Introduction
At present, there are ~38.0 million people living
with HIV/AIDS globally, with the annual number of deaths standing
at 700,000 in 2019 (1,2). Although HIV/AIDS affects all parts of
the world, infections are highest in developing countries. Africa
remains the most severely affected region, with the number of
infected people standing at 25.7 million in 2019(2). South Africa has the highest number of
infected individuals, with 7.5 million people infected (1). Therefore, it remains critical to
continue with HIV research to reduce the burden of disease,
especially in countries that are the most affected.
HIV has an RNA genome surrounded by p24 capsid
proteins that can be detected in the blood during infection
(3). The virus also comprises a p17
matrix and the protease, reverse transcriptase and integrase
enzymes used during viral replication in host cells (4). The viral envelope consists of a
trimeric complex made of the heterodimer proteins gp120 and gp41,
which make up the gp160 protein. These proteins are essential for
recognition by target cells and infection (5,6). More
specifically, the gp41 protein is responsible for fusing the virus
and cellular membranes, while gp120 binds to CD4 receptors
expressed on host cell surfaces (6).
Antibodies are used in flow cytometry for the
detection of cells infected with HIV-1. These include antibodies
specific to gp120 and those against p24 (3,6).
However, these reagents have several disadvantages. For example,
antibodies are produced by the immunization of animals with the
antigen of interest; as a result, they are expensive to make, and
their manufacture is time consuming (7). In addition, target molecules that do
not elicit an immune response cannot be detected using antibodies
(8,9). Furthermore, antibodies are large,
non-flexible, cannot be modified easily with functional groups and
are not stable, as they are redox-sensitive to pH and temperature
(8). These limitations can be
circumvented using aptamers that are cheaper, considerably smaller
and can be generated against almost any target molecule.
Aptamers are short, synthetic single-stranded
nucleic acid (DNA or RNA) oligomers. They fold into unique 3D
structures by intramolecular interactions. These structures are
characterized by stems, loops, bulges, hairpins, pseudoknots, and
triplexes or quadruplexes that allow them to bind with high
affinity and specificity to different target molecules (10,11).
Aptamers are isolated from a random single stranded-library using a
technique called systematic evolution of ligands by exponential
enrichment (SELEX) in as little as 5-9 rounds of selection
(10). SELEX employs a repetitive
process of selection and amplification of high binding
oligonucleotides, which results in the exponential increase of the
best-binding aptamers (12,13). The number of selection rounds in
this technique depends on the diversity of the nucleic acid
library, target features and concentration, ratio of target
molecules to oligonucleotides, the stringency of selection and bias
of amplification (14,15). Previously, aptamers have been
selected against HIV-1 gp120 to inhibit its interaction with the
CD4 receptor (11). Some aptamers
have been reported to inhibit or neutralize HIV-1 infection with
50% inhibition concentration (IC50) values in the
nanomolar range (10,11). Aptamers are known to be equivalent
to antibodies in their potential as research, diagnostic and
therapeutic reagents (16,17). Their advantages over antibodies also
include their lack of immunogenicity and ease of modification by
chemical substitutions (7,18). Together, these advantages make
aptamers attractive molecules for diagnostic purposes and suitable
for the replacement of antibodies in several research and
therapeutic uses.
Although aptamers are attractive molecules, they
have several limitations. RNA and DNA molecules are unstable in
cells or sera; however, this has been circumvented by chemically
modifying the aptamer for stability and nuclease resistance
(10,19). For example, it has been shown that
disulfide cross-links can be introduced into the nucleic acid
secondary structure, endowing significant increases in thermal
stability without compromising the structural integrity (20). The addition of 2'flouropyrimidine
groups to aptamers during selection has been reported to increase
the stability and protection against nucleases (10). Furthermore, chemical modification
increases not only the stability, but also the half-life of
aptamers (11).
The use of fluorescence-labelled aptamers has been
explored for application as detectors and inhibitors of target
molecules. For example, a fluorescence-labelled
5'-hexachlorofluorescein pseudoknot RNA aptamer and a
5'-carboxyfluorescein DNA aptamer have been isolated against HIV-1
reverse transcriptase (21,22). These aptamers were intended for use
as inhibitors of the viral reverse transcriptase. RNA and DNA
aptamers conjugated to fluorescent dyes have also been used as
reagents to detect or diagnose cancer and viral infections
(23,24).
Based on the above studies, it was hypothesized that
the conjugation of FITC to CSIR1.1 and UCLA1 aptamers, specific to
HIV-1 gp120, will not alter the aptamers' structures, binding
affinity and specificity for gp120. This will enable them to be
used as low-cost reagents in HIV-1 research and diagnostics that
can benefit resource-poor settings. Here, the labelling of HIV-1
envelope specific aptamers to the fluorescent dye FITC and the
subsequent testing of these aptamers for potential use in
applications involving flow cytometry and virus capture assays is
reported. The results showed that the dye conjugated better with
the aptamer that had no stability related modifications at its 5'
or 3' ends. In addition, the conjugated aptamer could detect the
recombinant HIV-1 gp120 coated on latex beads by flow cytometry. It
could also detect the glycoprotein in its natural environment when
expressed on the viral envelope. Lastly, this aptamer performed
better in its detection of HIV-1 gp120 when compared with an
antibody-based detection system.
Materials and methods
Reagents
The UCLA1 anti-gp120 RNA aptamer, a shortened
derivative (54 nucleotides in length) of the B40t77 aptamer
(11,25), and the CSIR1.1 anti-gp120 RNA
aptamer, made up of 112 nucleotides, were used in the present
study. UCLA1 was obtained from the University of California, Los
Angeles (California, USA). The CSIR1.1 aptamer was isolated at the
Council for Scientific and Industrial Research in Pretoria, South
Africa, by London et al (10). Both aptamers were previously shown
to have anti-HIV inhibitory activity (10,11).
Plasmids encoding HIV-1 envelopes ZM53M.PB12, CAP239.2.00.G3J,
CAP210.E8 and DU156.12 were obtained from the AIDS Virus Research
Unit of the National Institute for Communicable Diseases
(Johannesburg, South Africa). Biotinylated gp120 polyclonal
antibody, used as a positive control, was purchased from Abcam (cat
no. ab53937).
Cell lines used for the neutralization
assay
Two types of cell lines were used, TZM-bl and 293T
cells. The TZM-bl cells used for the neutralization assays were
obtained from the National Institutes of Health HIV/AIDS Reference
Reagent Program (niaid.nih.gov/research/nih-aids-reagent-program).
TZM-bl cells are HeLa cell clones engineered to express, CD4
receptors, and CCR5 and CXCR4 co-receptors. These cells also
contain integrated reporter genes for firefly luciferase regulated
by HIV-1 long terminal repeat (11). The 293T cells, used for transfection
to make pseudotyped viruses, were obtained from the American Type
Culture Collection. They are adherent cells originally derived from
human embryonic kidney cells. Both TZM-bl and 293T cell lines were
sub-cultured at a concentration of 1x106 cells/ml every
2 days using DMEM (Thermo Fisher Scientific, Inc.) supplemented
with 10% FBS (Thermo Fisher Scientific, Inc.), with gentamycin
(Thermo Fisher Scientific, Inc.) as the antibiotic at 37˚C, with 5%
CO2 and 95% humidity.
Pseudoviruses for neutralization and
capture assays
The pseudoviruses used in the present study were
generated as previously described (11). The Env-pseudotyped virus stocks were
produced for ZM53M.PB12, CAP210.E8, CAP239.G3J and DU156.12 viruses
and the 50% tissue culture infective doses (TCID50) were
quantified as previously described (26).
Conjugation of FITC to anti-gp120
UCLA1 and CSIR 1.1 aptamers
Using a commercially available kit. FITC is a
fluorophore that contains reactive isothiocyanate and carboxylic
acid groups that react with nucleophiles such as amines. This
fluorophore has a molecular weight of 389.38 Da. The anti-gp120
UCLA1 and CSIR1.1 aptamers to FITC, and the FluoroTag™-FITC
conjugation kit (Sigma-Aldrich; Merck KGaA) were used to conjugate
the aptamers according to the manufacturer's protocol. Briefly, a
0.1 M sodium carbonate-bicarbonate buffer, pH 9.4, was prepared
according to the manufacturer's instructions. FITC was
reconstituted in 2 ml buffer. Then 89 µl anti-gp120 UCLA1 and
CSIR1.1 aptamers dissolved in the same buffer were each added to a
1.5 ml Eppendorf tube, and the volume made up to 200 µl with the
buffer. This was followed by a dropwise addition of 50 µl FITC and
shaking for 2 h in the dark at room temperature.
