1. Introduction
The current COVID-19 pandemic poses a substantial
challenge for the entire medical community globally, and has
spurred numerous studies to improve our understanding of the immune
system. COVID-19, which can be detected by PCR and antibody titres
has resulted in a widespread increase in the number of infected
individuals, as well >5 million deaths, globally, and repeated
lockdown measures to prevent or reduce the severity of outbreaks.
This pandemic has highlighted a need to revisit some of the dogmas
of immunology. One such dogma states that the memory of past
infections is formed by T and B memory cells only. Several research
articles have shown the presence of memory in innate nonspecific
immunity, and this may provide a challenge to the prevailing point
of view (1,2). Conversely, the substantial amount of
literature concerning gene regulation by RNA makes it possible to
formulate a mechanism of human antiviral defence from a novel
angle: Every cell in the human body with a full complement of
chromosomes potentially has its own antiviral protection mechanism
based on RNA interference.
To prove this point, the following sections of this
review article describe the activity of RNA-guided gene regulation,
and the role of interferons and the central immune system in viral
invasion.
2. RNA-guided antiviral protection
To understand the mechanism of RNA-guided
protection, it is necessary to start with unicellular organisms, as
development of reliable mechanisms for countering viruses, the
evolutionary acquisition of viral infection immunity allowed for
the generation of multicellular organisms. It should be noted that
this transition took ~2 billion years of evolution (the first
multicellular organisms appeared 1.7 billion years ago (3,4), and
the first cells already existed 3.7-4.2 billion years ago (5,6), as a
result of which, multicellular organisms now possess the
intracellular protection systems initially developed by bacteria
and archaea, CRISPR-Cas. This system is adaptive and accumulates
memory about previous encounters with viruses. This memory is
stored in special DNA regions, which appear after a challenge with
a foreign genome of the virus or plasmid in CRISPR arrays-short
palindromic repeats, regularly arranged in groups (7). The virus entering a bacterial cell is
detected by the CRISPR-associated (Cas) proteins, a type of
nuclease that acts like a pair of scissors, to cut-out viral
nucleic acid sequences. CRISPR-Cas is a real-time adaptive immune
system with a memory of previous encounters with foreign viruses,
that is stored in the unique spacer sequences obtained from the
viral and plasmid genomes, and inserted into the CRISPR arrays
(7). Spacer transcripts, together
with other regions of the surrounding repeats, are used as guide
RNAs to recognize related sequences in foreign genomes, and thus
provide specific cleavage sites for Cas nucleases (8). This provides a simple and effective
protective mechanism against viruses, whereby a part of the viral
DNA (spacer) is inserted into the cell genome, and upon repeat
infection with the same virus, a copy of this spacer, in the form
of a small RNA, directs the nuclease enzymes to destroy the foreign
genome (Fig. 1).
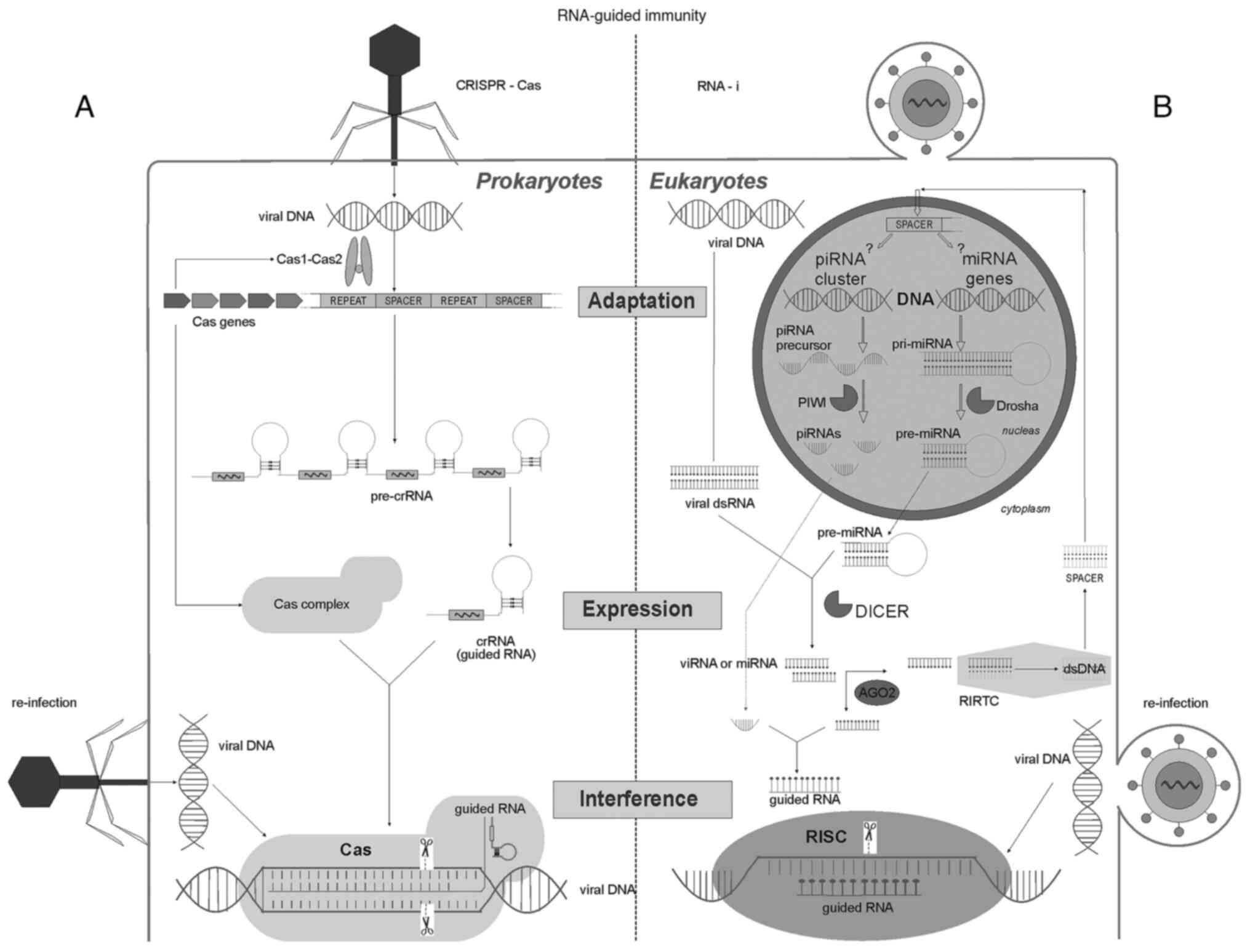 | Figure 1Key stages of RNA-guided
immunity-adaptation, expression and interference of the antiviral
