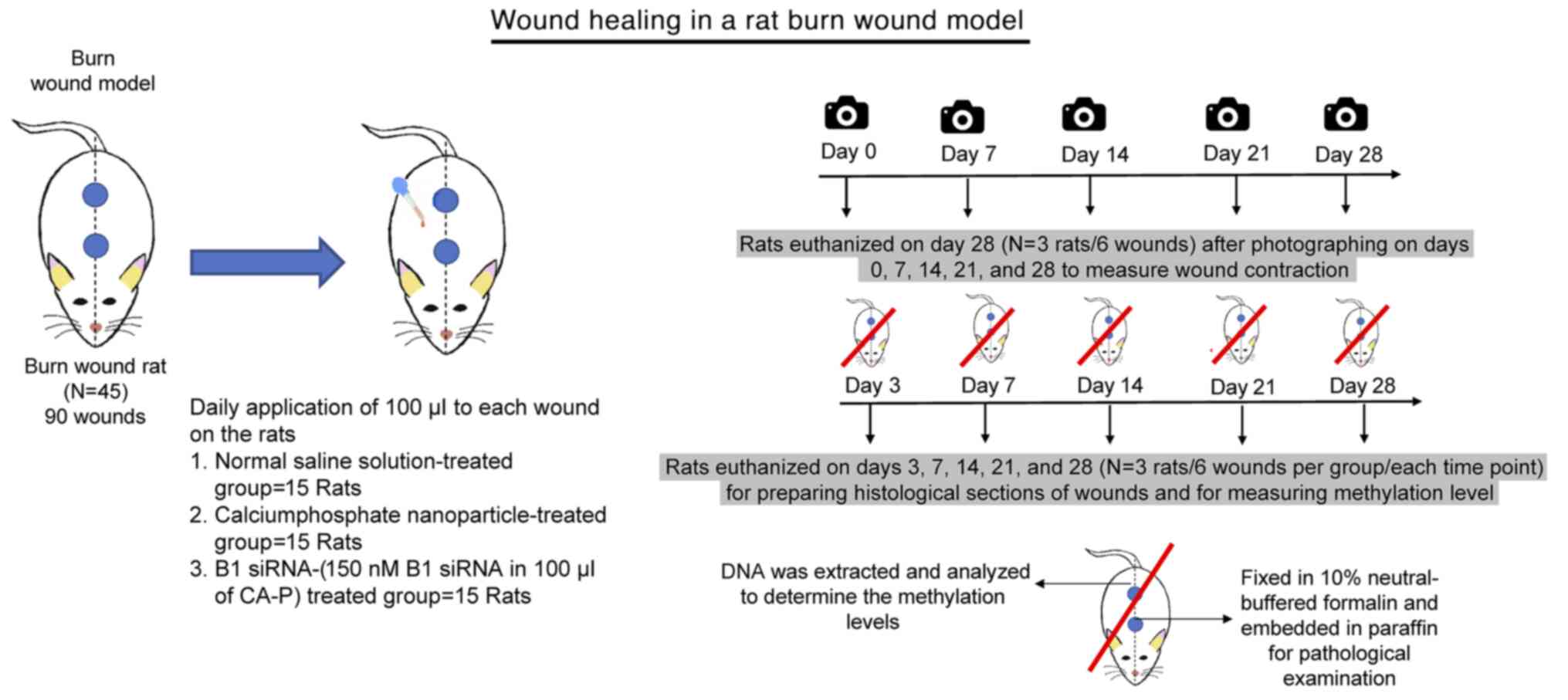
Introduction
Burn wounds can cause severe clinical injuries, and
delayed wound healing leads to infection, prolonged hospital stays,
increased morbidity and >250,000 deaths annually in the United
States of America alone (1).
Researchers and clinicians are actively studying the application of
new substances to improve burn-treatment outcomes.
Interspersed repetitive sequences (IRSs) are the
primary contributor to the genome (~45%), and their methylation
levels are crucial for preserving the stability of the genome
(2). Mutation rates are increased
in cells with IRS hypomethylation (3,4). A
short interspersed nuclear element (SINE) retrotransposon is a
repetitive element of 85-500 bp in length. Two SINE families exist,
namely the B1 (in mice, rats and hamsters) and Alu (in humans)
families (5,6). Similar to the Alu element, the B1
element in rodents stems from 7SL RNA, which is cytoplasmic RNA; it
facilitates protein excretion as a part of the signal recognition
particle (7). The B1 element
assists in transcriptional regulation and DNA stability by binding
to the Aryl hydrocarbon (dioxin) receptor, which is a
ligand-activated transcription factor (8,9);
however, to the best of our knowledge, no study has reported a
correlation between B1 element methylation and wound healing and
epithelialization.
Epigenetic changes associated with chromatin
re-organization can facilitate the transcription machinery and
promote wound repair (10). Genomic
hypomethylation is characterized by reduced methylation of the
methyl groups at the 5' position of cytosine and plays crucial
roles in important events such as aging, cancer and various skin
diseases (11-14).
Furthermore, current research shows that global hypomethylation is
associated with delayed proliferation; in contrast, following
increased Alu methylation, cells can tolerate toxic substances and
decreased DNA damage responses, and show an enhanced proliferation
rate (15). Regarding corneal
ulcers, a previous study has shown that the expression levels of
DNA methyltransferase 1 (Dnmt1) and Dnmt3a, which participate in
the methylation process, are significantly upregulated during
corneal epithelial healing (16).
Therefore, decreased Dnmt1 expression and genomic hypomethylation
defer corneal epithelial wound healing and block human corneal
epithelial cell proliferation and migration. The roles of DNA
methylation in rodents include considerably reducing the wound area
and increasing the wound healing rate by increasing DNA Dnmt3a
expression (16). Moreover,
decreased DNMT1 expression is associated with a low rate of
squamous skin cell proliferation, whereas increased DNMT1
expression is associated with rapid limb bud generation (17,18). A
previous study revealed an association between age and Alu
hypomethylation (11). Positive
associations have also been observed between Alu hypomethylation in
blood cells and several aging phenotypes (19,20).
In addition, treating cells with Alu small-interfering RNA (siRNA)
increased Alu methylation and enabled cells to better tolerate
toxic substances and proliferate faster (15).
RNA-directed DNA methylation is a natural process
through which RNA molecules, which are not translated into any
proteins, direct the addition of methyl groups at the 5' position
of cytosine specific to the RNA in CpG islands. These islands are
genomic regions where CpG sites, DNA regions in which a cytosine
nucleotide is followed by a guanine nucleotide, occur at a high
frequency. The CpG islands are predominantly found in SINE
repetitive sequences, such as Alu and B1 (6,21).