Using an in-house method. To conjugate UCLA1
and CSIR1.1 aptamers to FITC, 1,750 ng/µl, in 7.5 µl, of these
compounds were mixed with 1.25 mg 1-ethyl-3-(3-dimethylaminopropyl)
carbodiimide and 0.25 M (5 µl) FITC in imidazole (0.1 M, pH 6). The
mixture was thoroughly vortexed and centrifuged at 17,000 x g, 15˚C
for 5 min. To the reaction, 20 µl 0.1 M, pH 6 imidazole was added
and mixed using a shaker at room temperature for 48 h. This was
followed by centrifugation of each conjugated aptamer in 3,000 and
10,000 molecular weight cut-off (MWCO) Sartorius vivaspin columns
(Sigma-Aldrich; Merck KGaA) to remove unconjugated FITC. The
different MWCO columns were used for both aptamers to compare their
respective efficiency in eliminating unconjugated FITC. The
retained aptamers were washed three times with 0.1 M imidazole by
centrifuging for 20 min at 3,901 x g, 15˚C. The washed solution was
collected and stored at -20˚C. The two aptamer solutions were
transferred to different wells of a 96-well black plate, and
fluorescence was measured using the DTX 880 Multimode detector at a
wavelength of 485 nm. Unconjugated FITC was used as the positive
control.
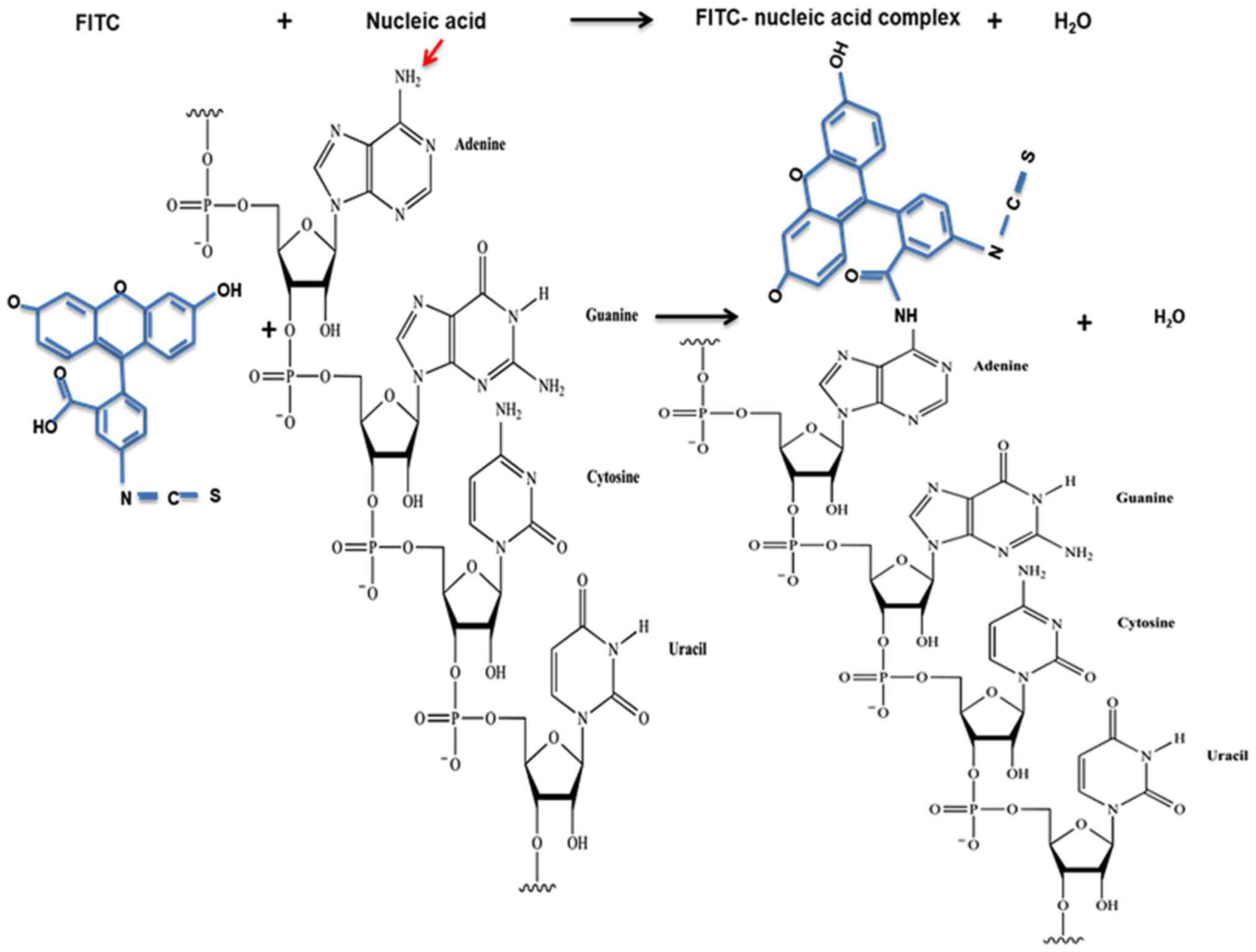
Expected reaction between FITC and aptamer.
The FITC isothiocyanate and carboxyl groups are reactive groups
that react with nucleophiles, such as amines and hydroxyls
(27). A free hydroxyl group is
found on the 3' end of nucleic acids when the sequence terminates
with an unmodified nucleotide. In contrast, a free amine is found
at the 5' end when it has an unmodified purine or pyrimidine
(Fig. 1). Of note, in a nucleic
acid molecule, there are also amine groups attached to different
bases (28). However, these are
unlikely to react with FITC due to steric hindrance caused by their
location within the RNA molecule. The inability of outside
molecules to react with bases within nucleic acids due to steric
hindrance has been previously described (29-31).
Detection of fluorescence emitted by
FITC-conjugated UCLA1 and CSIR1.1 aptamers
Fluorescence microscopy. Fluorescence
microscopy imaging was performed using TZM-bl cells infected with
HIV-1 ZM53M.PB12 to evaluate whether the conjugation was
successful. Cells were cultured in the presence of DEAE-dextran in
a flat-bottom 96-well plate at a concentration of 10,000
cells/well/100 µl DMEM supplemented with 10% FBS and gentamicin to
make them more susceptible to infection. This was followed by
infection with 200 TCID50 of ZM53M.PB12 pseudovirus. The
reaction was incubated for 48 h at 37˚C, 5% CO2 and 95%
humidity.
The infected cells were stained with the
FITC-conjugated anti-gp120 UCLA1 aptamer before imaging using the
Cytation 3 Cell Imaging Multimode reader (BioTek Instruments,
Inc.).
Flow cytometry. The conjugation of the
anti-gp120 CSIR1.1 aptamer to FITC was further confirmed using flow
cytometry. Latex beads (Sigma-Aldrich; Merck KGaA) were diluted to
a 1% working stock solution in double-distilled water. Then, 10 µl
recombinant gp120 at a concentration of 1.29 mg/ml was added to 4
µl 1% latex beads, and the solution was incubated overnight at 4˚C,
followed by washing the beads coupled to gp120 and blocking twice
with 500 µl 0.1% BSA-PBS using centrifugation for 2 min at 17,000 x
g, 4˚C. The beads were then resuspended in 700 µl the 0.1% BSA-PBS
buffer. The beads were then transferred to flow cytometry tubes,
centrifuged at 3,901 x g, 4˚C for 5 min and resuspended in 100 µl
1X BD binding buffer (Becton-Dickinson and Company). A total of 4
test tubes were used with gp120 coated (or coupled) beads stained
with a 3-fold serial dilution of FITC-conjugated CSIR1.1 aptamer
starting at 869 nM. The control tubes contained gp120 coated beads
stained with FITC only, starting at 869 nM, uncoated beads stained
with FITC-conjugated CSIR1.1 aptamer and other uncoated beads
stained with FITC only. Fluorescence was subsequently measured
using the BD LSR Fortessa cell analyzer (Becton-Dickinson and
Company) across the FITC channel with an excitation of 485 nm, and
analyzed using the BD FACSDiva version 9.0 (BD Biosciences). The
percentage fluorescence was calculated relative to the
concentration of the FITC and FITC-conjugated aptamer (869 µM)
using FlowJo version 10 (FlowJo, LLC) analysis with unstained beads
as a reference point. This was performed after instrument voltage
optimization to exclude electronic noise and background.
Capture assay. This assay was used to
determine the ability of CSIR1.1 FITC-conjugated anti-gp120 aptamer
to bind gp120 on whole HIV-1 particles and was performed as
previously described (32) with
some modifications. As CSIR1.1 aptamer has been shown to bind to
gp120(10), 100 µl gp41 10E8
monoclonal antibody (mAb) at a concentration of 1 µg/ml was used in
this assay to avoid competitive binding between the capturing
antibody and the aptamer. Thus, a high-binding 96-well black plate
was coated with 1 µg/ml anti-gp41 10E8 and incubated at 4˚C
overnight. The plate was then washed and blocked with 3% BSA, and
50 µl HIV-1 pseudoviruses were added and incubated for 1 h at 37˚C,
with 5% CO2 and 95% humidity to allow binding to 10E8
mAb. Pseudovirus ZM53.PB12, CAP210.E8 and Du156.12 were used as
they are known to bind the CSIR1.1 aptamer strongly (10). After incubation, the viruses were
washed with PBS followed by the addition of a 3-fold serial
dilution of FITC-conjugated CSIR1.1 aptamer, with an initial
concentration of 50 µg/ml. Biotinylated gp120 antibody and
FITC-conjugated streptavidin were used as positive controls at a
starting concentration of 50 µg/ml, and the negative control was
the unconjugated aptamer. The plates were incubated again as above
and then washed. Next, fluorescence was measured at 485 nm using
the DTX 880 multimode detector (Beckman Coulter, Inc.). The
excitation wavelength was 485 nm, and emission occurred at 535 nm.
