Introduction
Periodontitis is one of the most common inflammatory
diseases affecting the tissues supporting teeth and it is
characterized by progressive resorption of the alveolar bone
(1-3).
Periodontitis is also one of the primary causes of loss of teeth in
adults, and it can affect an individual's quality of life if left
untreated (4-6).
Periodontitis is initiated by bacterial plaque on the tooth surface
and can be caused by multiple factors, such as an imbalance of
periodontal pathogens, host immunity and other environmental, local
and systemic factors (7,8). The majority of studies on this topic
have emphasized on the role of genetic factors in disease
pathogenesis, and on their ability to influence the unique reaction
that characterizes the susceptibility of each individual (1,5).
Approximately 50% of the clinical severity of periodontitis may be
associated with host genetics (3,8).
However, the genetic effects may differ among different ethnicities
due to the diversity of the subjects in a population (3,7).
Therefore, understanding of the pathophysiology of this disease
from a genetic standpoint is essential for its early detection and
diagnosis.
Over the past decade, considerable efforts have been
made to identify the influence of various genetic factors, such as
genes that code for IL-1(9), TNF-α
(10), IL-10(11), IL-4(12), IL-4 receptor-α (13), Fc γ receptor (14), CD-14(15) and the vitamin D receptor (VDR). The
genetic predispositions associated with these genes are considered
to affect the severity of different diseases (16,17).
VDR, in particular, is a promising candidate in periodontitis since
it affects both bone metabolism and immunological function
(4,18). This receptor is present in various
cell types and can act as a transcriptional regulator (2). It is located on chromosome 12 q and
has 14 exons, 6 of which are located on the 5'region that is not
translated (1a-1f) (7,18). The untranslated 3'region of the
VDR gene comprises a polymorphism cluster in TaqI, ApaI and
BsmI (19,20). If the function of VDR is affected by
the polymorphisms of the VDR gene, the contribution of these
variants is considered critical for the pathogenesis of systemic
diseases related to bone tissues, such as periodontal disease
(19,20).
Several studies have attempted to elucidate an
association between the presence of VDR gene polymorphisms
and the pathogenesis of certain diseases. A series of characterized
VDR gene polymorphisms, including those for the FokI,
ApaI, TaqI and BsmI genes have been previously
reported (2,3,5). To
date, an association has been found between the susceptibility to
periodontal disease and a certain number of single-candidate gene
polymorphisms (21). Other studies
have also shown a connection between periodontal disease and
vitamin D levels (2,22,23).
Although extensive evidence has been obtained, the majority of the
studies were only focused on the investigation of the association
between periodontitis and the VDR gene polymorphism
(2,21-23).
In the present study, DNA sequencing and restricted fragment length
polymorphism (RFLP)-PCR analysis were used to further examine the
changes in nucleotide arrangement of the VDR gene. The
identification of genetic variants linked to the increased
susceptibility to certain diseases may be key to the development
and advancement of preventive medicine. Therefore, the aim of the
present study was to investigate possible correlations between the
VDR gene polymorphisms, specifically in exon 9 (TaqI), and
the severity of periodontitis. This study was a case-controlled
study and the subjects were recruited from a Makassar-based
population in Indonesia.