defence systems of protozoa and humans. (A) The left side shows the
immune system of bacteria and archaea, including the CRISPR-Cas
system. During adaptation, phage DNA is cleaved by Cas1/Cas2
enzymes into spacers and integrated into a special CRISPR locus. A
specific adaptive memory is formed based on a previous infection.
Upon re-infection with the same virus, RNA transcripts from these
spacers are directed to complexes formed by other Cas nucleases
(the CRISPR-Cas9 complex is shown here), where they serve as a
template for enzymes that are used to cut similar nucleotide
sequences in the viral genome. (B) Stages of RNA interference in
human cells. When cells are infected with RNA or DNA viruses (DNA
polymerases of viruses generate RNA sequences), long
double-stranded RNAs are formed in the cytoplasm, which are cut by
DICER nucleases into short double-stranded viRNAs. Thereafter, the
AGO2 enzyme unwinds the viRNA strands and loads them either into an
RISC or into an RIRTC. In the RISC, the viral RNAs are cleaved, and
in the RIRTC a spacer is synthesized-a double-stranded DNA molecule
from the RNA template. It is hypothesized that this involves a
reverse transcription complex similar to the telomerase TERT, in
which the AGO2 protein directs the RNA sequence to form new DNA
telomeres (62). The spacer
integrates into the DNA of the cell, forming a specific memory of a
past infection. It remains unclear where exactly in human
chromosomes spacer sequences are formed in piRNA clusters or at
miRNA loci. When viruses enter this cell again, the spacer is
transcribed into a pri-miRNA, which, under the action of DROSHA is
shortened to a pre-miRNA and shuttled to the cytoplasm after
DICER-mediated cutting, where it is loaded by AGO2 into a RISC, and
serves as a template for cleaving viral RNAs. piRNA,
piwi-interacting RNA; miRNA, microRNA; dsRNA, double stranded RNA;
dsDNA, double stranded DNA; viRNA, viral short РНК; PIWI, P-element
Induced WImpy testis; DROSHA, Drosha Ribonuclease III; DICER,
endoribonuclease Dicer; AGO-2, Argonaute RISC Catalytic Component
2; RISC, RNA induced silencing complex; RIRTC, RNA induced reverse
transcription complex; crRNA, CRISPR RNA; Cas, CRISPR
associated. |
In multicellular organisms, there is a similar
mechanism for regulating the activity of various genes termed RNA
interference. Here, it is hypothesized that RNA interference is an
essential component of the adaptive immune system in multicellular
organisms, including humans. This system was first discovered in
1998 in Caenorhabditis elegans by Fire et al
(9), who was subsequently awarded
the Nobel Prize in Physiology or Medicine (2006). The mechanism of
interference has already been studied in detail-it is widely used
in experimental biology for knocking down certain genes, and in
medicine for treatment of certain types of cancer (10-12).
The interference itself consists of halting the
translation of viral genes by cutting or modifying them (13,14).
For this, the cells have a special complex of nuclease enzymes,
which are controlled by small RNAs-the same transcript spacers.
Insertion of the spacer into the DNA of the cell itself is the
final ‘vaccination’ stage of the target cell after viral invasion.
When the virus enters the cell again, the small RNAs are
synthesized and loaded into the nuclease complex to direct cutting
of the foreign genome (Fig. 1).