SoftMaxPro version 6.4 (Molecular Devices, LLC) was used for data
acquisition and analysis.
HIV-1 infectivity assay
TZM-bl neutralization assay. The single-cycle
neutralization assay was used to test the infectivity of the
pseudovirus CAP239.G3J and the inhibitory effect of the aptamer
(CSIR1.1), as previously described (33). This assay used a 3-fold dilution
series of the inhibitor (CSIR1.1 FITC-conjugated aptamer or the
CSIR1.1 unconjugated aptamer). The dilution series started with an
11.9 nM concentration of each aptamer, followed by the addition of
50 µl HIV-1 CAP239.G3J pseudovirus. The negative control wells
included uninfected and untreated TZM-bl cells. Luminescence
readings at 510 nm were performed using the TECAN Infinite F500
Luminometer and i-control 1.5 software (Tecan Group, Ltd.). The
IC50 value of the aptamers was calculated as the
inhibitory concentration that caused a 50% reduction in relative
light units compared to the virus control (without inhibitor) after
subtraction of the background (without virus and inhibitor).
Statistical analysis
Statistical analysis was performed using SPSS
version 22 (IBM Corp.) and GraphPad Prism version 5.02 (GraphPad
Software, Inc.) using a one-way ANOVA with a post-hoc Tukey's test,
or an unpaired Student's t-test with 95% confidence. Each
experiment was repeated three times, and data are expressed as the
mean ± standard deviation. Graphs were constructed using GraphPad
Prism version 5. P<0.05 was considered to indicate a
statistically significant difference.
Results
Conjugation of FITC to gp120 aptamers
using two different methods
CSIR1.1 and UCLA1, which have been previously shown
to bind to the gp120 glycoprotein, was conjugated with FITC to
generate a fluorescence-labelled gp120 aptamer. This was achieved
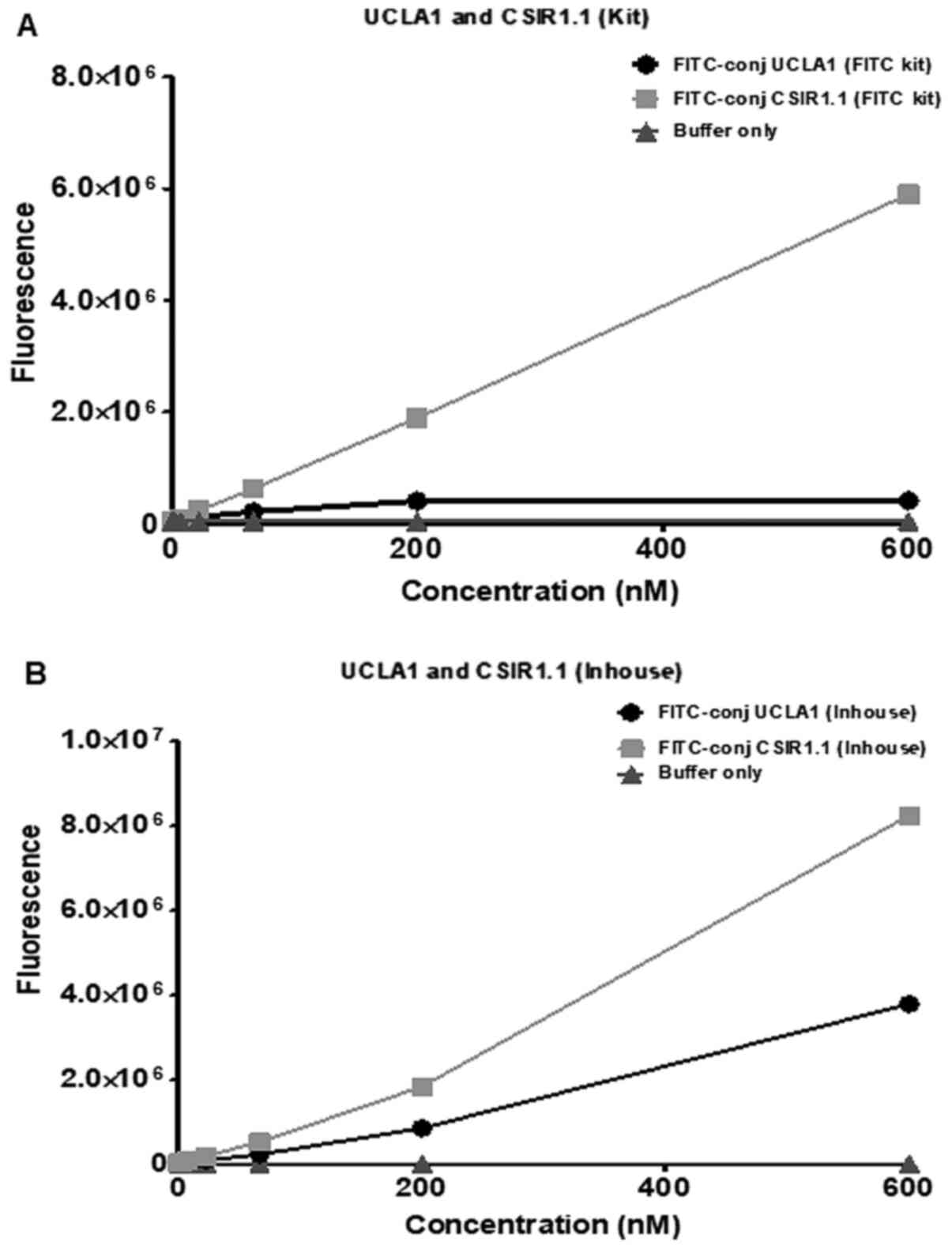
using a commercially available kit and an in-house method. As shown
in Fig. 2A, the conjugation of the
dye using the kit was more successful in the case of CSR1.1. The
fluorescence intensity for this aptamer after conjugation increased
from 0 to 6x106 RFU. At the same time, with UCLA1, the
value was very low at 4x105, resulting in the difference
of fluorescence emitted by the two conjugated aptamers to be
5.6x106. The fluorescence conjugation using the in-house
method was successful for both CSIR1.1 and UCLA1, although the
former still emitted significantly more fluorescence than the
latter (Fig. 2B). The difference in
the fluorescence emitted by the two aptamers was ~2-fold. Given
that the stoichiometry of the aptamer and FITC reaction was 1:1 for
CSIR1.1 and UCLA1, these results suggested that the former was more
efficient in interacting with FITC than the latter.