Materials and methods
Study population and data
collection
The present study was a case-controlled study
including patients who were enrolled at the Periodontology
Department of the Dental Hospital, Hasanuddin University,
Indonesia. The study protocol was approved by the Ethics Committee
for Biomedical Research in Humans, Faculty of Medicine, Hasanuddin
University (approval no. 0189/H.04.8.4.5.31/PP36-KOMETIK/2010,
approval date 04/06/2010). The current study was performed in
accordance with the Declaration of Helsinki (24). In total, 162 individuals were
recruited, and they were divided into two groups: Case group,
patients diagnosed with periodontitis (14 males and 67 females) and
the control group, healthy patients without periodontitis (38 males
and 43 females). The median age of the case group was 38 years (age
range 25-60) and the median age of the control group was 34 years
(age range 22-60). Determination of the sample size was based on
the Lemeshow formula for the case-controlled study design. The
actual odd ratio (OR) estimate was ~2. Consideration of OR=2 has
been used in previous research studies; an OR of 2 obtained in
these previous studies suggested that samples with VDR gene
polymorphisms were 2x more likely to suffer from chronic
periodontitis than those without VDR gene polymorphisms. Thus we
used this as minimum standard to calculate our sample size
(25,26). Since the number of populations was
infinite (unknown) P2*=0.5 was used with a value of 0.05. The
anticipated rate was 10% and the power of the test was 0.842.
The diagnostic criteria for the case group included
patients diagnosed with periodontitis based on the New
Classification of Periodontal Disease (2018) (27). The periodontitis stage in the
present study was divided into three categories as determined
previously (24). It was based on
the clinical attachment loss (CAL) and probing pocket depth (PPD):
Stage I, CAL 1-2 mm and maximum PPD ≤4 mm; stage II, CAL 3-4 mm and
maximum PPD≤5 mm; and stages III and IV, CAL≥5 mm and PPD≥6 mm. The
diagnostic criteria for the control group were healthy patients
without periodontitis, which was defined by the absence of
clinically detectable inflammation in the gingiva, such as an
intact periodontium without a gingival recession and CAL, ‘salmon’
or ‘coral pink’ in color, firm in consistency and firmly attached
to the underlying alveolar bone. The subjects that were suitable
for the present study were included in the case and the control
groups and were provided with the necessary information regarding
the research procedure. They were asked to sign the relevant
informed consent form for their agreement in participating in the
study. The recruitment of the patients was performed for 1 year by
two allocated researchers. The first researcher selected patients
from the medical record and completed the information in the
research form regarding age, sex, ethnicity, occupation, medical
history and history of drugs used. Subsequently, a Professor in the
Department of Periodontology performed the clinical examination for
the diagnosis of periodontal disease and suggested whether or not
each patient was suitable for inclusion in the study.
Inclusion and exclusion criteria
The inclusion criteria were: Indonesian patients,
age range of 25-60 years, and had at least 20 teeth. The patients
who exhibited 2 interproximal sites with CAL≥2 mm, and 6
interproximal sites with PPD≥4 mm were also included. The exclusion
criteria were the following: Patients with systemic diseases, such
as diabetes, tuberculosis, hepatitis or human immunodeficiency
virus, as well as patients with smoking habits or had used
prosthetics. Moreover, patients who were pregnant and/or
breastfeeding were also excluded from the study.
Clinical examination
Patients who met the inclusion criteria received a
comprehensive dental examination and a complete research chart was
recorded. It is well established that important clinical signs of
periodontitis include poor to moderate scores in the oral hygiene
index-simplified (OHI-S), PPD and CAL (27,28),
therefore these clinical signs were used to evaluate their
influence on the incidence of VDR gene polymorphisms. Initially,
the number of caries teeth, the number of edentulous and the OHI-S
score (29) were assessed.
Subsequently, scaling was performed on all subjects using an
ultrasonic scaler. The clinical examination was performed to
evaluate the PPD and the level of CAL. The measurements of PPD were
made at 6 sites on each tooth according to the charting form used
in the Department of Periodontology. The largest value from 6 tooth
surfaces in each tooth was obtained for analysis. CAL was measured
based on the distance from the cementoenamel junction to the base
of the pocket. For analysis, interdental CAL at the site of the
greatest loss was taken.
Laboratory analysis
Venous blood (0.5-1 ml) samples were obtained from
all subjects in both groups for laboratory analysis. The DNA from
the peripheral leukocytes was extracted and purified using the Boom
method (30). The analysis was
performed in the Laboratory of Immunology and Molecular Biology,
Faculty of Medicine, Hasanuddin University (Makassar, Indonesia).