Thus, there is a complete analogy between these two systems of
RNA-the guided antiviral immunity of cells by RNA. At present, it
is unclear how certain regions of the viral material are
incorporated into the cell's DNA. However, the very existence of
such mechanisms has been described in studies on retrotransposons
and pseudogenes (15,16), where intracellular reverse
transcriptase converts cytoplasmic RNA and transcribes
retroelements into complementary DNA. Human telomerase, which is
essentially a reverse transcriptase, actively uses proteins
involved in RNA interference to synthesize telomeres with their
subsequent integration into the DNA of chromosomes. It should be
noted that retroelements make up a half of the human DNA (17,18),
and it is logical to assume that a significant part of the human
genome has encoded some DNA fragments of previously encountered
viral genomes-those very spacers (19). Moreover, this assumption has already
been proven by the presence of SARS-CoV-2 spacers in DNA of
infected people (20).
The role of RNA interference has been proven to
occur in several infections caused by the human respiratory
syncytial virus (21), human
immunodeficiency virus type 1(22),
hepatitis B virus (23) hepatitis C
virus (24,25), influenza virus (26) and coronavirus SARS-CoV-1(27). The presence of such spacers
effectively prevents viral infection in mammals as well. It is
known that the spacers in the DNA of target cells inhibit the
reproduction of viruses (28,29).
Recent work on the suppression of SARS-CoV-2 viral reproduction
using specific siRNAs (30) leaves
no doubt regarding the validity of this hypothesis.
The data mentioned above directly indicate the
ability of cells themselves to resist viral invasion. Every cell in
the human body that contains a full complement of chromosomes may
potentially preserve an ancient system for counteracting viruses
using small RNAs. Moreover, this protection is adaptive and forms a
type of full-fledged intracellular immune memory.
3. The role of the interferon system
The interferon system is another important mechanism
for cellular protection, which is based on production of special
proteins preventing further infection (31,32).
It is hypothesized that this additional system is required for
highly organized organisms to respond quickly to a viral invasion.
The increase in the number of densely grouped cells of the same
type facilitates the spread of viruses-having multiplied in one
sensitive cell, virions can easily infect the nearby cells.
Accordingly, the innate RNA-guided protection may not be able to
cope with the high viral load. To prevent this possibility, an
early warning system exists and uses interferons as ‘alarms’.
All nucleated cells have receptors for interferon I
(33). Following activation of this
receptor upon ligand binding, the expression of several genes is
upregulated placing the cell in a state of ‘alarm’, halting almost
all protein synthesis, and endo- and exocytosis are inhibited,
which prevents both entry and exit of viral particles (34,35).
Interferon itself is produced by cells infected with viruses
(36). Each human cell possesses a
substantial profile of receptors that recognize certain pathogenic
motifs. In the case of viral infections, there are special
cytoplasmic RLR receptors that recognize viral double-stranded RNA
(37). Their activation triggers a
cascade of intracellular mechanisms, ultimately resulting in the
synthesis of interferons and pro-inflammatory cytokines (Fig. 2).