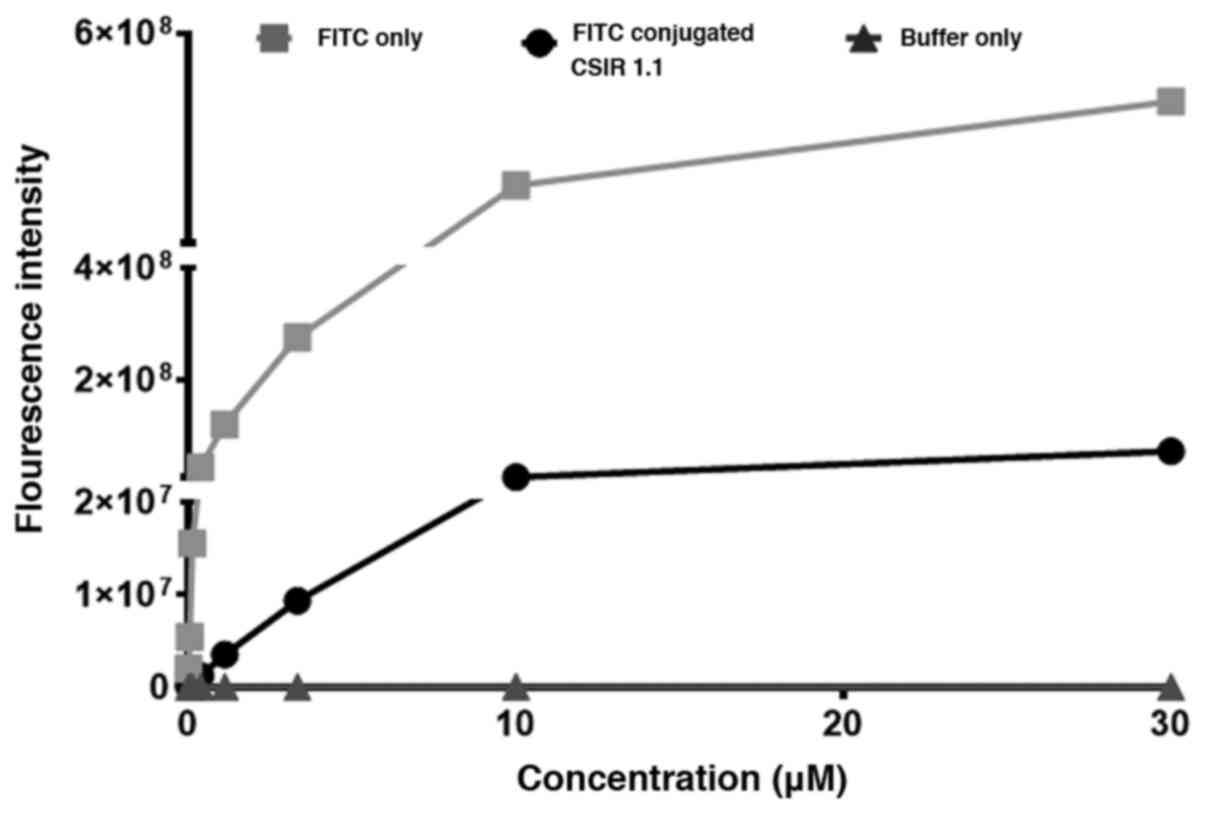
Next, the fluorescence emission of FITC conjugated
gp120 aptamer was compared with unconjugated FITC to determine the
percentage of the dye in the solution that became conjugated to the
aptamer after the conjugation reaction. The in-house conjugation
method was used for the CSIR1.1 aptamer in this experiment, as it
was the condition that emitted the highest fluorescence. It should
also be noted that in all subsequent experiments in the present
study, only FITC-conjugated CSIR1.1 was used because of its high
fluorescence emission. Here, a 0.25 M concentration of FITC was
conjugated with CSIR1.1 before filtration, using a Vivaspin column
to remove the dye that did not react with the aptamer. Then the
fluorescence emitted by the resulting FITC-CSIR1.1 conjugate was
compared to that emitted by the same amount of FITC used in the
conjugation reaction, which was used as the control. As shown in
Fig. 3, at any given concentration
of FITC reacted with the aptamer, the amount of fluorescence
emitted by FITC-CSIR1.1 was only 20% of that obtained from FITC
only. These data suggest a ~20% efficiency of the conjugation
reaction between FITC and CSIR1.1. The efficiency of this reaction
was lower (~1%) with the UCLA1 aptamer (Fig. S1).
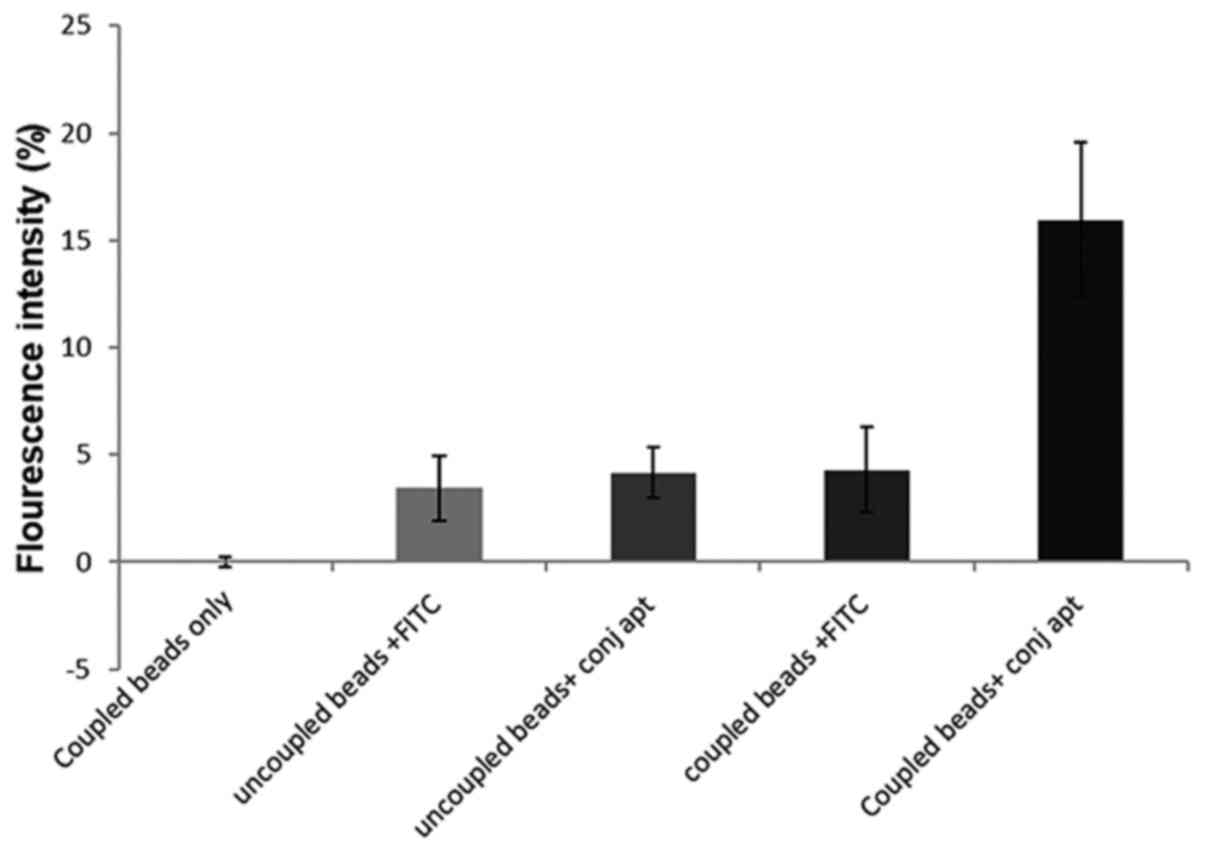
Detection of the interaction between
FITC-conjugated CSIR1.1 and HIV-1 gp120 by flow cytometry
Since one of the possible applications of
FITC-conjugated CSIR1.1 is its use in flow cytometry to detect
HIV-1 infected and gp120-expressing cells (34), its binding to the glycoprotein was
assessed using this technique. This was undertaken by coupling
latex beads with recombinant gp120 derived from the Consensus C
virus, a representative of all circulating HIV-1 subtype C viruses
(35). The gp120 coupled beads were
incubated with the FITC-conjugated CSIR1.1 aptamer (36). As the negative control, the beads
(uncoupled) were also stained with FITC (37). Other controls included unstained
gp120-coupled beads and uncoupled beads incubated with the
FITC-CSIR1.1. After acquiring 10,000 events on the flow cytometer,
no fluorescence was observed with the unstained gp120-coupled
beads. Only a 3.5±1.51% increase in fluorescence emission was
reported for the gp120-uncoupled beads stained with FITC alone.
Uncoupled beads with the FITC-conjugated CSIR1.1 showed a
4.14±1.18% increase in fluorescence emission (Figs. 4 and S2). These findings suggest some
background level/autofluorescence due to the interaction between
the dye and the beads. However, more importantly, a 16±3.6%
increase in fluorescence was reported for gp120-coupled beads
stained with FITC-CSIR1.1 (Figs. 4
and S2).