This process is facilitated by a cascade of enzymes, Dicer,
RNA-induced silencing complex and Argonaute proteins (21,22).
This alternative pathway is initiated in chromatin within the
nucleus, and leads to epigenetic modifications, including DNA
cytosine methylation and histone methylation (23,24).
Burns and heat stress can cause DNA damage in
various cells and animals (11,25-30)
and result in DNA damage responses (31). For example, in heated germline
cells, an increase in 8-hydroxy-2'-deoxyguanosine (8-OHdG) level
was observed in response to DNA damage (32). Keratinocytes experience DNA damage
and exhibit delayed proliferation and apoptosis after exposure to
temperatures >42˚C for 24 h (33). Furthermore, an increase in histone
H2AX phosphorylation on serine 139 (γH2AX) occurs in response to
reactive oxygen species-induced DNA double strand breaks, which
have multiple causes, including irradiation, burns and laser
exposure (34-36).
There is a positive correlation between increasing the temperature
from 41.5˚C to 45.5˚C with γH2AX levels in H1299 (human
non-small-cell lung carcinoma p53-deficient) cells (27). Moreover, high temperature-induced
formation of γ-H2AX foci may result in carcinogenesis (37).
The relationship between heat, burns and methylation
has been studied previously (38-40).
There is evidence that heat stress and high temperatures lower DNA
methylation levels in pig muscles and fish tissues, specifically in
the promoter regions of heat shock protein genes (41-43).
An increase in the ambient temperature can affect Alu methylation
in humans (44,45). However, to best of our knowledge, no
study has investigated the relationship between burn wound healing
and Alu methylation with markers of genomic instability, as has
been shown for 8-OHdG and γH2AX.
Therefore, it was hypothesized that DNA methylation
in IRSs can prevent DNA damage and accelerate burn wound healing in
rats. In the present study, the role of B1 methylation in genomic
instability in relation to wound healing was studied. The following
questions were addressed: How can B1 methylation be restored to
reduce genomic stability caused by heat, and how can the rate and
quality of wound healing and epithelialization be accelerated and
the quality improved.
Materials and methods
Animal experiments
Animal experiments were approved by the Animal Care
and Use Committee of Chulalongkorn University (approval no. 012;
March 03/2019).
A total of 45 8-week-old male Wistar rats (150-180
g) were obtained from Namura Animal Laboratory Center (Bangkok,
Thailand). The rats were acclimated for 7 days under a controlled
12-h light/dark cycle, fed standard mice chow, and provided ad
libitum access to water. To create second-degree burns, the
rats were anesthetized with 2% isoflurane (Sigma-Aldrich; Merck
KGaA) (45), and the dorsal skin
was shaved. Two burn wounds were created on the back of each rat
using a 10-mm-diameter aluminum rod, which was heated to 100˚C and
placed on the skin for 10 sec (46). The rats were divided into three
groups with 15 rats/group. Normal saline solution (NSS; 100 µl),
calcium phosphate (Ca-P) nanoparticles, or 150 nM B1 siRNA in 100
µl of a solution containing Ca-P nanoparticles was applied daily to
each wound on the rats in the control and treatment groups.
The wounds of three animals in each group (a total
of six) were imaged on days 0, 7, 14, 21 and 28 after injury, and
the wound areas were measured using ImageJ version 1.52t (National
Institutes of Health) with a freehand selection tool. Wound areas
are reported as a percentage of the initial wounded area. The rats
were euthanized by exposing them to 5% isoflurane for 5-10 min
until respiration ceased and they were confirmed dead on day
28(45). For 12 animals in each
group (a total of 24 wounds), three rats (a total of six wounds)
were euthanized with 5% isoflurane on days 3, 7, 14 and 21. On each
of the specified euthanasia days, wound tissues were excised, and
three of these samples (from the initial six wounds) were
immediately subjected to DNA extraction and analyzed to determine
the methylation levels. The remaining three wound tissues were
fixed in 10% neutral-buffered formalin at room temperature
(28-30˚C) for at least 48 h and embedded in paraffin for
pathological examination (Fig.
1).
Cells and cell culture
Rat epidermal keratinocytes (REK) (Cell
Applications) were cultured in DMEM supplemented with 10% FBS, 100
U/ml penicillin and 100 mg/ml streptomycin (Thermo Fisher
Scientific, Inc.). Cells were incubated at 37˚C in a humidified
incubator supplied with 95% air and 5% CO2. The cells
were cultured in T-75 Corning™ U-shaped cell culture
flasks and harvested at a confluency of ~80% using 0.05%
Trypsin-EDTA with phenol red (Thermo Fisher Scientific, Inc.).
siRNA delivery using a
nanoparticle-coating system
The B1 siRNA sequences were
5'-CGCACGCCUUUAAUCCCAGCACUCGUU-3' (sense) and
5'-CGAGUGCUGGGAUUAAAGGCGUGCGUU-3' (antisense). The scrambled siRNA
sequences were 5'-GGCCAUUUCCCCGACUGCACACCUAUU-3' (sense) and
5'-UAGGUGUGCAGUCGGGGAAAUGGCCUU-3' (antisense). The B1 siRNAs and
scrambled siRNA were purchased from Bioneer, Inc. The siRNA
sequences were designed using siRNA wizard software version 3.1
(InvivoGen). The day before transfection, 5x104 REK
cells/well were seeded in a 24-well plate in 0.5 ml DMEM
supplemented with 10% fetal bovine serum. After 24 h, the medium in
each well in the control groups was replaced with normal saline
solution (NSS; 100 µl) and experiment groups was replaced with 100
µl Ca-P nanoparticles, 150 nM B1 siRNA in 100 µl Ca-P nanoparticles
and 150 nM scramble siRNA in 100 µl Ca-P nanoparticles. The cells
were incubated at 37˚C in a CO2 incubator for 48 h,
following which, the cells were collected through
trypsinization.
To deliver siRNAs to target cells, the siRNAs were
coated with a nanoparticle solution before topical administration
on the burn wounds. The most effective siRNA:nanoparticle solution
ratio was 150 nM B1 siRNA in 100 µl CA-P nanoparticle solution,
based on the results of previous studies (15,47,48).
First, B1 siRNA was mixed with an appropriate
proportion and concentration of calcium solution (MilliporeSigma),
a B1 siRNA-binding reagent. Next, the B1 siRNA-calcium complex was
added to a mixture of sodium carbonate
(Na2CO3; MilliporeSigma) and sodium
dihydrogen phosphate monohydrate
(NaH2PO4·H2O; MilliporeSigma).