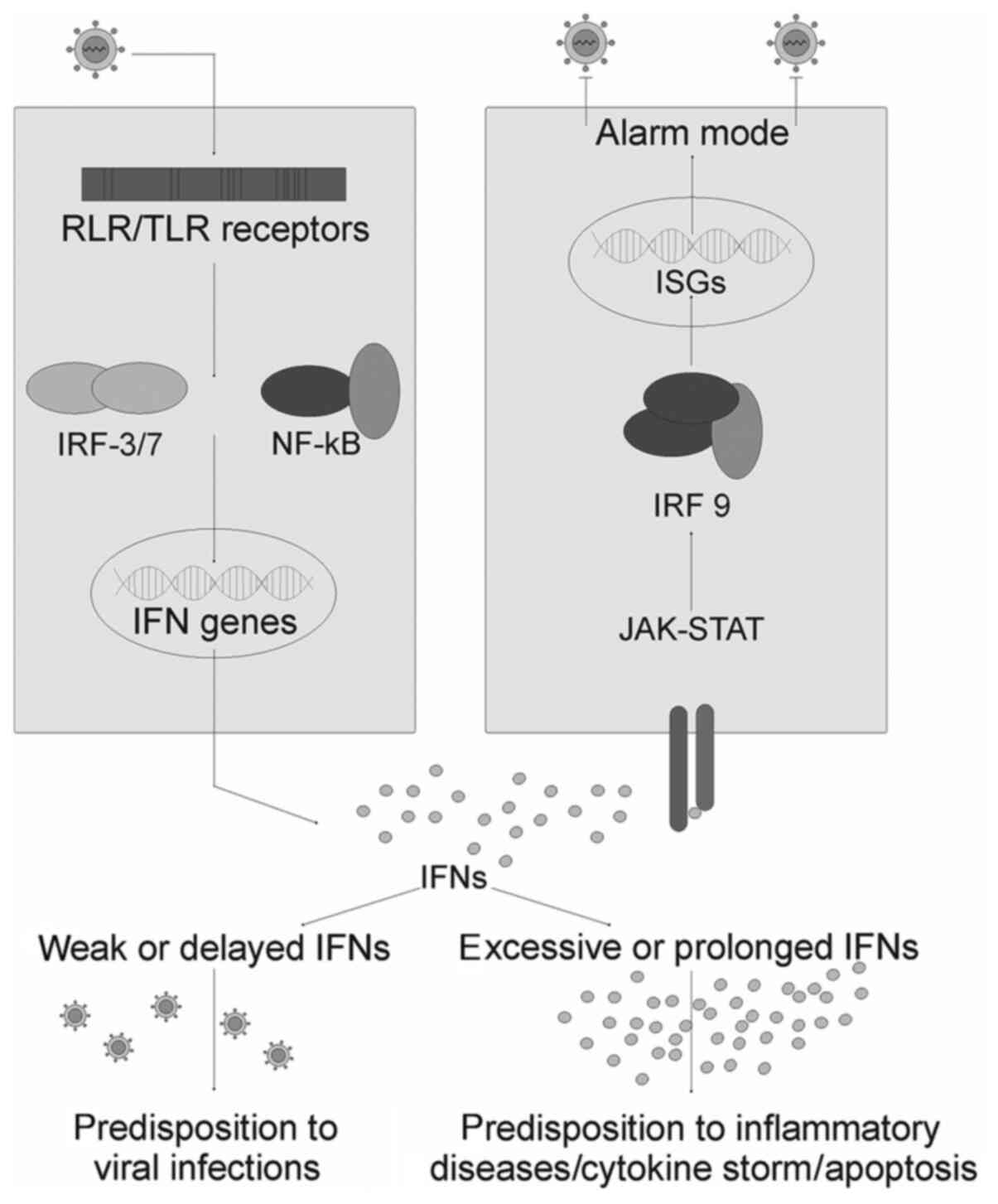 | Figure 2The Interferon alarm system: Adapted
from Onomoto et al (40).
Activation of the cytoplasmic receptors RIG-1 and MDA5 by viral
dsRNAs, as well as endosomal receptors TLR 7/8 by ssRNAs, lead to
phosphorylation of IRF3 and 7, and the pro-inflammatory factor
NF-kB. These transcription factors induce the expression of
interferon genes and inflammatory cytokines upon nuclear
translocation. Newly synthesized interferons bind to their
receptors on neighbouring cells and induce the expression of ISGs
through the Jak/STAT pathway. Under the influence of the
restriction factors encoded by these genes, the cell goes into an
‘alarm’ mode, and in this state, they halt protein synthesis,
including that of viral proteins, and marks all newly synthesized
proteins for degradation. Vesicular transport is slowed, which
leads to inhibition of virion assembly and release. Excessive
production of IFN induces cell apoptosis and is associated with the
development of a cytokine storm. During a cytokine storm, there is
the risk of the formation of autoimmune attacks. The lack of IFN
production does not prepare cells for viral invasion and, as a
result, exacerbates an infectious disease. RIG-1, retinoic acid
inducible gene-1; MDA5, melanoma differentiation associated
protein-5; TLR, toll-like receptor; IFN, interferon; IRF, IFN
regulatory factor; NF-κB, nuclear factor-κB; JAK, Janus kinase;
STAT, Signal Transducer and Activator of Transcription proteins;
ISG, IFN stimulated gene; ssRNA, single stranded RNA. |
Interferons themselves, in turn, regulate the
functions of >2,000 interferon stimulated genes (ISGs); there is
a database of these ISGs, which highlights their effect on
metabolism (interferome.org/interferome/home.jspx). In the context
of this review, only a few points will be highlighted. First, the
epithelial cells, which are the first to meet various pathogens,
possess receptors for interferon III. This emphasizes the need for
prompt and coordinated activity during viral invasions of these
barrier forming cells. Second, prolonged contact with interferons
induces the cells to initiate apoptosis. Finally, several enzymes
are produced by interferon blocking nucleases, which are required
for the full function of the RNA-guided system (38). Thus, an additional safety interferon
system is able to interfere with the RNA-controlled protection and,
moreover, induce cell death. This particular kind of antagonism in
the interferon and RNA-guided systems in antiviral protection has
been studied in several studies, and is extensively reviewed here
(39). The first cell lines that
exhibit viral entry are well-differentiated surface epithelial or
endothelial cells. When the virus enters these cells, DICER
nucleases begin immediately cutting the viral genome, the TLR and
RLR receptors, and in turn, trigger a cascade of interferon
synthesis (37,38). This is the first non-specific phase
of countering the viral invasion. The following steps are dependent
on the viral load; if it is small, then the infected cells cope on
their own and ‘warn’ the neighbouring intact cells about the
infection, assisting the latter in reducing their susceptibility to
viral infection. The infection is interrupted without a pronounced
clinical effect. As the load increases, the infected cells undergo
apoptosis, and the neighbouring cells, under the influence of
increasing doses of interferon, halt protein synthesis, exo- and
endocytosis, and the activity of almost all enzymes; these cells
metaphorically freeze (33,34,40).