Binding of FITC-conjugated CSIR1.1 to
gp120 on intact HIV-1 particles
Given that the gp120 coupled on latex beads was a
recombinant glycoprotein, whether FITC-CSIR1.1 could recognize the
gp120 when expressed in its natural environment was assessed. To
achieve this, a HIV-1 capture assay was performed (32) with three HIV-1 subtype C
pseudoviruses, ZM53M.PB12, DU156.12 and CAP210.E8, known to
interact with CSIR1.1(10). The
virus was captured with the 10E8 mAb (38) that binds gp41 and detected using
different concentrations of the FITC-CSIR1.1 aptamer ranging from
0.00-0.05 mg/ml. The choice of 10E8 mAb for capture was because it
binds the membrane-proximal external region of the viral
glycoprotein, thus making it less likely to compete with the
aptamer for binding to the envelope (39). FITC-CSIR1.1 was observed to bind and
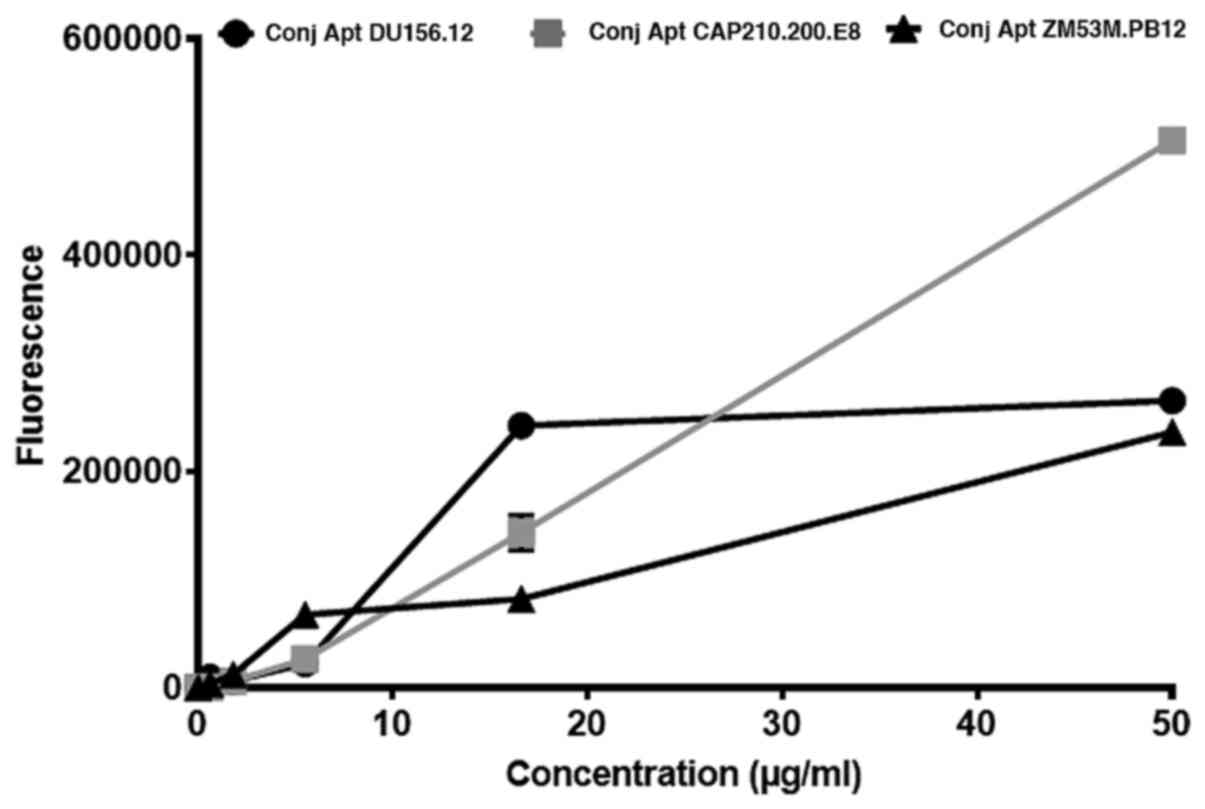
detect all three viruses as expected (Fig. 5). When comparing capture of the
viruses by the highest concentration of aptamer, CAP210.E8
pseudovirus was the most captured with a fluorescence intensity of
506,000±3.71. The other two viruses, DU156.12 and ZM53M.PB12 had
fluorescence intensities of 265,333±1.67 and 236,333±1.47,
respectively. The P-value of the comparison of the three viruses'
fluorescence intensities was 0.859, suggesting that they were not
significantly different.
Previous studies have shown that antibodies specific
to gp120 are used in flow cytometry to detect HIV-1 infected cells
(40). The fluorescence emission of
the FITC-conjugated CSIR1.1 aptamer was therefore compared to a
commercially available polyclonal biotin-conjugated anti-gp120
antibody using the capture assay, one of the three HIV-1 subtypes C
mentioned above, ZM53M.PB12, was chosen for this experiment, given
that statistically, all three viruses had similar fluorescence
intensity. The same concentration of the aptamer and
biotin-conjugated antibody was used. However, this antibody had to
be detected with FITC-conjugated streptavidin. Thus, the
FITC-streptavidin/biotin-antibody interaction was first optimized;
the strongest fluorescence signal was detected when the
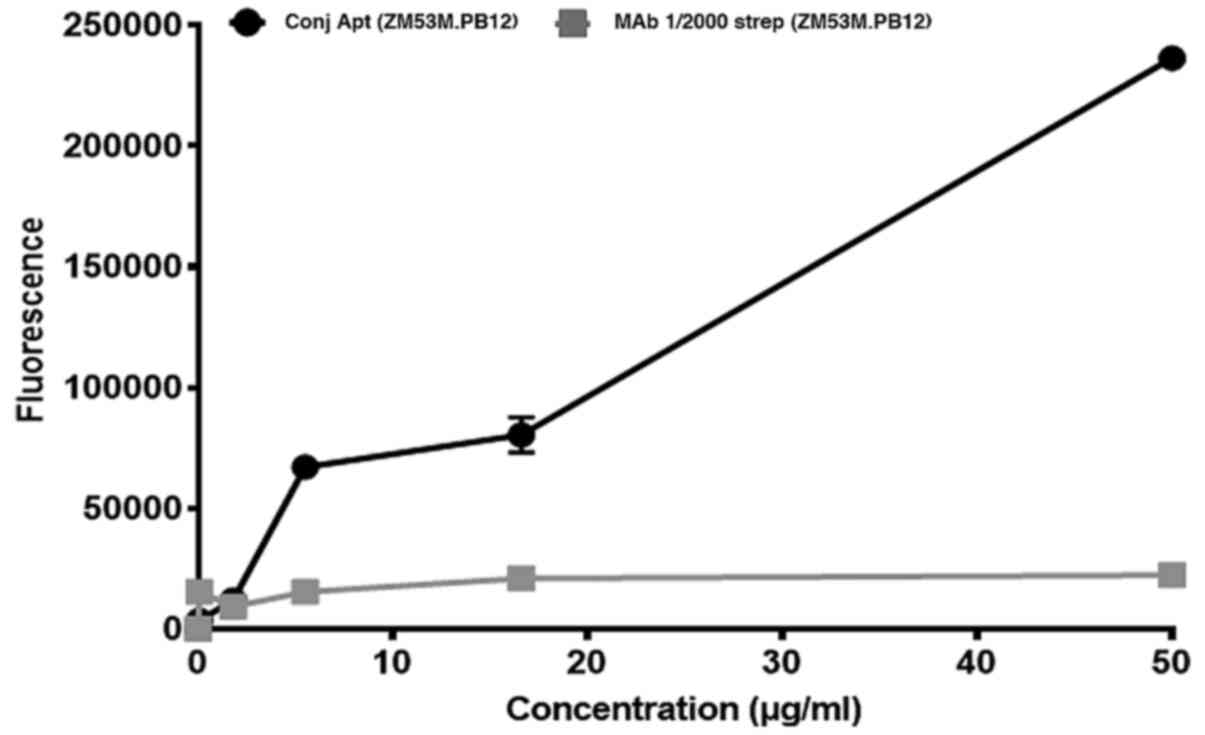
FITC-streptavidin was used at a dilution of 1:2,000 (Fig. S3). When the FITC-CSIR1.1 and the
biotin-antibody were compared to detect the captured HIV-1, the
fluorescence emitted by the aptamer was markedly stronger (Fig. 6). This difference was also observed
to increase significantly as the concentrations of the aptamer and
antibody were increased. For example, at <20 µg/ml of both
aptamer and antibody, the difference was ~3-fold. This increased to
8-fold at 50 µg/ml, where the aptamer produced an average
fluorescence of 236,333±0.063, while the antibody had an average of
22,333±1.53. The difference between the aptamer and antibody was
statistically significant (P<0.01) determined using an upraised
t-test. Thus, FITC-CSIR1.1 performance was greater than that of the
antibody destined for the same application.
HIV-1 neutralization with the
FITC-conjugated CSIR1.1 aptamer
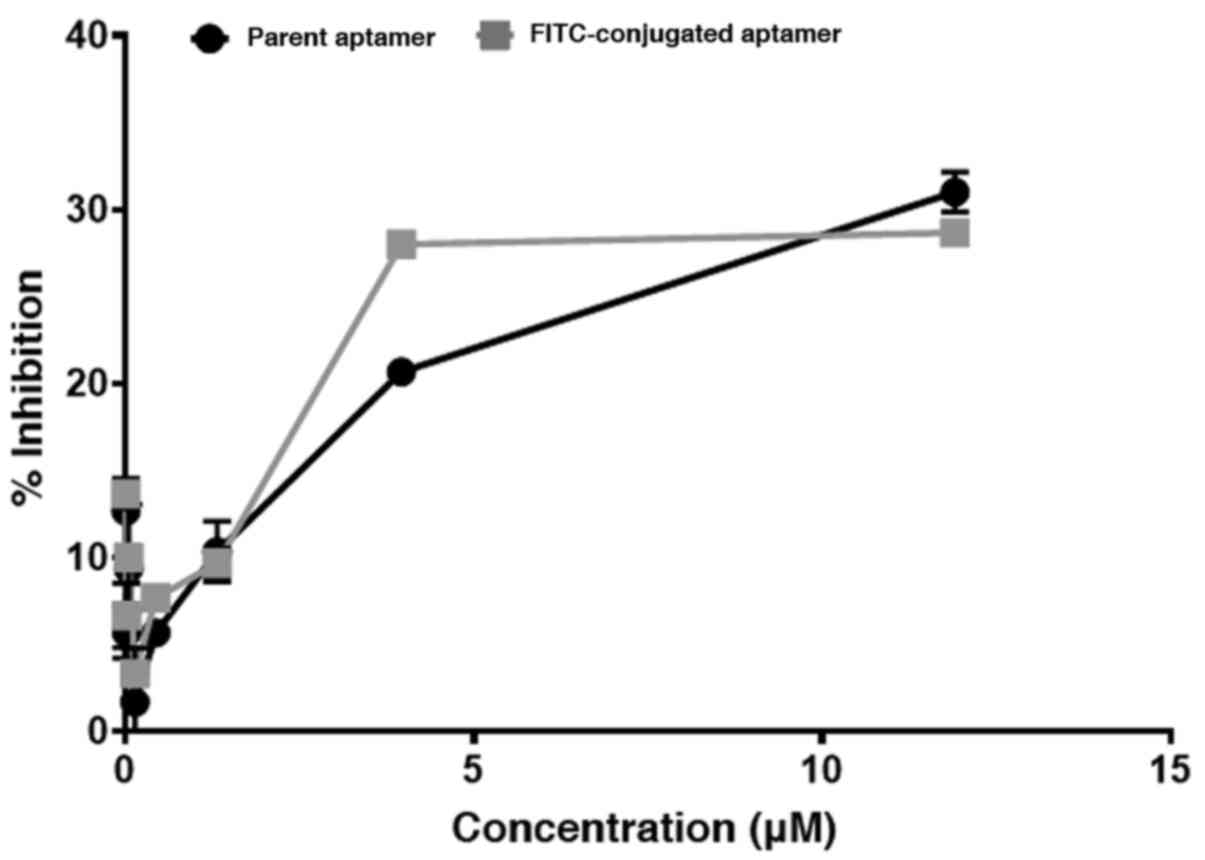
The CSIR1.1 aptamer has been shown to neutralize
HIV-1(10). HIV-1 neutralization
efficiency of the conjugate was compared with the unconjugated
(parent) aptamer to determine whether the conjugation of CSIR1.1
with FITC altered the aptamer's structure and/or functionality.