The gene polymorphisms were determined based on endonuclease
restriction in exon 9 of the examined VDR gene using
RFLP-PCR and direct sequencing.
Amplification by PCR
In all subjects, VDR gene polymorphisms were
detected in exon 9 using the following specific primers: Forward,
5'-CTGGGGAGCGGGGAGTATGAAGGA-3' and reverse,
5'-GGGTGGCGGCAGCGGATGTA-3' (https://omim.org/entry/601769). DNA amplification was
conducted for RFLP using the TaqI restriction enzyme. The PCR
amplification was performed using 32 cycles, consisting of
denaturation for 1 min at 94˚C, annealing for 1.5 min at 59˚C, and
extension for 2 min at 72˚C. Following the completion of 32 cycles,
the samples were heated at 72˚C for 7 min. The amplification
products were analyzed by agarose gel electrophoresis.
RFLP-PCR analysis
Following amplification, 5 µl PCR amplification
products and 2 µl loading buffer were mixed and loaded onto 1.5%
agarose gels along with ethidium bromide. The gel was soaked in a
container with Tris borate EDTA buffer, and subsequently,
electrophoresis was performed at a steady voltage of 80 V for 1 h.
The gel was removed and observed under UV light. The fragment bands
observed at different distances from the sample indicated genotypic
differences of the VDR gene polymorphism. The differences
were marked with the letter t (a restriction area is present) or
the letter T (no restriction area is present). The genotypes based
on the bands of the agarose gels were classified as follows: i) TT:
Absence of fragment sizes at 1,398 bp; ii) Tt: Presence of fragment
sizes at (946 + 452 bp) and 1,398 bp and iii) tt: Presence of
fragment sizes at 946 bp and 452 bp.
DNA sequencing
In all obtained samples, direct sequencing was
performed by Macrogen, Inc. Sequencing confirmed the changes in the
nucleotide base arrangement in exon 9 of the VDR gene.NCBI
BLAST was used to analyze the sequencing results.
Statistical analysis
SPSS version 11.5 (SPSS, Inc.) was used for data
analysis. The mean ± SD were calculated for all descriptive
variables. An independent samples t-test was performed to detect
phenotypic differences in subjects with periodontitis. P<0.05
was considered to indicate a statistically significant difference.
Hardy-Weinberg equilibrium was verified manually using a standard
calculation formula (31).
Results
Distribution and characteristics of
the subjects
In total, the data were collected for 162 patients
from the Dental Hospital, Hasanuddin University. The age was known
for all 162 patients. The detailed characteristics of the subjects
are shown in Table I. Based on the
independent samples t-test, it was found that both edentulous and
OHI-S were significantly higher is the case group compared with the
control groups (P<0.05; Table
I). This difference indicated that patients with periodontitis
exhibited worse oral hygiene than healthy patients without
periodontitis.
 | Table IClinicopathological characteristics
of the recruited cohort. |
Table I
Clinicopathological characteristics
of the recruited cohort.