Under the influence of interferons and other cytokines, the central
immune system is primed; the first antibodies and symptoms of
inflammation appear. The infected cells carrying viral antigens on
their surface to attract T-killers, these cells are opsonized with
antibodies, and the complement system is activated (41,42).
The classical signs of an infection attributed to each specific
virus manifest. Note, while the viral proteins are presented at the
plasma membrane of infected cells, the immune system is in an
activated state. Moreover, the cells presenting these viral
antigens are eventually destroyed.
Concurrently, in the basal layers of these tissues,
where actively proliferating and unipotent cells are located, when
the viruses and/or spacers themselves enter from the infected
cells, an adaptive intracellular antiviral protection begins to
form. As mentioned above, poorly differentiated cells, under the
influence of interferons, do not halt RNA and protein synthesis
(43). Accordingly, there remains a
place for full-fledged RNA interference, which is not disturbed by
blocking interferon signals. Therefore, immune memory is formed in
unipotent progenitor cells when a virus or spacer RNA penetrates
them through extracellular vesicles or nucleoprotein complexes with
AGO 2 (44,45). After maturation, these cells will
possess specific antiviral memory, providing them a powerful tool
to effectively eliminate any further infections with the same virus
(28,29). The newly formed layer of cells
readily copes with the residual viral load and no longer carries
the antigens of the virus; the central immune system returns to a
normal mode once stimulation is gradually decreased. This change in
endothelial and epithelial cells usually takes several days, which
is the time required for formation of specific local antiviral
memory.
Thus, it is hypothesized that every cell of the
human body with a full complement of chromosomes potentially
possesses this form of antiviral protection; and it is the RNA
interference response that initially determines the course of a
viral infection.
4. Discussion
The CRISPR-Cas system has proven to be incredibly
effective in combating mobile genetic elements, and thus has
retained its role in multicellular organisms, having slightly
changed, taking into account the presence of a nuclear membrane and
terminal chromosomes. The primary goal of the central immune system
possessed by higher order animals, based on T and B cells, is to
maintain the integrity of the organism and counter foreign
organisms. To accomplish this, the immune system possesses
phagocytic, regulatory, antigen-presenting and killer cells, as
well as a complement system, and of course, various antibodies. The
complement system is a group of proteins, which, after being
activated, promote membranolytic cascades that destroy target
cells. The classical pathway is activated when the C1q complement
protein binds to the Fc-fragment of the antibody (this is the
invariable part of all antibodies in the body) attached to the
antigen (42). It should be
emphasized that antibodies do not destroy a foreign object by
themselves, even in high concentrations, they perform a diagnostic
and guiding role only. In a multi-trillion cell human body, the
antibodies help immune cells to identify foreign agents by binding
to specific foreign antigens (opsonization) (46). Namely, the Fc-fragment of antibodies
is a ‘black mark’ for immune cells and complement, which perceive
it as a signal to destroy this object (41). In the case of bacteria, protozoa,
fungi and helminths, this is a working strategy by which they are
destroyed through the use of antibodies. This also works to counter
mutant cells, including tumour cells, where antibodies bind to
cancer antigens, and such cells are also discarded. But what
happens in the case of a viral infection? A priori, viruses
multiply inside cells only, and when viral antigens present on the
cell membrane are detected by antibodies, these infected cells are
also destroyed. For the entire organism, this is incredibly
beneficial; when something foreign is identified, even inside their
own cells, the foreign body and/or affected cell is destroyed,
sacrificing a part for the greater good of the whole. Note that
these infected cells have no other fate-they are all destroyed.
Until the viruses have penetrated the cells, they
are just a set of nucleic acids and proteins; they do not multiply,
and do not possess any pathological effects on the body. When
viruses bind with antibodies, they of course lose their ability to
target cells, making it difficult for them to bind to cellular
receptors. This is the rationale behind the vaccination strategy.
However, let us consider a further course of events; the resulting
virus-antibody complex must be destroyed by immune cells carrying
the receptor for the Fc fragment. In several cases, this happens,
but if phagocytosis is not effective, and the virus remains viable
inside these cells, the infection intensifies further, since the
virus has infected cells that do not carry receptors to it, and
this phenomenon is called antibody-dependent enhancement (ADE)
(47-49).
In the case of ADE, the virus infects susceptible cells through the
corresponding receptor and penetrates monocytes/macrophages,
granulocytes, platelets, mast cells and several other host cells by
interacting with receptors for the Fc fragment or complement
(50). There are several examples
of ADE caused by α and β coronaviruses (51,52).
The current clinical data on the course of COVID-19 indicate
involvement of antibodies in the enhancement of clinical
manifestations of the disease. The most severe patients appear to
possess the highest antibody titres (53). A specific symptom of COVID-19,
coagulopathy, clearly indicates complement hyperactivity (54); and this process of cell destruction
does not stop as long as they carry viral antigens on their
surface. Only as a result of RNA-guided nucleases does the cell
achieve clearance of the viral genome, and, accordingly, the viral
proteins. Further activation of the central immune system stops,
antibody titres fall, and the affected individual recovers.
