Material
Comparative genomic
hybridization-array showing a normal female pattern. No gains or
losses were detected in the breakpoints of the chromosomes involved
in the translocation t(X; 9; 22) or in other chromosomes.
Acknowledgements
Not applicable.
Funding
This work was supported in part by the Hay Esperanza Foundation
(grant no. EAM.C03FHE) Madrid, Spain.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding authors on reasonable
request.
Authors' contributions
AI, RO, CC and EA performed experiments and
participated in writing the manuscript. EA edited the manuscript.
All authors have read and approved the final manuscript. All
authors confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Human material and clinical information were
obtained in accordance with the Declaration of Helsinki, informed
consent was obtained from the patient and the present study was
approved by the Hospital Clínico San Carlos Ethics Committee.
Patient consent for publication
The patient provided written informed consent for
the publication of the data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H and Thiele J (eds): WHO classification of tumour
of haematopoietic and lymphoid tissues. 4th edition. Vol 2. Lyon,
pp30-36, 2017.
|
|
2
|
Höglund M, Sandin F and Simonsson B:
Epidemiology of chronic myeloid leukaemia: An update. Ann Hematol.
94 (Suppl 2):S241–247. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chereda B and Melo JV: Natural course and
biology of CML. Ann Hematol. 94 (Suppl 2):S107–S121.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cho SY, Kim SY, Jeon YL, Oh SH, Cho EH,
Lee WI, Cho KS and Park TS: A novel three-way Ph variant t(8;9;22)
in adult acute lymphoblastic leukemia. Ann Clin Lab Sci. 41:71–78.
2011.PubMed/NCBI
|
|
5
|
Jabbour E and Kantarjian H: Chronic
myeloid leukemia: 2018 update on diagnosis, therapy and monitoring.
Am J Hematol. 93:442–459. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Costa D, Grau J, Espinet B, Arias A, Gómez
C, López-Guerra M, Nomdedeu M and Cervantes F: Conventional and
molecular cytogenetic studies to characterize 32 complex variant
Philadelphia translocations in patients with chronic myeloid
leukemia. Oncol Lett. 17:5705–5710. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Marzocchi G, Castagnetti F, Luatti S,
Baldazzi C, Stacchini M, Gugliotta G, Amabile M, Specchia G,
Sessarego M, Giussani U, et al: Variant Philadelphia
translocations: molecular-cytogenetic characterization and
prognostic influence on frontline imatinib therapy, a GIMEMA
Working Party on CML analysis. Blood. 117:6793–6800.
2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Stagno F, Vigneri P, Del Fabro V, Stella
S, Cupri A, Massimino M, Consoli C, Tambè L, Consoli M, Agostino
Antolino and Francesco Di Raimondo: Influence of complex variant
chromosomal translocations in chronic myeloid leukemia patients
treated with tyrosine kinase inhibitors. Acta Oncol. 49:506–508.
2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Manabe M, Yoshii Y, Mukai S, Sakamoto E,
Kanashima H, Inoue T and Teshima H: A rare t(9;22;16)(q34;q11;q24)
translocation in chronic myeloid leukemia for which imatinib
mesylate was effective: A case report. Leuk Res Treatment.
2011(592519)2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
World Medical Association Declaration of
Helsinki. Ethical Principles for Medical Research Involving Human
Subjects. Bulletin of the World Health Organization. 9:373–374.
2001.
|
|
11
|
Vermeesch JR, Brady PD, Sanlaville D, Kok
K and Hastings RJ: Genome-wide arrays: quality criteria and
platforms to be used in routine diagnostics. Hum Mutat. 33:906–915.
2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
van Dongen JJ1, Macintyre EA, Gabert JA,
Delabesse E, Rossi V, Saglio G, Gottardi E, Rambaldi A, Dotti G,
Griesinger F, et al: Standardized RT-PCR analysis of fusion gene
transcripts from chromosome aberrations in acute leukemia for
detection of minimal residual disease. Report of the BIOMED-1
Concerted Action: investigation of minimal residual disease in
acute leukemia. Leukemia. 13:1901–1928. 1999.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gabert J, Beillard E, van der Velden VH,
Bi W, Grimwade D, Pallisgaard N, Barbany G, Cazzaniga G, Cayuela
JM, Cavé H, et al: Standardization and quality control studies of
‘real-time’ quantitative reverse transcriptase polymerase chain
reaction of fusion gene transcripts for residual disease detection
in leukemia - a Europe Against Cancer program. Leukemia.