After the B1 siRNA-coating process, the nanoparticle-coated B1
siRNAs were stored at room temperature (28-30˚C) and prepared on
the day of use. The nanoparticle solution was composed 50 µl of a
mixture containing 0.5 M calcium chloride (CaCl2)
solution (MilliporeSigma) and 150 nM B1 siRNA + 50 µl of a mixture
of 0.01 M Na2CO3 solution (MilliporeSigma)
and 0.01 M NaH2PO4·H2O solution
(MilliporeSigma). A 31:1 molar ratio of
CO32-:PO43- was used.
The B1 siRNA was mixed with 16 µl 0.5 M CaCl2 solution
and adjusted to a final volume of 50 µl using sterile
dH2O. Thereafter, the B1 siRNA-calcium complex was added
to 50 µl of a mixture of Na2CO3 solution and
NaH2PO4·H2O solution (16 µl) and
34 µl sterile dH2O (47). All steps in the preparation of the
nanoparticle solution were performed using sterile techniques. Each
nanoparticle solution preparation was used to validate the
transfection efficiency with rat dermal keratinocytes (Cell
Applications) before each experiment (48-52).
Histopathology and
immunohistochemistry
Formalin-fixed wound tissues were dehydrated and
embedded in paraffin, after which, 3-µm-thick sections were
prepared using a microtome. Hematoxylin and eosin (H&E)
staining was performed for scoring, using hematoxylin solution for
6 h and eosin Y ethanol solution for 48 h at a temperature of 60˚C.
Immunohistochemical staining of the tissue sections was performed
using monoclonal antibodies against γH2AX (1:100; cat. no. Ab2893;
Abcam) and 8-OHdG (1:100; cat. no. Ab48508; Abcam) (53). The sections and antibodies were
incubated at 37˚C for 32 min using the Ventana®
Benchmark XT (Ventana Medical Systems, Inc; Roche Diagnostics)
automated slide strainer in combination with the Ventana ultraView
DAB v3 IHC Detection Kit before mounting, according to the
manufacturer's protocol. Thereafter, the slides were counterstained
at 25˚C with Hematoxylin II for 8 min and Bluing Reagent for 4 min
(53).
The histological sections of the wounds were graded
using a histopathological scoring system, following a previously
published method with some modifications (54-58)
Three dermatopathologists who were blinded to the treatment regimen
independently performed H&E scoring, based on five criteria:
Epithelialization, polymorphonuclear leukocyte (PMNL) infiltration,
collagen formation, number of fibroblasts and presence of new blood
vessels. A score of 0 was assigned when there was no evident
epithelialization and no increase in the number of fibroblasts,
PMNLs or newly formed blood vessels. A score of 1 indicated an
increased thickness of the edges of the cut epithelial tissue
sections, or the presence of a few fibroblasts, PMNLs and newly
formed blood vessels. A score of 2 indicated migration of
epithelial cells, or the presence of a moderate number of
fibroblasts, PMNLs and newly formed blood vessels. A score of 3
indicated epithelial bridging of the incision, or the presence of
several fibroblasts, PMNLs and newly formed blood vessels. A score
of 4 was assigned for sections where there was complete
regeneration of the epithelium, or presence of excessive
fibroblasts, PMNLs and newly formed blood vessels. The mean scores
of the three scorers were determined and combined to obtain an
overall pathology score. Sections positive for anti-cytokeratin
immunohistochemical staining were scored in a similar manner; three
dermatopathologists scoring the sections on a scale of 0-4,
starting from no staining to strong staining.
DNA preparation
DNA extraction from rat skin was performed using the
phenol-chloroform method, which is the most commonly used method to
purify and concentrate DNA (59),
followed by the addition of phenol-chloroform to a solution of
lysed cells, and mixing and separation of the two phases by
centrifugation for 5 min at 16,000 x g. The resulting solution was
separated into two phases: A lower organic phase and an upper
aqueous phase. The top aqueous phase containing DNA was then
carefully removed. A total of 1 µl 20 mg/ml Glycogen solution, 0.1X
3 M sodium acetate and 2.5X volume of absolute ethanol (100%) were
added to the precipitated DNA, and the mixture was centrifuged for
1 min at 8,000 x g at 25˚C. Finally, the DNA was washed with 70%
ethanol, air-dried and dissolved in dH2O (59).
Quantitative combined bisulfite
restriction analysis for B1 element (COBRA B1)
A total of 1 µl bisulfited DNA taken from an EZ DNA
methylation-GoldTM kit (Zymo Research Corp.) was
subjected to 45 cycles of PCR, using the PCR mastermixTM
(Thermo Scientific, Inc.). The following thermocycling conditions
were used: 95˚C for 15 min; followed by 40 cycles at 95˚C for 45
sec, 53˚C for 45 sec and 72˚C for 45 sec; with a final extension
step of 72˚C for 10 min using the B1 COBRA forward primer,
5'-YGYAYGYYTTTAATYYYAGYAAT-3' and reverse primer,
5'-CCCTRRCTRTCCTRRAACTCAC-3', which were designed using Primer 3
version 4.1.0 (2,60), and had an annealing temperature of
70˚C. Primer sequences with ‘Y’ represent pyrimidine bases (C or T)
that have the ability to bind to G or A bases, and ‘R’ represents
purine bases (A or G) that have the ability to bind to T or C bases
in DNA sequences following bisulfite conversion (Fig. 2C). The amplified PCR products were
digested with two units of Taq1 and Tas1 endonuclease
in NE buffer II (MBI Fermentas; Thermo Fisher Scientific, Inc.) at
37˚C overnight. The digested PCR products were identified using 8%
non-denaturing polyacrylamide gel electrophoresis and stained with
SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich; Merck KGaA). All
samples were assessed in duplicate. After enzyme digestion, six
products of different lengths were detected after COBRA B1 (98, 78,
54, 44, 34 and 20 bp). The band intensities of the COBRA B1
products were determined using Image Quant version 8.2 (Molecular
Dynamics; GE Healthcare).
 | Figure 2Effect of B1 siRNA treatment on the
contracture rate of second-degree burn wounds. (A) Images of
second-degree burn wounds in rats whose wounds were treated daily
with a saline control, Ca-P nanoparticles or B1 siRNA. The wounds
treated with B1 siRNA exhibited increased contraction and less
inflammation compared with both controls. (B) Wound healing plotted
as a percentage of the original wound area. Wound healing was
significantly enhanced in the B1 siRNA-treated group compared with
that in the control groups, especially on days 14 and 28
post-injury (Kruskal-Wallis test). (C) The methylation patterns of
B1 detected by COBRA-B1, and the restriction enzyme digest sites.