While discussing human antiviral immunity, one
cannot fail to mention the mechanisms of protection observed in
early childhood. As recent data have shown, along with antibodies,
breast milk contains ~1,400 different types of microRNAs (55). Given the ability of each of these
molecules to alter the activity of an average of 15-20 genes, there
is a tremendous opportunity to suppress or enhance the activity of
genes in infants. It has been shown that these microRNAs, after
absorption, are present in the bloodstream and all tissues of the
body, including the brain (56,57).
Thus, with regards to antiviral immunity, it is necessary to
emphasize the presence of such a transfer of protection through
milk to a child.
The role of vitamin A in prevention and treatment of
viral diseases should also be mentioned. There is a positive
association between vitamin A administration and the management of
measles. During a measles infection, it has been shown that vitamin
A deficiency clearly correlates with the severity of the course,
and timely treatment of measles with two doses of retinol (200,000
IU) dramatically reduces both morbidity and mortality rates
(58-60).
We hypothesize that there may be a possibility of wider use of this
simple and cheap drug for other viral diseases, including COVID-19.
This vitamin is undoubtedly important for the synthesis of RLR
receptors, but we were interested in the fact that the DICER
nuclease and the RLR receptor have a similar structure, they both
possess a DECH box domain that recognizes viral RNA (61). DICER nuclease is a key player in
RNA-driven gene regulation, and further research is required on the
possible relationship between Vitamin A and RNA interference.
From the above concept of cellular adaptive
antiviral immunity, several assumptions can follow regarding
interpretation of clinical indicators during the course of a viral
infection. Taking COVID-19 as an example, they are as follows: i)
The presence or absence of specific antibodies to SARS-CoV-2 is not
a predictor of the disease. The presence of antibodies in the blood
reflects only the fact that a person has been in contact with the
virus. Lack of antibodies does not mean any contact, and people
with high titres of specific antibodies are not protected from
re-infection with SARS-Co2. ii) PCR tests for those who have had
COVID may return false positives if the swab sample is taken from
the point of the initial spread of the virus (usually from the
nasopharynx). We suggest that a negative PCR result for COVID in
the blood plasma and urine may be a more reliable indicator of a
lack on infection, even when a swab sample from the nasopharynx
returns a positive result.
5. Conclusions
Here, a novel concept is proposed-the antiviral
protection of all organisms based on intracellular RNA-guided
mechanisms. A simple and effective defence against viruses is
contained as part of the virus's DNA (spacer) in the chromosomes.
Following a reinfection, the RNA transcribes the incorporated
spacer and directs nuclease enzymes to cut the viral genome. This
is a real-time adaptive immune response potentially possessed by
every cell that contains a nucleus. Thus, antiviral immunity may
not only be mediated by neutralizing antibodies and memory B- and
T-cells, but also through the incorporation of specific spacers
into the DNA of the cells genome.
Acknowledgements
We would like to thank Dr S. Muratkhodjaeva
(Institute of Immunology and Human Genomics, Academy of Sciences of
Uzbekistan, Tashkent, Uzbekistan) for her skilful drawings.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
TA and JM both wrote and revised the manuscript.
Both authors have read and approved the final manusript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Reimer-Michalski EM and Conrath U: Innate
immune memory in plants. Semin Immunol. 28:319–327. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Netea MG, Quintin J and van der Meer JW:
Trained immunity: A memory for innate host defense. Cell Host
Microbe. 9:355–361. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rosing MT: 13C-Depleted carbon
microparticles in >3700-Ma sea-floor sedimentary rocks from west
greenland. Science. 283:674–676. 1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Dodd MS, Papineau D, Grenne T, Slack JF,
Rittner M, Pirajno F, O'Neil J and Little CT: Evidence for early
life in Earth's oldest hydrothermal vent precipitates. Nature.
543:60–64. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Knoll AH, Javaux EJ, Hewitt D and Cohen P:
Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond
B Biol Sci. 361:1023–1038. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Fedonkin MA: The origin of the Metazoa in
the light of the Proterozoic fossil record. Paleontol Res. 7:9–41.
2003.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Barrangou R: The roles of CRISPR-Cas
systems in adaptive immunity and beyond. Curr Opin Immunol.
32:36–41. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Koonin EV and Makarova KS: Mobile genetic
elements and evolution of CRISPR-Cas systems: All the way there and
back. Genome Biol Evol. 9:2812–2825. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Fire A, Xu S, Montgomery MK, Kostas SA,
Driver SE and Mello CC: Potent and specific genetic interference by
double-stranded RNA in Caenorhabditis elegans. Nature.
391:806–811. 1998.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Han H: RNA interference to knock down gene
expression. Methods Mol Biol. 1706:293–302. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Abdelrahim M, Safe S, Baker C and
Abudayyeh A: RNAi and cancer: Implications and applications. J RNAi
Gene Silencing. 2:136–145. 2006.PubMed/NCBI
|
|
13
|
Ghildiyal M and Zamore PD: Small silencing
RNAs: An expanding universe. Nat Rev Genet. 10:94–108.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Maillard PV, Ciaudo C, Marchais A, Li Y,
Jay F, Ding SW and Voinnet O: Antiviral RNA interference in
mammalian cells. Science. 342:235–238. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Habibi L and Salmani H: Pivotal impacts of
retrotransposon based invasive RNAs on evolution. Front Microbiol.