17:2318–2357. 2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Beillard E, Pallisgaard N, van der Velden
VH, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E,
Gottardi E, Saglio G, et al: Evaluation of candidate control genes
for diagnosis and residual disease detection in leukemic patients
using ‘real-time’ quantitative reverse-transcriptase polymerase
chain reaction (RQ-PCR) - a Europe against cancer program.
Leukemia. 17:2474–2486. 2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Baccarani M, Deininger MW, Rosti G,
Hochhaus A, Soverini S, Apperley JF, Cervantes F, Clark RE, Cortes
JE, Guilhot F, et al: European LeukemiaNet recommendations for the
management of chronic myeloid leukemia: 2013. Blood. 122:872–884.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sokal JE, Cox EB, Baccarani M, Tura S,
Gomez GA, Robertson JE, Tso CY, Braun TJ, Clarkson BD, Cervantes F,
et al: Prognostic discrimination in ‘good-risk’ chronic
granulocytic leukemia. Blood. 63:789–799. 1984.PubMed/NCBI
|
|
17
|
Hasford J, Baccarani M, Hoffmann V,
Guilhot J, Saussele S, Rosti G, Guilhot F, Porkka K, Ossenkoppele
G, Lindoerfer D, et al: Predicting complete cytogenetic response
and subsequent progression-free survival in 2060 patients with CML
on imatinib treatment: the EUTOS score. Blood. 118:686–692.
2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hasford J, Pfirrmann M, Hehlmann R, Allan
NC, Baccarani M, Kluin-Nelemans JC, Alimena G, Steegmann JL and
Ansari H: A new prognostic score for survival of patients with
chronic myeloid leukemia treated with interferon alfa. J Natl
Cancer Inst. 90:850–858. 1998.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Cross NC, White HE, Colomer D, Ehrencrona
H, Foroni L, Gottardi E, Lange T, Lion T, Machova Polakova K,
Dulucq S, et al: Laboratory recommendations for scoring deep
molecular responses following treatment for chronic myeloid
leukemia. Leukemia. 9:999–1003. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen Z, Morgan R, Berger CS, Pearce-Birge
L, Stone JF and Sandberg AA: Identification of masked and variant
Ph (complex type) translocations in CML and classic Ph in AML and
ALL by fluorescence in situ hybridization with the use of bcr/abl
cosmid probes. Cancer Genet Cytogenet. 70:103–107. 1993.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bennour A, Sennana H, Laatiri MA, Elloumi
M, Khelif A and Saad A: Molecular cytogenetic characterization of
variant Philadelphia translocations in chronic myeloid leukemia:
genesis and deletion of derivative chromosome 9. Cancer Genet
Cytogenet. 194:30–37. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gorusu M, Benn P, Li Z and Fang M: On the
genesis and prognosis of variant translocations in chronic myeloid
leukemia. Cancer Genet Cytogenet. 173:97–106. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Koshiyama DB, Capra ME, Paskulin GA, Rosa
RF, Oliveira CA, Vanelli T, Fogliatto LM and Zen PR: Cytogenetic
response to imatinib treatment in Southern Brazilian patients with
chronic myelogenous leukemia and variant Philadelphia chromosome.
Ann Hematol. 92:185–189. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hossfeld DK and Köhler S: New
translocations in chronic granulocytic leukaemia: t(X;22)(p22;q11)
and t(15;22)(q26;q11). Br J Haematol. 41:185–191. 1979.PubMed/NCBI View Article : Google Scholar
|
|
25
|
El-Zimaity MMT, Kantarjian H, Talpaz M,
O'Brien S, Giles F, Garcia-Manero G, Verstovsek S, Thomas D,
Ferrajoli A, Hayes K, et al: Results of imatinib mesylate therapy
in chronic myelogenous leukaemia with variant Philadelphia
chromosome. Br J Haematol. 125:187–195. 2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mitelman F and Levan G: Clustering of
aberrations to specific chromosomes in human neoplasms. Hereditas.