| | Case, n=81 | Control, n=81 | |
|---|
| Parameters | Mean | SD | Mean | SD | P-value |
|---|
| Age, year | 38.9 | 9.24 | 37.61 | 11.82 | 0.443 |
| Height, cm | 155.53 | 6.55 | 157.33 | 7.33 | 0.101 |
| Weight, kg | 55.46 | 6.45 | 56.19 | 9.22 | 0.618 |
| Body mass index,
kg/m2 | 22.47 | 2.9 | 23.18 | 3.4 | 0.936 |
| Edentulous | 2.48 | 2.69 | 1.06 | 1.07 |
<0.001b |
| Caries | 2.17 | 2.3 | 2.04 | 1.69 | 0.673 |
| Oral hygiene
index-simplified | 2.63 | 0.96 | 2.29 | 0.69 | 0.011a |
| Probing pocket
depth, mm | 4.54 | 1.27 | - | - | |
| Stage I (≤4
mm) | 4 | 0 | | | |
| Stage II (≤5
mm) | 5 | 0 | | | |
| Stage III/IV (≥6
mm) | 7.2 | 1.98 | | | |
| Clinical attachment
loss, mm | 3.02 | 1.48 | - | - | |
| Stage I (1 to 2
mm) | 2 | 0 | | | |
| Stage II (3 to 4
mm) | 3.55 | 0.54 | | | |
| Stage III/IV (≥6
mm) | 5.91 | 1.16 | | | |
VDR gene polymorphism
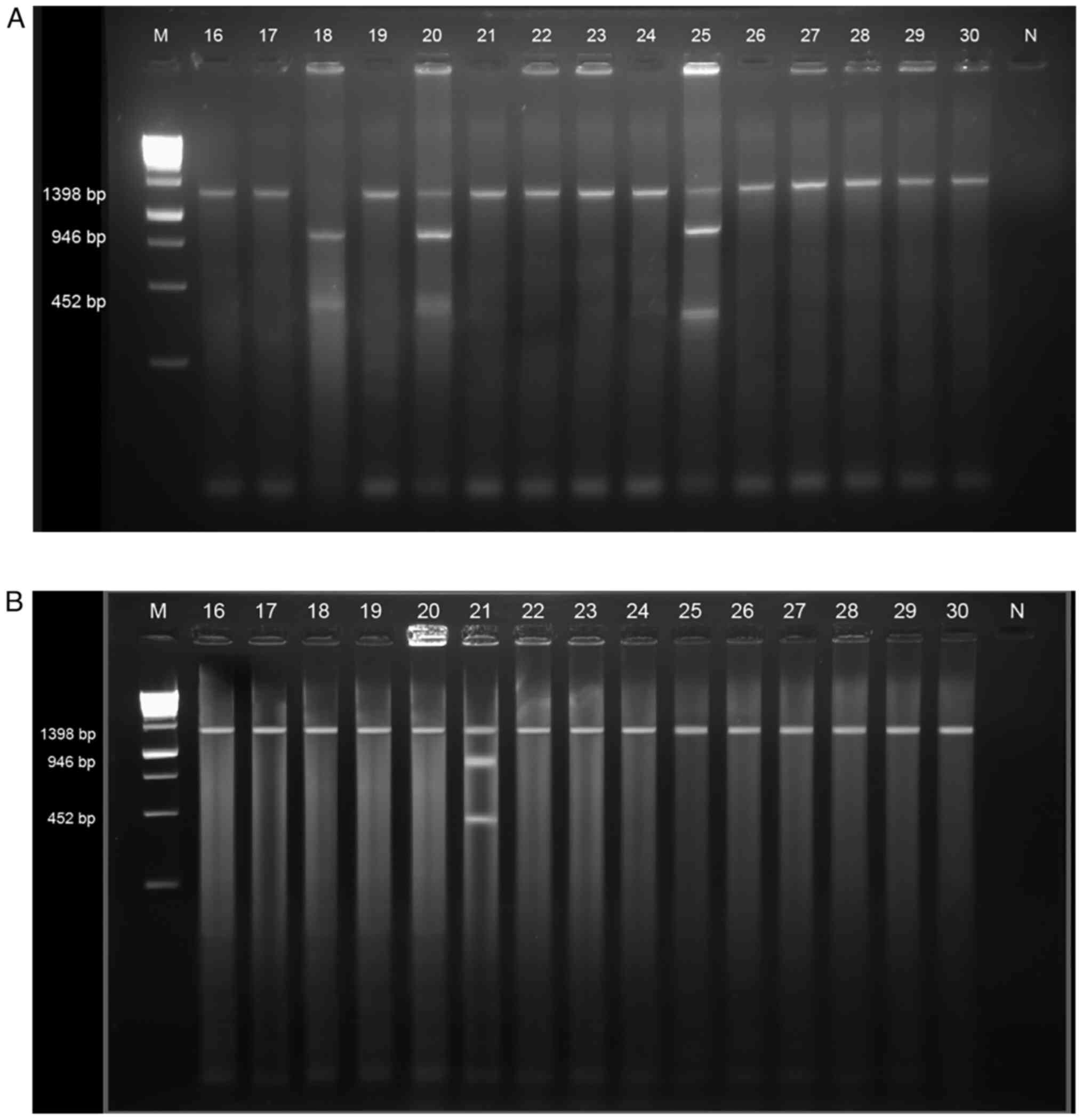
The VDR gene polymorphism consists of the
following three genotypes: TT (1 band), tt (2 bands), and Tt (3
bands) (Fig. 1A and B). The TT genotype (1,398 bp) indicated no
fragment sizes, whereas fragment sizes of 946, 452 and 1,398 bp
were obtained for the Tt genotype and fragment sizes of 946 and 452
bp for the tt genotype. The genotype distribution of the VDR
gene in the case and control groups is highlighted in Table II. It was noted that the case group
exhibited a higher percentage of the TT genotype (86.4%) compared
with either the Tt (12.3%) or the tt (1.2%) genotypes. The control
group had a TT genotype percentage of 98.8%, a Tt genotype