8(1957)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wei W, Morrish TA, Alisch RS and Moran JV:
A transient assay reveals that cultured human cells can accommodate
multiple LINE-1 retrotransposition events. Anal Biochem.
284:435–438. 2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Wicker T, Sabot F, Hua-Van A, Bennetzen
JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O,
et al: A unified classification system for eukaryotic transposable
elements. Nat Rev Genet. 8:973–982. 2007.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Javdat M and Tamara A: RNA interference:
Antiviral defense mechanism and immune memory. Adv Appl Physiol.
5:24–29. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang L, Richards A, Barrasa MI, Hughes
SH, Young RA and Jaenisch R: Reverse-transcribed SARS-CoV-2 RNA can
integrate into the genome of cultured human cells and can be
expressed in patient-derived tissues. Proc Natl Acad Sci USA.
118(e2105968118)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bitko V and Barik S: Phenotypic silencing
of cytoplasmic genes using sequence-specific double-stranded short
interfering RNA and its application in the reverse genetics of wild
type negative-strand RNA viruses. BMC Microbiol.
1(34)2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Coburn GA and Cullen BR: Potent and
specific inhibition of human immunodeficiency virus type 1
replication by RNA interference. J Virol. 76:9225–9231.
2002.PubMed/NCBI View Article : Google Scholar
|
|
23
|
McCaffrey AP, Nakai H, Pandey K, Huang Z,
Salazar FH, Xu H, Wieland SF, Marion PL and Kay MA: Inhibition of
hepatitis B virus in mice by RNA interference. Nat Biotechnol.
21:639–644. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Yokota T, Sakamoto N, Enomoto N, Tanabe Y,
Miyagishi M, Maekawa S, Yi L, Kurosaki M, Taira K, Watanabe M and
Mizusawa H: Inhibition of intracellular hepatitis C virus
replication by synthetic and vector-derived small interfering RNAs.
EMBO Rep. 4:602–608. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Krönke J, Kittler R, Buchholz F, Windisch
MP, Pietschmann T, Bartenschlager R and Frese M: Alternative
approaches for efficient inhibition of hepatitis C virus RNA
replication by small interfering RNAs. J Virol. 78:3436–3446.
2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ge Q, McManus MT, Nguyen T, Shen CH, Sharp
PA, Eisen HN and Chen J: RNA interference of influenza virus
production by directly targeting mRNA for degradation and
indirectly inhibiting all viral RNA transcription. Proc Natl Acad
Sci USA. 100:2718–2723. 2003.PubMed/NCBI View Article : Google Scholar
|
|
27
|
He ML, Zheng B, Peng Y, Peiris JS, Poon
LL, Yuen KY, Lin MC, Kung HF and Guan Y: Inhibition of
SARS-associated coronavirus infection and replication by RNA
interference. JAMA. 290:2665–2666. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Fujino K, Horie M, Honda T, Merriman DK
and Tomonaga K: Inhibition of Borna disease virus replication by an
endogenous bornavirus-like element in the ground squirrel genome.
Proc Natl Acad Sci USA. 111:13175–13180. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Honda T and Tomonaga K: Endogenous
non-retroviral RNA virus elements evidence a novel type of
antiviral immunity. Mob Genet Elements. 22(e1165785)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Idris A, Davis A, Supramaniam A, Acharya
D, Kelly G, Tayyar Y, West N, Zhang P, McMillan CLD, Soemardy C, et
al: A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19.
Mol Ther. 29:2219–2226. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Stetson DB and Medzhitov R: Type I
interferons in host defense. Immunity. 25:373–381. 2006.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Levy DE: Whence interferon? Variety in the
production of interferon in response to viral infection. J Exp Med.
195:15–18. 2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
de Weerd NA and Nguyen T: The interferons
and their receptors-distribution and regulation. Immunol Cell Biol.
90:483–491. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Houglum JE: Interferon: Mechanisms of
action and clinical value. Clin Pharm. 2:20–28. 1983.PubMed/NCBI
|
|
35
|
McNab F, Mayer-Barber K, Sher A, Wack A
and O'Garra A: Type I interferons in infectious disease. Nat Rev
Immunol. 15:87–103. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Katze MG, He Y and Gale M Jr: Viruses and
interferon: A fight for supremacy. Nat Rev Immunol. 2:675–687.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
37
|
Wu J and Chen ZJ: Innate immune sensing
and signaling of cytosolic nucleic acids. Annu Rev Immunol.