89:207–232. 1978.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Levan G and Mitelman F: The different
origin of primary and secondary chromosome aberrations in cancer.
Haematol Blood Transfus. 26:160–166. 1981.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fitzgerald PH, McEwan C, Fraser J and
Beard ME: A complex Ph1 translocation in a patient with primary
thrombocythaemia. Br J Haematol. 47:571–575. 1981.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Morris CM, Rosman I, Archer SA, Cochrane
JM and Fitzgerald PH: A cytogenetic and molecular analysis of five
variant Philadelphia translocations in chronic myeloid leukemia.
Cancer Genet Cytogenet. 35:179–197. 1988.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kanakasetty GB, Kuntejowdahalli L, Thanky
AH, Dasappa L, Jacob LA, Mallekavu SB and Kumari P: Predictive and
Prognostic implications of variant Philadelphia translocations in
CML: Experience from a tertiary oncology center in Southern India.
Clin Lymphoma Myeloma Leuk. 17:52–59. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Albano F, Anelli L, Zagaria A, Coccaro N,
Casieri P, Rossi AR, Vicari L, Liso V, Rocchi M and Specchia G: Non
random distribution of genomic features in breakpoint regions
involved in chronic myeloid leukemia cases with variant t(9;22) or
additional chromosomal rearrangements. Mol Cancer.
9(120)2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Becher R, Qiu JY, Parr A, Wendehorst E and
Schmidt CG: Seven variants including four new Philadelphia
translocations. Cancer Genet Cytogenet. 44:181–186. 1990.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Bonifacio M, Elena Ch, D'Adda M, Scaffidi
L, Pucci M, Aprili F, Ferrari J, Tucci A, Stagno F, Scortechini AR,
et al: Do not miss karyotyping at chronic myeloid leukemia
diagnosis: An Italian Campus CML study on the role of complex
variant translocations. Blood. 136 (Suppl 1):43–44. 2020.
|
|
34
|
Brahmbhatt MM, Trivedi PJ, Dalal EN, Patel
DM, Purani SS, Shukla SN, Shah PM and Patel PS: ABL/BCR gene
variant with two-step mechanism: Unusual localization and
rare/novel chromosomal rearrangements in CML patients. J Assoc
Genet Technol. 37:69–75. 2011.PubMed/NCBI
|
|
35
|
Sessarego M, Fugazza G, Bruzzone R and
Patrone F: Variant Philadelphia Chromosome Translocations are
Frequently Associated with Additional Structural Abnormalities.
Cancer Genet Cytogenet. 73:57–59. 1994.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Trivedi P, Varma P, Patel D, Ladani D,
Patel D, Kazi M, Patel N and Patel P: Clinical Implications of
simultaneous occurrence of variant Philadelphia translocations in
chronic myeloid leukemia. J Assoc Genet Technol. 45:61–65.
2019.PubMed/NCBI
|
|
37
|
Hochhaus A, Baccarani M, Silver RT,
Schiffer C, Apperley JF, Cervantes F, Clark RE, Cortes JE,
Deininger MW, Guilhot F, et al: European LeukemiaNet 2020
recommendations for treating chronic myeloid leukemia. Leukemia.
34:966–984. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jabbour E, Kantarjian HM, O'Brien S, Shan
J, Quintás-Cardama A, Garcia-Manero G, Rios MB and Cortes JE:
Front-line therapy with second-generation tyrosine kinase
inhibitors in patients with early chronic phase chronic myeloid
leukemia: what is the optimal response? J Clin Oncol. 29:4260–4265.
2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Eiring AM, Khorashad JS, Morley K and
Deininger MW: Advances in the treatment of chronic myeloid
leukemia. BMC Med. 9(99)2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hehlmann R: The New ELN recommendations
for treating CML. J Clin Med. 9(3671)2020.PubMed/NCBI View Article : Google Scholar
|