percentage of 1.2%, and an absence of the tt genotype. Both case
and control groups exhibited a genotype distribution within the
Hardy-Weinberg equilibrium [p2+2pq+q2=1;
(0.96)2+2 (0.96) (0.04)+(0.04)2=1.0016].
 | Table IIVitamin D receptor gene genotype
frequency in the case and control groups. |
Table II
Vitamin D receptor gene genotype
frequency in the case and control groups.
| | Case | Control |
|---|
| Genotype | n | % | n | % |
|---|
| TT | 70 | 86.42 | 80 | 98.77 |
| Tt | 10 | 12.35 | 1 | 1.23 |
| tt | 1 | 1.23 | 0 | 0 |
| Total | 81 | 100 | 81 | 100 |
The comparison between the VDR genotype of
the subjects with periodontitis was based on the mean value of
OHI-S, PPD and CAL (Table III).
The results confirmed that the mean of OHI-S was higher for the
patients with periodontitis and the TT genotype (mean score, 2.71)
than that of the patients with periodontitis and the Tt/tt genotype
(mean score, 2.12). However, the difference was not statistically
significant (P>0.05). Both the PPD status and CAL were higher
for the Tt/tt genotype (PPD mean score, 5.14; CAL mean score, 4.41)
compared with those of the TT genotype (PPD mean score, 3.52; CAL
mean score, 2.80). Both of these parameters indicated significant
differences, with P-values of 0.006 and 0.001,
respectively.
 | Table IIIComparison of the Vitamin D receptor
genotype in patients with periodontitis based on OHI-S, PPD and
CAL. |
Table III
Comparison of the Vitamin D receptor
genotype in patients with periodontitis based on OHI-S, PPD and
CAL.