(D) The B1 methylation level of second-degree burn wounds. B1
methylation was significantly enhanced in the treated group
compared with that in the control group, especially on days 7, 14
and 21 post-injury (one-way ANOVA). *P<0.05,
**P<0.01, ***P<0.001. n=6 wounds/group.
siRNA, small interfering RNA; COBRA B1, combine bisulfite
restriction analysis for B1 element; ns, not significant; Ca-P,
calcium phosphate. |
Sequences of B1 repetitive elements in
rats and locations of the Taq1 and Tas1 restriction enzyme
sites
According to the B1 analysis and calculations, two
types of COBRA B1 products could be classified based on the
methylation status of the CpG dinucleotides, namely methylated loci
(mC) and unmethylated loci (uC). The percentage of B1 methylation
was calculated as follows. The letters demonstrated the normalized
scores of each band, and the percentage of intensity of each band
was divided by the amplicon length in terms of bp: %98/98=A,
%78/74=B, %54/54=C, %44/42=D, %34/34=E, and %20/20=F. Subsequently,
the percentage of methylation was calculated by bring the
normalized scores of the digested methylation fragment divided by
the sum of the normalized scores of the undigested and digested
products as following formula: % total methylation (%mC)=100 x (A +
C + D)/2A + 2B + 2D. As an internal control, 25% methylated rat
genomic DNA (EpigenDX) was used for the experiments and inter-assay
adjustment.
Statistical analysis
Distribution of data were determined using a
Shapiro-Wilk test. For normally distributed data, a one-way ANOVA
followed by Bonferroni post-hoc corrections were used to compare
the groups. For non-normally distributed data, a Kruskal-Wallis
analysis followed by a Dunn's post-hoc test was used to test the
differences between groups. Data are reported as the mean ±
standard deviation. GraphPad Prism version 9.0.0 (GraphPad
Software, Inc.) was used for all statistical analyses. P<0.05
was considered to indicate a statistically significant
difference.
Results
Treatment with B1 siRNA increases the
rate of healing of second-degree burn wounds as well as the
methylation levels in the wounds
Second-degree burn wounds were treated each day with
150 nM B1 siRNA and the wound areas were measured using photographs
taken on days 0, 7, 14, 21 and 28 post-injury (Fig. 2A). The improvements in wound healing
were greater in the B1 siRNA group. The respective percentage wound
healing ± standard deviation in the groups treated with saline or
Ca-P nanoparticles, relative to that in the B1 siRNA-treated group,
on different days was as follows: i) Day 7, 15.81±3.59 and
15.66±2.36 vs. 20.01±1.97%, both P<0.01; ii) day 14, 56.58±1.24
and 56.73±1.56 vs. 60.38±1.39%, both P<0.001; iii) day 21,
73.14±2.27 and 73.39±2.62 vs. 77.15±1.32%, both P<0.01; and iv)
day 28, 90.63±1.39 and 90.81±0.86 vs. 97.21±0.84%, both P<0.001.
There were no differences in wound areas between the NSS-treated
control and Ca-P nanoparticle groups. Furthermore, significant
differences were observed amongst the control and Ca-P nanoparticle
compared with the B1 siRNA treatment group, particularly on days 14
and 28 post-injury (Fig. 2B).
B1 siRNA promotes methylation through siRNA directed
DNA methylation (21,23); it improves DNA stability and
enhances cell proliferation (15).
The methylation levels of the B1 siRNA-treated group were
significantly higher than those of the saline-treated control and
Ca-P nanoparticle groups, especially on days 7 and 14. The
respective percentage methylation ± standard deviation in the NSS
control, Ca-P nanoparticle and B1 siRNA-treated wounds on different
days were: i) Day 3, 30.33±1.52 and 29.0±1.0 vs. 33.00±1.0%, both
P<0.05; ii) day 7, 30.0±2.0 and 31.0±2.0 vs. 35.0±1.0%, both
P<0.001; iii) day 14, 33.67±1.52 and 33.0±1.73 vs. 45.0±1.0%,
both P<0.001; iv) day 21, 33.67±2.08 and 34.67±0.58 vs.
41.0±1.0%, both P<0.001; and v) day 28, 34.33±0.58 and
34.33±0.58 vs. 38.00±1.00%, both P<0.05 (Fig. 2D). Significant differences were
observed in the B1 siRNA treated group compared with the control
groups 3 days post-injury.
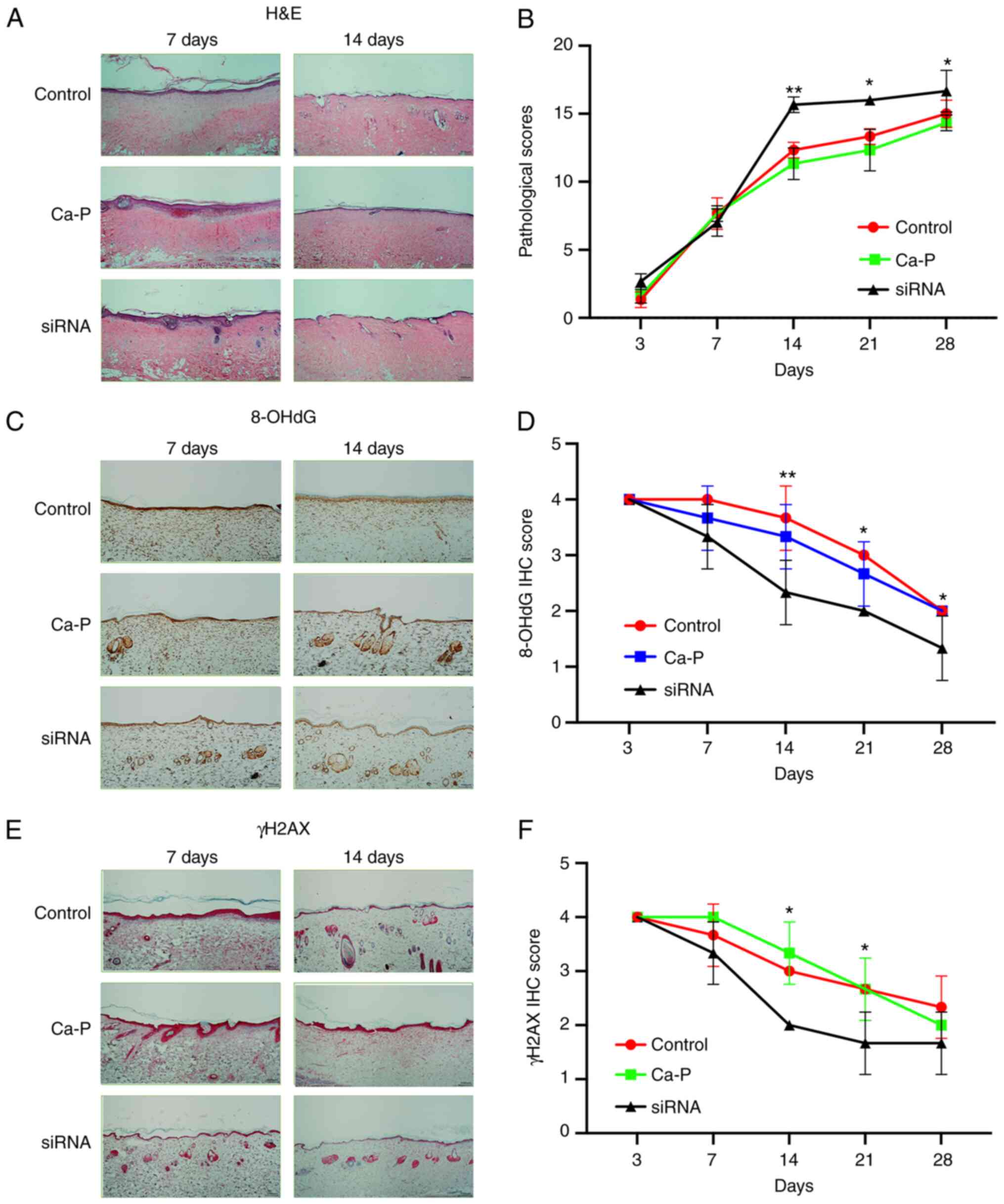
Pathological and immunohistochemical
skin changes in the rat model of burn wounding
Individual histopathological scores were added