32:461–488. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
van der Veen AG, Maillard PV, Schmidt JM,
Lee SA, Deddouche-Grass S, Borg A, Kjær S, Snijders AP and Reis e
Sousa C: The RIG-I-like receptor LGP2 inhibits Dicer-dependent
processing of long double-stranded RNA and blocks RNA interference
in mammalian cells. EMBO J. 37(e97479)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Maillard PV, van der Veen AG, Poirier EZ
and Reis e Sousa C: Slicing and dicing viruses: Antiviral RNA
interference in mammals. EMBO J. 38(e100941)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Onomoto K, Onoguchi K and Yoneyama M:
Regulation of RIG-I-like receptor-mediated signaling: Interaction
between host and viral factors. Cell Mol Immunol. 18:539–555.
2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Flesch BK and Neppert J: Functions of the
Fc receptors for immunoglobulin G. J Clin Lab Anal. 14:141–156.
2000.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Duncan AR and Winter G: The binding site
for C1q on IgG. Nature. 332:738–740. 1988.PubMed/NCBI View
Article : Google Scholar
|
|
43
|
Eggenberger J, Blanco-Melo D, Panis M,
Brennand KJ and tenOever BR: Type I interferon response impairs
differentiation potential of pluripotent stem cells. Proc Natl Acad
Sci. 116:1384–1393. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Holtzman J and Lee H: Emerging role of
extracellular vesicles in the respiratory system. Exp Mol Med.
52:887–895. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Geekiyanage H, Rayatpisheh S, Wohlschlegel
JA, Brown R Jr and Ambros V: Extracellular microRNAs in human
circulation are associated with miRISC complexes that are
accessible to anti-AGO2 antibody and can bind target mimic
oligonucleotides. Proc Natl Acad Sci USA. 117:24213–24223.
2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Mevorach D: Opsonization of apoptotic
cells. Implications for uptake and autoimmunity. Ann N Y Acad Sci.
926:226–235. 2000.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hawkes RA: Enhancement of the infectivity
of arboviruses by specific antisera produced in domestic fowls.
Aust J Exp Biol Med Sci. 42:465–482. 1964.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Smatti MK, Al Thani AA and Yassine HM:
Viral-induced enhanced disease illness. Front Microbiol.
9(2991)2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Tirado SM and Yoon KJ: Antibody-dependent
enhancement of virus infection and disease. Viral Immunol.
16:69–86. 2003.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Khandia R, Munjal A, Dhama K, Karthik K,
Tiwari R, Malik YS, Singh RK and Chaicumpa W: Modulation of
dengue/zika virus pathogenicity by antibody-dependent enhancement
and strategies to protect against enhancement in zika virus
infection. Front Immunol. 9(597)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Wan Y, Shang J, Sun S, Tai W, Chen J, Geng
Q, He L, Chen Y, Wu J, Shi Z, et al: Molecular mechanism for
antibody-dependent enhancement of coronavirus entry. J Virol.
94:e02015–19. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yip MS, Cheung CY, Li PH, Bruzzone R,
Peiris JSM and Jaume M: Investigation of antibody-dependent
enhancement (ADE) of SARS coronavirus infection and its role in
pathogenesis of SARS. BMC Proc. 5 (Suppl 1)(P80)2011.
|
|
53
|
Chen X, Pan Z, Yue S, Yu F, Zhang J, Yang
Y, Li R, Liu B, Yang X, Gao L, et al: Disease severity dictates
SARS-CoV-2-specific neutralizing antibody responses in COVID-19.
Signal Transduct Target Ther. 5(180)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lo MW, Kemper C and Woodruff TM: COVID-19:
Complement, coagulation, and collateral damage. J Immunol.
205:1488–1495. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Benmoussa A and Provost P: Milk MicroRNAs
in health and disease. Compr Rev Food Sci Food Saf. 18:703–722.
2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Manca S, Upadhyaya B, Mutai E, Desaulniers
AT, Cederberg RA, White BR and Zempleni J: Milk exosomes are
bioavailable and distinct microRNA cargos have unique tissue
distribution patterns. Sci Rep. 8(11321)2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zempleni J: Milk exosomes: Beyond dietary
microRNAs. Genes Nutr. 12(12)2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Soye KJ, Trottier C, Richardson CD, Ward
BJ and Miller WH Jr: RIG-I is required for the inhibition of
measles virus by retinoids. PLoS One. 6(e22323)2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
D'Souza RM and D'Souza R: Vitamin A for
the treatment of children with measles-a systematic review. J Trop
Pediatr. 48:323–327. 2002.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Huiming Y, Chaomin W and Meng M: Vitamin A
for treating measles in children. Cochrane Database Syst Rev.
2005(CD001479)2005.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Paro S, Imler JL and Meignin C: Sensing
viral RNAs by Dicer/RIG-I like ATPases across species. Curr Opin
Immunol. 32:106–113. 2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Laudadio I, Orso F, Azzalin G, Calabrò C,
Berardinelli F, Coluzzi E, Gioiosa S, Taverna D, Sgura A, Carissimi
C and Fulci V: AGO2 promotes telomerase activity and interaction
between the telomerase components TERT and TERC. EMBO Rep.
20(e45969)2019.PubMed/NCBI View Article : Google Scholar
|
