| | TT genotype,
n=70 | Tt/tt genotype,
n=11 | |
|---|
| Parameter | Mean | SD | Mean | SD | P-value |
|---|
| OHI-S | 2.71 | 0.95 | 2.12 | 0.93 | 0.06 |
| PPD | 3.52 | 1.49 | 5.14 | 2.98 | 0.006a |
| CAL | 2.8 | 1.26 | 4.41 | 2.06 | 0.001a |
Polymorphism analysis of the VDR gene
by sequencing
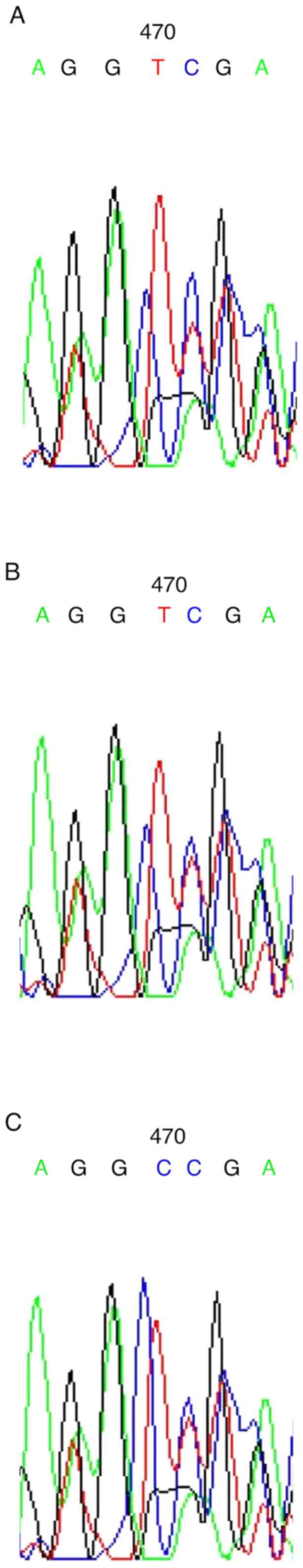
DNA sequencing confirmed a change in the sequence of
the VDR gene nucleotides (Fig.
2). The data indicated the sequencing results of the three
genotypes. The nucleotide sequence for the tt genotype at codon 352
was altered compared with that of the initial sequence of AGGTCGA
(Fig. 2A and B); the final sequence was: AGGCCGA
(Fig. 2C). This indicated a
nucleotide substitution from T to C (GTC to GCC) due to the change
in the amino acid valine (coded by GTC) to alanine (coded by
GCC).
Discussion
The aim of the present study was to identify the
presence of the VDR gene polymorphisms in patients with
periodontitis (32). Subsequently,
the VDR gene polymorphism was assessed as a risk factor
associated with periodontitis between patients with periodontitis
and healthy subjects (33). The
present study examined the change in the nucleotide sequence of the
VDR gene to explore the potential association of the VDR
gene polymorphism with the severity of the disease. Limited studies
have been performed utilizing DNA sequencing for the identification
of genetic variants that are linked to susceptibility to
periodontitis. In addition, to the best of our knowledge, the
present study is the first study in this context conducted in
Indonesia.
Current research conducted in the field of
periodontology has particularly focused on the influence of genetic
factors on individual susceptibility to periodontitis (34-36).
The tendency for early-onset periodontitis can be inherited with
either an autosomal recessive or autosomal dominant trait (37,38).
In the present study, it was found that the frequency of the Tt and
tt genotypes (t allele) was lower than that of the TT genotype (T
allele) in both periodontitis and healthy subjects. This result is
reasonable owing to the fact that the population in Makassar,
Indonesia, as a part of the Asian race, has a minor presentation of
the Tt and tt genotypes in the VDR gene. A similar result
was noted in a study by Zmuda et al (39) that reported a frequency of the Tt
and tt genotypes of only 2% in Asians, 5% in African Americans and
17% in Caucasians. In accordance with these findings, two previous
studies conducted by Sun et al (37) and Tachi et al (25) also reported that the t allele was
present in only 4% of a Chinese ethnic group and in 11% of a
Japanese ethnic group. Moreover, a small distribution of the t
alleles in the Asian race was also noted by Wang et al
(40); only in 5% of the population
examined.
The differences in the frequency of the genotypes or
alleles in a population can be explained by the following concept:
All polymorphisms begin as mutations occurring as a result of DNA
damage. As the frequency of the allele increases in the population,
the mutation is converted to a polymorphism (41,42).