together to provide an overall score for each wound. Blinded
scoring of the wounds by the three dermatopathologists confirmed
that the rats treated with B1 siRNA displayed considerably higher
pathological scores than those treated with NSS or Ca-P
nanoparticles, with the greatest improvement observed on days 14,
21 and 28 post injury. The pathological scores ± standard deviation
in the NSS and Ca-P nanoparticle-treated groups (respectively),
relative to that in the B1 siRNA-treated group, on different days
were as follows: i) Day 3, 1.33±0.58 and 1.67±0.58 vs. 2.67±0.58%,
both P>0.05; ii) day 7, 7.67±1.16 and 7.67±0.58 vs. 7.0±1.0%,
both P>0.05; iii) day 14, 12.33±0.58 and 11.33±1.16 vs.
15.67±0.58%, both P<0.01; iv) day 21, 13.33±0.58 and 12.33±1.52
vs. 16.0±0.0%, P<0.05; and v) day 28, 15.0±1.0 and 14.33±0.58
vs. 16.67±1.53%, P<0.05 (Fig. 3A
and B).
8-OHdG and γH2AX scores were determined to measure
DNA damage response after introducing burn injuries. A low
immunohistochemistry score for 8-OHdG and γH2AX reflected a low DNA
damage response. The B1 siRNA-treated group exhibited lower levels
of 8-OHdG than the NSS and Ca-P nanoparticle groups on days 14 and
21. In addition, the wounds in the B1 siRNA-treated group showed
significantly lower γH2AX levels from days 7 to 21 than the NSS-
and Ca-P nanoparticle-treated groups, with the highest level
observed 14 days post-injury. The respective 8-OHdG
immunohistochemistry score ± standard deviation in the NSS and Ca-P
nanoparticle-treated groups (respectively), relative to that in the
B1 siRNA-treated group was as follows: i) Day 3; 4.0±0.0 and
4.0±0.0 vs. 4.0±0.0%, both P>0.05; ii) day 7; 4.0±0.0 and
3.67±0.58 vs. 3.33±0.58%, both P>0.05; iii) day 14, 3.0±0.0 and
3.33±0.58 vs. 2.67±0.58%; both P<0.01; iv) day 21, 3.0±0.0 and
2.67±0.58 vs. 2.0%±0.0%, both P<0.05; and v) day 28, 2.0±0.0 and
2.0±0.0 vs. 1.67±0.58%, P<0.05 (Fig.
3C and D). Furthermore, the
γH2AX immunohistochemistry scores ± standard deviation in the NSS
and Ca-P nanoparticle groups, relative to that in the B1
siRNA-treated group, was as follows: i) day 3, 4.0±0.0 and 4.0±0.0
vs. 4.0±0.0%, both P>0.05, ii) day 7, 3.67±0.58 and 4.0±0.0 vs.
3.33±0.58%, both P>0.05, iii) day 14, 3.0±0.0 and 3.33±0.58 vs.
2.67±0.58%, both P<0.05, iv) day 21, 2.67±0.58 and 2.67±0.58 vs.
2.0±0.0%, both P<0.05; and v) day 28, 2.0±0.0 and 2.0±0.0 vs.
1.67±0.58%; both P>0.05 (Fig. 3E
and F).
Discussion
The healing of burn wounds requires the synchronous
activity of numerous intricate molecular processes, such as cell
creeping and epithelialization, neoangiogenesis and fibroblast
remodeling (61). Global DNA
hypomethylation causes delayed wound healing; therefore, we induced
hypermethylation of IRSs to promote the wound healing process
(62).
A limitation of this study is that we performed only
in vitro experiments with the scrambled siRNA; an in
vivo scrambled siRNA control group was not included. B1 siRNA
significantly increased B1 methylation, whereas the scrambled siRNA
marginally increased B1 methylation. There were no off-target
effects in the scrambled siRNA group in the in vitro
experiment Fig. S1).
In the animal model experiments, 7-28 days
post-injury, the B1 siRNA-treated group showed faster
wound-contracture and re-epithelialization rates than the control
group. An examination of wound tissue sections also revealed that
treatment with B1 siRNA induced favorable pathological alterations
in the repaired tissues. The ability of B1 siRNA to increase the B1
methylation levels and heal thermally-induced injury corresponded
with a significant decrease in DNA damage responses, based on
8-OHdG and γH2AX scores in the wound tissues.
A previous in vitro study showed that
increased Alu methylation increases a cells tolerance to toxic
substances and also increases the proliferation of these cells
(15). A molecular substance that
can manage DNA methylation at a precise target, reduce DNA damage
responses, stabilize the genome and improve second-degree burn
wound healing in a rat model was developed by in the present study.
B1 siRNA specifically increased the methylation of B1 repetitive
sequences by RNA-directed DNA methylation (23,63).
DNA damage responses can lead to mutations and delay cell cycle
progression or cause cell cycle arrest (64,65).