CAP239.G3J, HIV-1 subtype C pseudovirus, known to be sensitive to
CSIR1.1(41), was used in the
neutralization assay. As shown in Fig.
7, the inhibitory activity of both the conjugated and
unconjugated aptamers was similar, judging by the consistency of
inhibition. The P-value, determined using an unpaired t-test, was
0.784; therefore, the difference in inhibition between the two
aptamers was not statistically significant. Thus, these data
suggested that the conjugation with FITC did not alter the aptamer
interaction with the viral envelope.
Discussion
Aptamers are nucleic acid molecules that can be
synthesized to bind to various targets (42,43).
As a result, these molecules are being studied and synthesized as
alternative reagents to antibodies in research, and for diagnosis
and treatment of different diseases (42,44).
The present study investigated the possibility of converting
previously isolated and studied aptamers against HIV-1 gp120
(10,11), fluorescence-labelled alternative
reagents for use in applications requiring antibodies. It was shown
that the CSIR1.1 aptamer could be conjugated efficiently to FITC
fluorescent dye and used to detect gp120 in different assay formats
involving flow cytometry and HIV capture assay. In addition, this
fluorescence-labelled aptamer performed better than the
antibody-based detection system designed for similar use. Given the
low costs associated with synthesizing aptamers (11), it is suggested that FITC-CSIR1.1 is
potentially a more viable alternative to antibodies for use in
these applications.
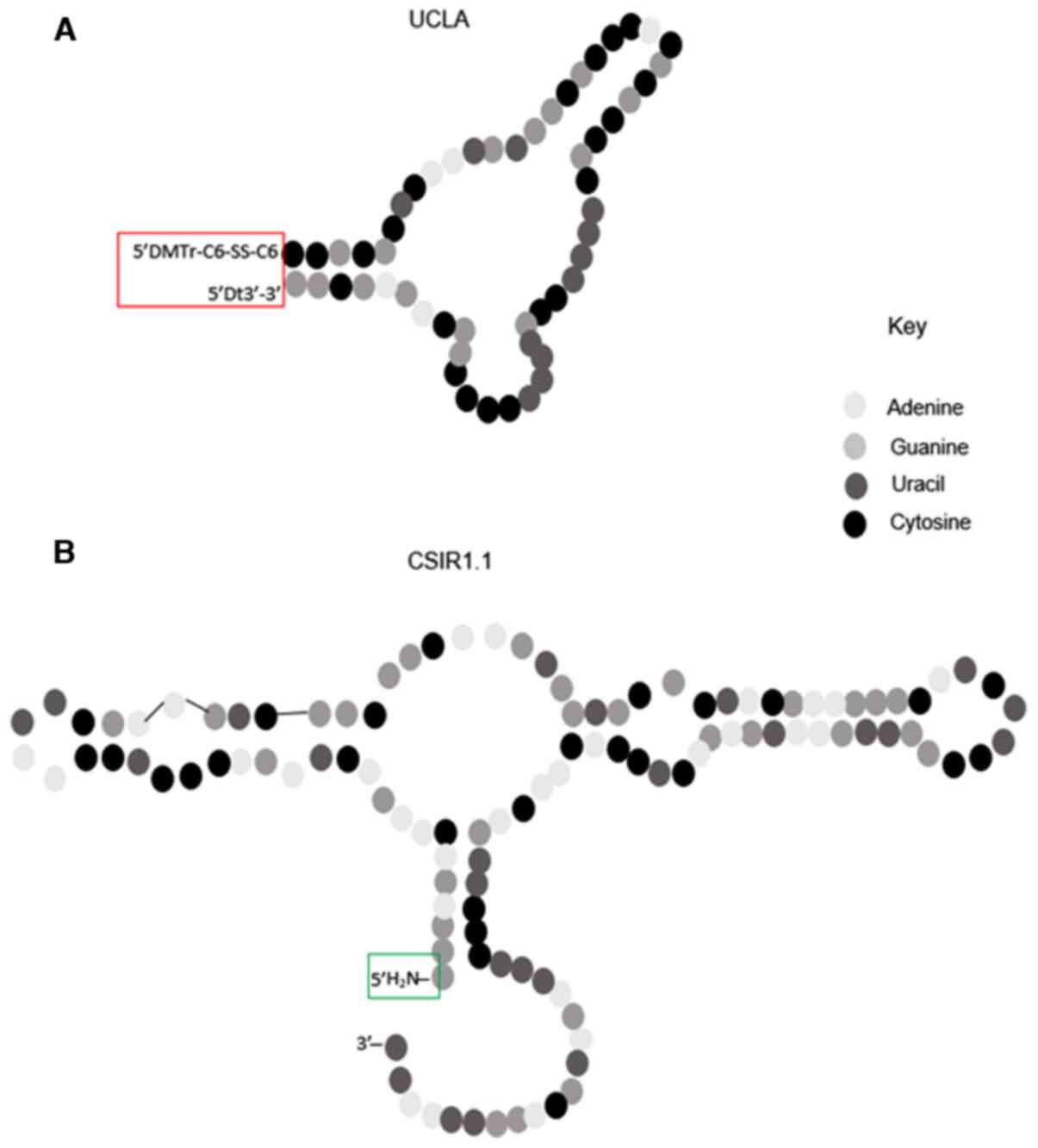
The conjugation of FITC with the aptamers was more
efficient with CSIR1.1 compared to UCLA1. This is likely due to the
differences in the structure of the two aptamers. As shown in
Fig. 8, CSIR1.1 has an amine group
at its 5' end where it is expected to react with the carboxyl group
of FITC. However, in UCLA1, there are extensive modifications at
the 5' and 3' ends that possibly prevent effective conjugation with
FITC (25). To be more specific,
the 5' end of UCLA1 is modified with a
dimethoxyltrityloxy-(CH2)6-SS-(CH2)6-phospho
linker, whereas its 3' end is modified with an inverted dT for
stability (11) (Fig. 8). Since these extensive
modifications are at the sites where the aptamer is expected to
bind FITC, these may be the cause of obstruction to the reaction,
thus resulting in a decreased conjugation efficiency observed for
this aptamer.
The fact that the FITC-CSIR1.1 neutralization of
HIV-1 mirrored that of the unconjugated parent aptamer showed that
the conjugation reaction did not alter the aptamer's interaction
with gp120. This is very important, given that CSIR1.1 was shown in
our laboratory to have high affinity and specificity for HIV-1
gp120(10). It was essential that
this property be maintained after the conjugation reaction if it is
intended for use as an alternative to high affinity and high
specificity molecules, such as antibodies (45,46).
Infected and gp120 expressing cells and latex beads
coupled to gp120 were used to investigate the possibility of using
this aptamer to detect HIV-1 by flow cytometry. The FITC-CSIR1.1
molecule bound to the HIV-1 gp120 and could be detected by
fluorescence. The ~16% increase in fluorescence intensity observed
by flow cytometry was significantly higher than the values obtained
by other investigators using fluorescence-labelled antibodies
against gp120. For example, when H9 cells were used to detect HIV
by flow cytometry using gp120 monoclonal antibodies, the percentage
fluorescence was reported to be 2.2 and 0.59% for infected and
uninfected cells, respectively (47).