Therefore, the reduction in DNA damage responses, as assessed using
8-OHdG and γH2AX scores, induced by increased B1 methylation using
B1 siRNA, led to increased wound contraction rates and improved
overall pathological scores in the wounded sections. Reduced
inflammation, greater epithelialization, new collagen deposition,
fibroblast migration and new blood vessel growth were also observed
(data not shown).
Following radiation exposure, DNA methylation is
reduced in mice heart muscles owing to DNA damage responses and
release of free radicals; how B1 methylation decreases DNA damage
responses remains to be determined (66). It is not clear how B1 siRNA can
accelerate burn wound healing and reduce DNA damage responses;
based the results of the present study, it is hypothesized that IRS
methylation is associated with other epigenetic phenomena, such as
the formation of heterochromatin (67,68).
Therefore, heterochromatin may protect DNA from damage and damage
responses, and downregulate DNA replication and transcription
processes (69). Another
possibility is that a change in the chromatin form could ameliorate
DNA repair activity (70,71). This process might reduce DNA damage
response in the heterochromatin region altered by B1
hypermethylation. Enhanced B1 methylation might lower oxidative
stress and, consequently, DNA damage. Alu hypermethylation is
correlated with weight gain in infants during the first year of
birth (72). Conversely, the
association between Alu hypomethylation with DNA damage and lesions
demonstrates that Alu hypomethylation increases the risk of
non-communicable diseases, such as diabetes mellitus, hypertension
and osteoporosis (73-75).
In summary, B1 siRNA can promote healing of
second-degree burn wounds in rats by increasing the
epithelialization rate, improving pathological scores and
accelerating the wound healing rate. These results indicate that B1
siRNA promotes wound healing by reducing the DNA damage response
and enhancing global DNA methylation. B1 siRNA assisted in genomic
recovery and improved the healing process of second-degree burn
wounds in rats; this suggests that B1 siRNA may serve as a novel
treatment method for burn wounds in clinical practice, perhaps as a
topical agent against repetitive sequences. Further studies to
evaluate the efficacy of B1 siRNA against different types of burns
with different severities will be highly beneficial for determining
the clinical potential of this treatment.
Supplementary Material
The B1 methylation levels of rat
epidermal keratinocytes cells. B1 methylation was significantly
enhanced in the B1 siRNA-treated group compared with that in the
control group and scramble siRNA group: i) Control, 28.83±0.86%;
ii) Ca-P, 29.0±0.70%; iii) scramble siRNA, 29.22±1.09; iv) B1
siRNA, 43.72±1.25% (Kruskal-Wallis test). **P<0.01,
***P<0.001. n=9/group. siRNA, small interfering RNA;
Ca-P, calcium phosphate; ns, not significant.
Acknowledgements
We would like to thank Dr Manita Attasuriyanan and
Dr Apasee Sooksamran, Division of Dermatology, Department of
Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok
for assistance with the pathological scoring.
Funding
Funding: This work was supported by the Faculty of medicine,
Chulalongkorn University, the Thailand Research Fund (grant no. RSA
6280053) and the National Science and Technology Development Agency
(grant no. FDA-CO-2561-8477-TH).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JM, AA, KJH and AM conceived the study. JM, PN, AA
and AM curated the data. JM, JW, NK, KJH, AA and AM analyzed the
data. JM, PN, JW, AM and AA performed the investigation. JM, JW,
NK, KJH, AM and AA designed the study. JM, JW, NK drafted the
manuscript. JM, JW, NK, KJH, AM and AA review and edited the
manuscript. All authors have read and approved the manuscript. KJH,
AM and AA confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Animal experiments were approved by the Animal Care
and Use Committee of Chulalongkorn University (approval no.
003/2562).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen L, He X, Xian J, Liao J, Chen X, Luo
Y, Wang Z and Li N: Development of a framework for managing severe
burns through a 17-year retrospective analysis of burn epidemiology
and outcomes. Sci Rep. 11(9374)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Veniaminova NA, Vassetzky NS and Kramerov
DA: B1 SINEs in different rodent families. Genomics. 89:678–686.
2007.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gaudet F, Hodgson JG, Eden A,
Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H and Jaenisch R:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Slimen IB, Najar T, Ghram A, Dabbebi H,
Ben Mrad M and Abdrabbah M: Reactive oxygen species, heat stress
and oxidative-induced mitochondrial damage. A review. Int J
Hyperthermia. 30:513–523. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gebhard W, Meitinger T, Höchtl J and
Zachau HG: A new family of interspersed repetitive DNA sequences in
the mouse genome. J Mol Biol. 157:453–471. 1982.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Batzer MA and Deininger PL: Alu repeats
and human genomic diversity. Nat Rev Genet. 3:370–379.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Tsirigos A and Rigoutsos I: Alu and b1
repeats have been selectively retained in the upstream and intronic
regions of genes of specific functional classes. PLoS Comput Biol.
5(e1000610)2009.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Roman AC, Benitez DA, Carvajal-Gonzalez JM
and Fernandez-Salguero PM: Genome-wide B1 retrotransposon binds the
transcription factors dioxin receptor and Slug and regulates gene
expression in vivo. Proc Natl Acad Sci USA. 105:1632–1637.
2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Román AC, González-Rico FJ and
Fernández-Salguero PM: B1-SINE retrotransposons: Establishing
genomic insulatory networks. Mob Genet Elements. 1:66–70.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ti D, Li M, Fu X and Han W: Causes and
consequences of epigenetic regulation in wound healing. Wound
Repair Regen. 22:305–312. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mutirangura A: A hypothesis to explain how
the DNA of elderly people is prone to damage: Genome-wide
hypomethylation drives genomic instability in the elderly by
reducing youth-associated genome-stabilizing DNA gaps. In:
Epigenetics. Meccariello R (ed). IntechOpen, London, 2018.
|
|
12
|
Bollati V, Schwartz J, Wright R, Litonjua
A, Tarantini L, Suh H, Sparrow D, Vokonas P and Baccarelli A:
Decline in genomic DNA methylation through aging in a cohort of
elderly subjects. Mech Ageing Dev. 130:234–239. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li Y, Sawalha AH and Lu Q: Aberrant DNA
methylation in skin diseases. J Dermatol Sci. 54:143–149.
2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Udomsinprasert W, Kitkumthorn N,
Mutirangura A, Chongsrisawat V, Poovorawan Y and Honsawek S: Global
methylation, oxidative stress, and relative telomere length in
biliary atresia patients. Sci Rep. 6(26969)2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Patchsung M, Settayanon S, Pongpanich M,
Mutirangura D, Jintarith P and Mutirangura A: Alu siRNA to increase
Alu element methylation and prevent DNA damage. Epigenomics.