It is noteworthy that the use of FITC conjugated
aptamers as flow cytometry reagents is not unprecedented. For
example, a study by Rong et al (48) using a FITC-conjugated aptamer showed
that this molecule bound specifically to cancer cells with no or
little binding to non-cancerous cells. Similarly, Song et al
(17) demonstrated that a single
stranded DNA aptamer conjugated to FITC could differentiate target
cancer cells from a mixture of cells in media using flow cytometry.
Therefore, these data confirmed the potential of using this
aptamer-based reagent to detect HIV-1 gp120 using flow cytometry
and also showed that it could perform better than antibodies in
some cases.
Given that aptamers are destined for use as
alternatives in applications that usually require antibodies, it
was important that FITC-CSIR1.1 be compared to an antibody-based
assay. As shown by the capture assay, the signal emitted by the
FITC-conjugated aptamer after binding gp120 expressed on HIV-1
envelope was markedly higher than that emitted using a
FITC-conjugated streptavidin/biotin-antibody system. One possible
explanation for this is that the size of antibodies, ~150 kDa
(49), is much larger than that of
the CSIR1.1 aptamer, at 55 kDa, (10). Thus, it is possible that fewer
molecules of the antibody were bound on the surface of the virus
compared to the aptamer, resulting in a stronger signal for the
latter. The relatively small size of aptamers is a well-known
advantage of these molecules over their competitors. It allows them
to bind with high specificity and affinity to their targets. This
was previously shown where aptamers could penetrate and be retained
by tumor cells longer than antibodies, both in vitro and
in vivo (50). As a result,
they could be employed as molecular imaging probes (44).
Generally, a capture assay utilizes a minimum of two
antibodies, one for capture and the other for detection (26). However, an added advantage of the
aptamer capture assay is that if two aptamers that bind gp120 at
different sites are selected (by SELEX), one aptamer will be used
to capture the virus, and the other will be used for detection
thus, completely removing the need for use of an antibody in the
assay. Moreover, the flow cytometry data indicated that there was
no need for use of an antibody together with the aptamer. Unlike
antibodies, the cost of synthesizing aptamers has been greatly
reduced through several years of modifications and refinements
leading to production time of weeks compared to antibodies with
longer in vivo procedures that span months (8,11). Of
note, the cost to perform a capture assay using a pair of gp120
specific antibodies is ~136 USD per reaction compared to only 40
USD for a pair of aptamers, as performed in the present study,
resulting in a 3-fold cost reduction.
Despite the advantages of the aptamer-based method
and potential ease of application in a hospital setting, the study
does not come without limitations. For example, the FITC conjugated
aptamer could have been tested against several HIV-1 strains to
determine whether it retained its broad spectrum of binding against
the virus (10). In addition, the
FITC-aptamer duration of conjugation was not determined in the
present study. It is important that the half-life of this bond be
determined in future studies since chemical bonds are known to
degrade; furthermore, fluorophores can be bleached, thus reducing
or abrogating their signal (51,52).
In conclusion, the results of the present study
showed that the CSIR1.1 aptamer can be conjugated to FITC and used
to detect gp120 by flow cytometry or HIV-1 capture assay. This
reagent can potentially perform at a higher level than its
antibody-based counterpart, employed in the same applications. More
importantly, these findings support the view that aptamers can be
used as cheaper, but equally, if not more suitable substitutes for
antibodies. Such reagents will be beneficial for research-poor
settings, especially in developing countries worldwide, where the
cost of reagents is one of the biggest obstacles to research. They
will also be valuable in diagnostics involving techniques such as
ELISA and imaging. In addition, the high cost of producing
antibodies in animals (7), and the
inability to produce them against non-immunogenic targets (53), further supports the development of
aptamer-based reagents.
Supplementary Material
Graph of fluorescence emission against
serial dilutions of FITC-conjugated UCLA1 and CSIR1.1 aptamers.
Buffer was used as a negative control. The conjugated aptamers were
filtered through the 3,000 molecular weight cut-off vivaspin
columns to remove unbound FITC. FITC, fluorescein
isothiocyanate.
Representative dot plots and
corresponding histogram plots showing binding of FITC-conjugated
CSIR1.1 to gp120 on latex beads. (A) Coupled beads alone; (B)
coupled beads stained with FITC only; (C) coupled beads stained
with conjugated aptamer; (D) uncoupled beads stained with FITC
only; (E) uncoupled beads stained with conjugated aptamer. FITC,
fluorescein isothiocyanate; FSC, forward scatter.
Optimization of the commercial
biotinylated anti-gp120 antibody for detection of gp120. HIV-1
subtype C ZM53M.PB12 was captured with the 10E8 mAb, followed by
the addition of biotin-conjugated anti-gp120 antibody. This was
later detected using different dilutions of FITC-conjugated
streptavidin. The experiment was repeated two times. FITC,
fluorescein isothiocyanate; strept, streptavidin.
Acknowledgements
We would like acknowledge the generous donation of
HIV-1 envelope plasmids for the production of pseudoviruses by the
AIDS Research Unit at the National Institute for Communicable
Diseases (NICD).
Funding
Funding: This work was supported by the Council for Scientific
and Industrial Research Parliamentary Grant (grant no. V1YAT74),
and the Council for Scientific and Industrial Research Masters
Studentship.
Availability of data and materials
The data sets used and/or analyzed during the
present study are available from the corresponding authors on
request.
Authors' contributions
KA and HM conceptualized and designed the study. KM
performed the experiments and collected the data. KM, PNF, and KA
analyzed and interpreted the data. KM wrote the draft manuscript.
PNF, HM, KA supervised the findings and provided critical and
intellectual revisions and feedback. HM, PNF and KA confirmed the
authenticity of all the raw data. All authors revised the
manuscript as well as read and approved the final version.
Ethics approval and consent to
participate
Ethics approval for this study was obtained from the
Council for Scientific and Industrial Research Ethics Committee
(approval no. 184/2016) and the waiver from the Human Research
Ethics (Medical) Committee (approval no. M1611138) of the
University of the Witwatersrand. Albeit the study did not require
the use of animal samples, both institutions require that
ethics/ethics waiver be obtained prior to conducting any work.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
UNAIDS: Global HIV & AIDS statistics -
2019 fact sheet. 2020.
|
|
2
|
World Health Organisation: Latest HIV
estimates and updates on HIV policies uptake. In: Global HIV,
Hepatitis and STI Programmes. 2020.
|
|
3
|
Zeng JY, Li X, Yuan HX, Ma ML, Li DD, Ma J
and Liao S: Screening ssDNA aptamers against HIV P24 antigen using
agarose beads as carriers. Bio Web Conf. 8(10)2017.
|
|
4
|
Freed EO: HIV-1 gag proteins: Diverse
functions in the virus life cycle. Virology. 251:1–15.
1998.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wyatt R, Kwong PD, Desjardins E, Sweet RW,
Robinson J, Hendrickson WA and Sodroski JG: The antigenic structure
of the HIV gp120 envelope glycoprotein. Nature. 393:705–711.
1998.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Chan DC and Kim PS: HIV entry and its
inhibition. Cell. 93:681–684. 1998.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Guo KT, Ziemer G, Paul A and Wendel HP:
CELL-SELEX: Novel perspectives of aptamer-based therapeutics. Int J
Mol Sci. 9:668–678. 2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen A and Yang S: Replacing antibodies
with aptamers in lateral flow immunoassay. Biosens Bioelectron.
71:230–242. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Toh SY, Citartan M, Gopinath SC and Tang
TH: Aptamers as a replacement for antibodies in enzyme-linked
immunosorbent assay. Biosens Bioelectron. 64:392–403.
2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
London GM, Mayosi BM and Khati M:
Isolation and characterization of 2'-F-RNA aptamers against whole
HIV-1 subtype C envelope pseudovirus. Biochem Biophys Res Commun.
456:428–433. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mufhandu HT, Gray ES, Madiga MC, Tumba N,
Alexandre KB, Khoza T, Wibmer CK, Moore PL, Morris L and Khati M:
UCLA1, a synthetic derivative of a gp120 RNA aptamer, inhibits
entry of human immunodeficiency virus type 1 subtype C. J Virol.
86:4989–4999. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tuerk C and Gold L: Systematic evolution
of ligands by exponential enrichment: RNA ligands to bacteriophage
T4 DNA polymerase. Science. 249:505–510. 1990.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Walker LM, Phogat SK, Chan-Hui PY, Wagner
D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al:
Protocol G Principal Investigators: Broad and potent neutralizing
antibodies from an African donor reveal a new HIV-1 vaccine target.
Science. 326:285–289. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Glökler J, Schütze T and Konthur Z:
Automation in the high-throughput selection of random combinatorial
libraries - different approaches for select applications.
Molecules. 15:2478–2490. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Stoltenburg R, Reinemann C and Strehlitz
B: SELEX - a (r)evolutionary method to generate high-affinity
nucleic acid ligands. Biomol Eng. 24:381–403. 2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rohloff JC, Gelinas AD, Jarvis TC, Ochsner
UA, Schneider DJ, Gold L and Janjic N: Nucleic acid ligands with
protein-like side chains: Modified aptamers and their use as
diagnostic and therapeutic agents. Mol Ther Nucleic Acids.