10:175–185. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Luo G, Jing X, Yang S, Peng D, Dong J, Li
L, Reinach PS and Yan D: DNA methylation regulates corneal
epithelial wound healing by targeting miR-200a and CDKN2B. Invest
Ophthalmol Vis Sci. 60:650–660. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Plikus MV, Guerrero-Juarez CF, Treffeisen
E and Gay DL: Epigenetic control of skin and hair regeneration
after wounding. Exp Dermatol. 24:167–170. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Aguilar C and Gardiner DM: DNA methylation
dynamics regulate the formation of a regenerative wound epithelium
during axolotl limb regeneration. PLoS One.
10(e0134791)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Erichsen L, Beermann A, Arauzo-Bravo MJ,
Hassan M, Dkhil MA, Al-Quraishy S, Hafiz TA, Fischer JC and
Santourlidis S: Genome-wide hypomethylation of LINE-1 and Alu
retroelements in cell-free DNA of blood is an epigenetic biomarker
of human aging. Saudi J Biol Sci. 25:1220–1226. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Mutirangura A: Is global hypomethylation a
nidus for molecular pathogenesis of age-related noncommunicable
diseases? Epigenomics. 11:577–579. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Erdmann RM and Picard CL: RNA-directed DNA
methylation. PLoS Genet. 16(e1009034)2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Castanotto D, Tommasi S, Li M, Li H, Yanow
S, Pfeifer GP and Rossi JJ: Short hairpin RNA-directed cytosine
(CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol
Ther. 12:179–183. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Matzke MA and Mosher RA: RNA-directed DNA
methylation: An epigenetic pathway of increasing complexity. Nat
Rev Genet. 15:394–408. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Mathieu O and Bender J: RNA-directed DNA
methylation. J Cell Sci. 117:4881–4888. 2004.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Reis AH, Vargas FR and Lemos B: Biomarkers
of genome instability and cancer epigenetics. Tumour Biol.
37:13029–13038. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kongruttanachok N, Phuangphairoj C,
Thongnak A, Ponyeam W, Rattanatanyong P, Pornthanakasem W and
Mutirangura A: Replication independent DNA double-strand break
retention may prevent genomic instability. Mol Cancer.
9(70)2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Takahashi A, Matsumoto H, Nagayama K,
Kitano M, Hirose S, Tanaka H, Mori E, Yamakawa N, Yasumoto J, Yuki
K, et al: Evidence for the involvement of double-strand breaks in
heat-induced cell killing. Cancer Res. 64:8839–8845.
2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Mah LJ, El-Osta A and Karagiannis TC:
gammaH2AX: A sensitive molecular marker of DNA damage and repair.
Leukemia. 24:679–686. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Mataix M, Rodríguez-Luna A,
Gutiérrez-Pérez M, Milani M, Gandarillas A, Espada J and Pérez-Davó
A: Deschampsia antarctica extract (Edafence®) as a
powerful skin protection tool against the aging exposome. Plast
Aesthet Res. 7(69)2020.
|
|
30
|
Korkmaz KS, Debelec Butuner B and
Roggenbuck D: Detection of 8-OHdG as a diagnostic biomarker. J Lab
Precis Med. 3(95)2018.
|
|
31
|
Purschke M, Laubach HJ, Anderson RR and
Manstein D: Thermal injury causes DNA damage and lethality in
unheated surrounding cells: Active thermal bystander effect. J
Invest Dermatol. 130:86–92. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Houston BJ, Nixon B, Martin JH, De Iuliis
GN, Trigg NA, Bromfield EG, McEwan KE and Aitken RJ: Heat exposure
induces oxidative stress and DNA damage in the male germ line. Biol
Reprod. 98:593–606. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hintzsche H, Riese T and Stopper H:
Hyperthermia-induced micronucleus formation in a human keratinocyte
cell line. Mutat Res. 738-739:71–74. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Firsanov DV, Solovjeva LV and Svetlova MP:
H2AX phosphorylation at the sites of DNA double-strand breaks in
cultivated mammalian cells and tissues. Clin Epigenetics.
2:283–297. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kinner A, Wu W, Staudt C and Iliakis G:
Gamma-H2AX in recognition and signaling of DNA double-strand breaks
in the context of chromatin. Nucleic Acids Res. 36:5678–5694.
2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kaneko H, Igarashi K, Kataoka K and Miura
M: Heat shock induces phosphorylation of histone H2AX in mammalian
cells. Biochem Biophys Res Commun. 328:1101–1106. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Dewhirst MW, Lora-Michiels M, Viglianti
BL, Dewey WC and Repacholi M: Carcinogenic effects of hyperthermia.
Int J Hyperthermia. 19:236–251. 2003.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Xu R, Li S, Guo S, Zhao Q, Abramson MJ, Li
S and Guo Y: Environmental temperature and human epigenetic
modifications: A systematic review. Environ Pollut.
259(113840)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bind MC, Coull BA, Baccarelli A, Tarantini
L, Cantone L, Vokonas P and Schwartz J: Distributional changes in
gene-specific methylation associated with temperature. Environ Res.
150:38–46. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hao Y, Cui Y and Gu X: Genome-wide DNA
methylation profiles changes associated with constant heat stress
in pigs as measured by bisulfite sequencing. Sci Rep.
6(27507)2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Varriale A and Bernardi G: DNA methylation
and body temperature in fishes. Gene. 385:111–121. 2006.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Vinoth A, Thirunalasundari T, Shanmugam M,
Uthrakumar A, Suji S and Rajkumar U: Evaluation of DNA methylation
and mRNA expression of heat shock proteins in thermal manipulated
chicken. Cell Stress Chaperones. 23:235–252. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bind MA, Zanobetti A, Gasparrini A, Peters
A, Coull B, Baccarelli A, Tarantini L, Koutrakis P, Vokonas P and
Schwartz J: Effects of temperature and relative humidity on DNA
methylation. Epidemiology. 25:561–569. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Dridi S: Alu mobile elements: From junk
DNA to genomic gems. Scientifica (Cairo).
2012(545328)2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Marquardt N, Feja M, Hünigen H, Plendl J,
Menken L, Fink H and Bert B: Euthanasia of laboratory mice: Are
isoflurane and sevoflurane real alternatives to carbon dioxide?
PLoS One. 13(e0203793)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Cai EZ, Ang CH, Raju A, Tan KB, Hing EC,
Loo Y, Wong YC, Lee H, Lim J, Moochhala SM, et al: Creation of
consistent burn wounds: A rat model. Arch Plast Surg. 41:317–324.
2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhao D, Wang CQ, Zhuo RX and Cheng SX:
Modification of nanostructured calcium carbonate for efficient gene
delivery. Colloids Surf B Biointerfaces. 118:111–116.
2014.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Wu X, Yamamoto H, Nakanishi H, Yamamoto Y,
Inoue A, Tei M, Hirose H, Uemura M, Nishimura J, Hata T, et al:
Innovative delivery of siRNA to solid tumors by super carbonate
apatite. PLoS One. 10(e0116022)2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Mostaghaci B, Loretz B and Lehr CM:
Calcium phosphate system for gene delivery: Historical background
and emerging opportunities. Curr Pharm Des. 22:1529–1533.
2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xie Y, Chen Y, Sun M and Ping Q: A mini
review of biodegradable calcium phosphate nanoparticles for gene
delivery. Curr Pharm Biotechnol. 14:918–925. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Xu X, Li Z, Zhao X, Keen L and Kong X:
Calcium phosphate nanoparticles-based systems for siRNA delivery.
Regen Biomater. 3:187–195. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Levingstone TJ, Herbaj S, Redmond J,
McCarthy HO and Dunne NJ: Calcium phosphate nanoparticles-based
systems for RNAi delivery: Applications in bone tissue
regeneration. Nanomaterials (Basel). 10(146)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wang F, Zieman A and Coulombe PA: Skin
keratins. Methods Enzymol. 568:303–350. 2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Tanideh N, Rokhsari P, Mehrabani D,
Mohammadi Samani S, Sabet Sarvestani F, Ashraf MJ, Koohi
Hosseinabadi O, Shamsian S and Ahmadi N: The healing effect of
licorice on Pseudomonas aeruginosa infected burn wounds in
experimental rat model. World J Plast Surg. 3:99–106.
2014.PubMed/NCBI
|
|
55
|
Farghali HA, Abdelkader NA, Khattab MS and
Abubakr HO: Evaluation of subcutaneous infiltration of autologous
platelet-rich plasma on skin-wound healing in dogs. Biosci Rep.
37(BSR20160503)2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Tavares Pereira Ddos S, Lima-Ribeiro MH,
De Pontes-Filho NT, Carneiro-Leão AM and Correia MT: Development of
animal model for studying deep second-degree thermal burns. J
Biomed Biotechnol. 2012(460841)2012.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Haghdoost F, Baradaran Mahdavi MM,
Zandifar A, Sanei MH, Zolfaghari B and Javanmard SH: Pistacia
atlantica resin has a dose-dependent effect on angiogenesis and
skin burn wound healing in rat. Evid Based Complement Alternat Med.
2013(893425)2013.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Edraki M, Akbarzadeh A, Hosseinzadeh M,
Tanideh N, Salehi A and Koohi-Hosseinabadi O: Healing effect of sea
buckthorn, olive oil, and their mixture on full-thickness burn
wounds. Adv Skin Wound Care. 27:317–323. 2014.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Ahmed I, Islam M, Arshad W, Mannan A,
Ahmad W and Mirza B: High-quality plant DNA extraction for PCR: An
easy approach. J Appl Genet. 50:105–107. 2009.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Untergasser A, Cutcutache I, Koressaar T,
Ye J, Faircloth BC, Remm M and Rozen SG: Primer3-New capabilities
and interfaces. Nucleic Acids Res. 40(e115)2012.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Rowan MP, Cancio LC, Elster EA, Burmeister
DM, Rose LF, Natesan S, Chan RK, Christy RJ and Chung KK: Burn
wound healing and treatment: Review and advancements. Crit Care.
19(243)2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Lewis CJ, Mardaryev AN, Sharov AA, Fessing
MY and Botchkarev VA: The epigenetic regulation of wound healing.
Adv Wound Care (New Rochelle). 3:468–475. 2014.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Chalertpet K, Pin-On P, Aporntewan C,
Patchsung M, Ingrungruanglert P, Israsena N and Mutirangura A:
Argonaute 4 as an effector protein in RNA-directed DNA methylation
in human cells. Front Genet. 10(645)2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Kantidze OL, Velichko AK, Luzhin AV and
Razin SV: Heat stress-induced DNA damage. Acta Naturae. 8:75–78.
2016.PubMed/NCBI
|
|
65
|
Velichko AK, Petrova NV, Razin SV and
Kantidze OL: Mechanism of heat stress-induced cellular senescence
elucidates the exclusive vulnerability of early S-phase cells to
mild genotoxic stress. Nucleic Acids Res. 43:6309–6320.
2015.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Koturbash I, Miousse IR, Sridharan V,
Nzabarushimana E, Skinner CM, Melnyk SB, Pavliv O, Hauer-Jensen M,
Nelson GA and Boerma M: Radiation-induced changes in DNA
methylation of repetitive elements in the mouse heart. Mutat Res.
787:43–53. 2016.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Baylin SB, Esteller M, Rountree MR,
Bachman KE, Schuebel K and Herman JG: Aberrant patterns of DNA
methylation, chromatin formation and gene expression in cancer. Hum
Mol Genet. 10:687–692. 2001.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Kim JH: Chromatin remodeling and
epigenetic regulation in plant DNA damage repair. Int J Mol Sci.
20(4093)2019.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Grewal SI and Jia S: Heterochromatin
revisited. Nat Rev Genet. 8:35–46. 2007.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Xu Y, Xu C and Price BD: Mechanistic links
between ATM and histone methylation codes during DNA repair. Prog
Mol Biol Transl Sci. 110:263–288. 2012.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Jakob B, Splinter J, Conrad S, Voss KO,
Zink D, Durante M, Löbrich M and Taucher-Scholz G: DNA
double-strand breaks in heterochromatin elicit fast repair protein
recruitment, histone H2AX phosphorylation and relocation to
euchromatin. Nucleic Acids Res. 39:6489–6499. 2011.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Rerkasem K, Rattanatanyong P, Rerkasem A,
Wongthanee A, Rungruengthanakit K, Mangklabruks A and Mutirangura
A: Higher Alu methylation levels in catch-up growth in
twenty-year-old offsprings. PLoS One. 10(e0120032)2015.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Jintaridth P, Tungtrongchitr R,
Preutthipan S and Mutirangura A: Hypomethylation of Alu elements in
post-menopausal women with osteoporosis. PLoS One.
8(e70386)2013.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Thongsroy J, Patchsung M and Mutirangura
A: The association between Alu hypomethylation and severity of type
2 diabetes mellitus. Clin Epigenetics. 9(93)2017.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Han L, Liu Y, Duan S, Perry B, Li W and He
Y: DNA methylation and hypertension: Emerging evidence and
challenges. Brief Funct Genomics. 15:460–469. 2016.PubMed/NCBI View Article : Google Scholar
|