3(e201)2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Song KM, Lee S and Ban C: Aptamers and
their biological applications. Sensors (Basel). 12:612–631.
2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ulrich H, Martins AH and Pesquero JB: RNA
and DNA aptamers in cytomics analysis. Cytometry A. 59:220–231.
2004.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Osborne SE, Matsumura I and Ellington AD:
Aptamers as therapeutic and diagnostic reagents: Problems and
prospects. Curr Opin Chem Biol. 1:5–9. 1997.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Osborne SE, Volker J, Stevens SY,
Breslauer KJ and Glick GD: Design, synthesis, and analysis of
disulfide cross-linked DNA duplexes. J Am Chem Soc.
118:11993–12003. 1996.
|
|
21
|
Zhang H, Wang Z, Li XF and Le XC:
Ultrasensitive detection of proteins by amplification of affinity
aptamers. Angew Chem Int Ed. 45:1576–1580. 2006.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pavski V and Le XC: Detection of human
immunodeficiency virus type 1 reverse transcriptase using aptamers
as probes in affinity capillary electrophoresis. Anal Chem.
73:6070–6076. 2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cao Y, Dong H, Yang Z, Zhong X, Chen Y,
Dai W and Zhang X: Aptamer-conjugated graphene quantum
dots/porphyrin derivative theranostic agent for intracellular
cancer-related microrna detection and fluorescence-guided
photothermal/photodynamic synergetic therapy. ACS Appl Mater
Interfaces. 9:159–166. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Moutsiopoulou A, Broyles D, Dikici E,
Daunert S and Deo SK: Molecular aptamer beacons and their
applications in sensing, imaging, and diagnostics. Small.
15(e1902248)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Cohen C, Forzan M, Sproat B, Pantophlet R,
McGowan I, Burton D and James W: An aptamer that neutralizes R5
strains of HIV-1 binds to core residues of gp120 in the CCR5
binding site. Virology. 381:46–54. 2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Alexandre KB, Gray ES, Mufhandu H, McMahon
JB, Chakauya E, O'Keefe BR, Chikwamba R and Morris L: The lectins
griffithsin, cyanovirin-N and scytovirin inhibit HIV-1 binding to
the DC-SIGN receptor and transfer to CD4(+) cells. Virology.
423:175–186. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Koniev O and Wagner A: Developments and
recent advancements in the field of endogenous amino acid selective
bond forming reactions for bioconjugation. Chem Soc Rev.
44:5495–5551. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Shapiro R, Ellis S, Hingerty BE and Broyde
S: Effect of ring size on conformations of aromatic amine-DNA
adducts: The aniline-C8 guanine adduct resides in the B-DNA major
groove. Chem Res Toxicol. 11:335–341. 1998.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Castronovo M, Radovic S, Grunwald C,
Casalis L, Morgante M and Scoles G: Control of steric hindrance on
restriction enzyme reactions with surface-bound DNA nanostructures.
Nano Lett. 8:4140–4145. 2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Xu F, Pellino AM and Knoll W:
Electrostatic repulsion and steric hindrance effects of surface
probe density on deoxyribonucleic acid (DNA)/peptide nucleic acid
(PNA) hybridisation. Thin Solid Films. 516:8634–8639. 2008.
|
|
31
|
Yue Y, Li G, Yang Y, Zhang W, Pan H, Chen
R, Shi F and Jin Y: Regulation of Dscam exon 17 alternative
splicing by steric hindrance in combination with RNA secondary
structures. RNA Biol. 10:1822–1833. 2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Alexandre KB, Gray ES, Lambson BE, Moore
PL, Choge IA, Mlisana K, Karim SS, McMahon J, O'Keefe B, Chikwamba
R, et al: Mannose-rich glycosylation patterns on HIV-1 subtype C
gp120 and sensitivity to the lectins, Griffithsin, Cyanovirin-N and
Scytovirin. Virology. 402:187–196. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Alexandre KB, Moore PL, Nonyane M, Gray
ES, Ranchobe N, Chakauya E, McMahon JB, O'Keefe BR, Chikwamba R and
Morris L: Mechanisms of HIV-1 subtype C resistance to GRFT, CV-N
and SVN. Virology. 446:66–76. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou J, Swiderski P, Li H, Zhang J, Neff
CP, Akkina R and Rossi JJ: Selection, characterization and
application of new RNA HIV gp 120 aptamers for facile delivery of
Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res.
37:3094–3109. 2009.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gray ES, Madiga MC, Hermanus T, Moore PL,
Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C,
et al: CAPRISA002 Study Team: The neutralization breadth of HIV-1
develops incrementally over four years and is associated with
CD4+ T cell decline and high viral load during acute
infection. J Virol. 85:4828–4840. 2011.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lekkerkerker AN, Ludwig IS, van Vliet SJ,
van Kooyk Y and Geijtenbeek TB: Potency of HIV-1 envelope
glycoprotein gp120 antibodies to inhibit the interaction of DC-SIGN
with HIV-1 gp120. Virology. 329:465–476. 2004.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kojima T, Takei Y, Ohtsuka M, Kawarasaki
Y, Yamane T and Nakano H: PCR amplification from single DNA
molecules on magnetic beads in emulsion: Application for
high-throughput screening of transcription factor targets. Nucleic
Acids Res. 33(e150)2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Guttman M, Padte NN, Huang Y, Yu J,
Rocklin GJ, Weitzner BD, Scian M, Ho DD and Lee KK: The influence
of proline isomerization on potency and stability of anti-HIV
antibody 10E8. Sci Rep. 10(14313)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Huang J, Kang BH, Pancera M, Lee JH, Tong
T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, et al: Broad
and potent HIV-1 neutralization by a human antibody that binds the
gp41-gp120 interface. Nature. 515:138–142. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Rigg RJ, Dando JS, Escaich S, Plavec I and
Böhnlein E: Detection of intracellular HIV-1 Rev protein by flow
cytometry. J Immunol Methods. 188:187–195. 1995.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Grace Mothepane London: Isolation,
Characterization and efficacy studies of 2'-F-RNA aptamers against
HIV-1 subtype C enveloped pseudovirus. p292, 2013.
|
|
42
|
Wang YK, Zou Q, Sun JH, Wang HA, Sun X,
Chen ZF and Yan YX: Screening of single-stranded DNA (ssDNA)
aptamers against a zearalenone monoclonal antibody and development
of a ssDNA-based enzyme-linked oligonucleotide assay for
determination of zearalenone in corn. J Agric Food Chem.
63:136–141. 2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Torrini F, Palladino P, Brittoli A,
Baldoneschi V, Minunni M and Scarano S: Characterization of
troponin T binding aptamers for an innovative enzyme-linked
oligonucleotide assay (ELONA). Anal Bioanal Chem. 411:7709–7716.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xiang D, Zheng C, Zhou SF, Qiao S, Tran
PH, Pu C, Li Y, Kong L, Kouzani AZ, Lin J, et al: Superior
performance of aptamer in tumor penetration over antibody:
Implication of aptamer-based theranostics in solid tumors.
Theranostics. 5:1083–1097. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Margolis DM, Koup RA and Ferrari G: HIV
antibodies for treatment of HIV infection. Immunol Rev.
275:313–323. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
McSharry JJ: Analysis of virus-infected
cells by flow cytometry. Methods. 21:249–257. 2000.PubMed/NCBI View Article : Google Scholar
|
|
47
|
McSharry JJ, Costantino R, Robbiano E,
Echols R, Stevens R and Lehman JM: Detection and quantitation of
human immunodeficiency virus-infected peripheral blood mononuclear
cells by flow cytometry. J Clin Microbiol. 28:724–733.
1990.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Rong Y, Chen H, Zhou XF, Yin CQ, Wang BC,
Peng CW, Liu SP and Wang FB: Identification of an aptamer through
whole cell-SELEX for targeting high metastatic liver cancers.
Oncotarget. 7:8282–8294. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Singh AA, Pooe O, Kwezi L, Lotter-Stark T,
Stoychev SH, Alexandra K, Gerber I, Bhiman JN, Vorster J, Pauly M,
et al: Plant-based production of highly potent anti-HIV antibodies
with engineered posttranslational modifications. Sci Rep.
10(6201)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ying G, Zhu F, Yi Y, Chen J, Mei J, Zhang
Y and Chen S: Selecting DNA aptamers for endotoxin separation.
Biotechnol Lett. 37:1601–1605. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Bhatt N, Huang PJ, Dave N and Liu J:
Dissociation and degradation of thiol-modified DNA on gold
nanoparticles in aqueous and organic solvents. Langmuir.
27:6132–6137. 2011.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zheng Q, Jockusch S, Zhou Z and Blanchard
SC: The contribution of reactive oxygen species to the
photobleaching of organic fluorophores. Photochem Photobiol.
90:448–454. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Jayasena SD: Aptamers: An emerging class
of molecules that rival antibodies in diagnostics. Clin Chem.
45:1628–1650. 1999.PubMed/NCBI
|