The difference in the allele frequencies between ethnicities tends
to be influenced by evolutionary processes and genetic traits of a
single population. Similar association studies regarding VDR
polymorphisms and periodontal disease have been performed in other
ethnic populations and races (34,43,44). A
study by Borges et al (45)
reported an OR of 4.57 for the association between VDR and
periodontitis in a Brazilian population. In 2004, de Brito et
al (26) also reported that the
genotype and haplotype of the VDR gene polymorphism may be
associated with the incidence of periodontal disease. The
corresponding OR values were 2.41 and 4.32, respectively.
Similarly, Brett et al (46)
demonstrated an association between VDR gene polymorphism
and periodontitis in the Caucasian race. Similarly, Tachi et
al (25) reported an OR value
as high as 2.3 in the Japanese population.
In our previous study, an OR value of 12.57 was
noted indicating that subjects with VDR gene polymorphisms
exhibited a 12.57-fold higher probability of suffering from
periodontitis than those without VDR gene polymorphism
(33). This is expected considering
that the VDR gene is involved in various processes ranging
from bone metabolism to the regulation of the immune response. The
fundamental etiology of periodontitis is inflammation caused by
bacterial infection, which promotes alveolar bone resorption;
therefore, the VDR gene appears to be an optimal candidate
for predicting periodontitis susceptibility. Previously published
studies demonstrated that VDR plays an important role in
trabecular bone compared with its role in cortical bone (26,46).
Moreover, the variation in the VDR allele is responsible for
the variation in bone mineral density (BMD) (26,46).
VDR, which is involved in controlling calcium and phosphate
concentrations in the blood, is disrupted by the presence of
variations in the DNA sequence or the presence of polymorphisms,
resulting in a decrease in BMD across the body, including the
mandible and maxilla (47). A
decrease in jaw bone density will increase the alveolar porosity by
altering the trabecular pattern, which will eventually increase the
bone resorption rate following the invasion of periodontal
pathogens (46,48).
In the current study, the clinical features of
periodontal tissue damage accompanying periodontitis were
investigated by examining the periodontitis phenotype. The results
indicated that patients with periodontitis who had the Tt/tt
genotype exhibited a higher severity of periodontal tissue damage
than those with the TT genotype. It was assumed that the Tt/tt
genotype of exon 9 of the VDR gene affected mRNA stability
or decreased the expression levels of mRNA of osteoblast-associated
genes. This would decrease osteoblast function and increase
osteoclast function, thereby leading to severe alveolar trabecular
bone resorption. These findings are in accordance with a previous
study conducted by Sun et al (37) who demonstrated that the presence of
the t allele in the VDR TaqI may be a risk indicator for the
susceptibility to early-onset periodontitis. Tachi et al
(25) supported the findings of Sun
et al (37) by demonstrating
that the incidence of the TaqI polymorphism was associated with the
susceptibility of developing aggressive periodontitis. In another
study, the incidence of the homozygous (tt) genotype of TaqI in
patients with osteoporosis was significantly higher in the
osteoporosis group compared with that of the control group
(18).
In addition to assessing the incidence of the
VDR gene polymorphism, the present study detected a change
in the sequence of the nucleotides corresponding to the amino acid
valine. The nucleotide substitution in exon 9 (TaqI) of the
VDR gene results in the conversion of valine to alanine in
patients with periodontitis. Another published study evaluated the
levels of VDR gene expression in intron 8 and demonstrated
that the detection of ApaI C/T single nucleotide polymorphism
#rs731236 in patients with chronic periodontitis was an important
factor that was often overlooked in the prevention of this disease
(43). Accumulated evidence from
the previously published systematic review and meta-analysis showed
that VDR is a biological effector of vitamin D that controls
bone metabolism and inflammatory gene expression (5). Accordingly, in another review, it was
also found that genetic polymorphisms could modify gene expression
or function. Therefore, they may affect biological pathways and the
susceptibility of the subjects to a variety of diseases (49). The likely explanation for these
observed associations is to assume the existence of a truly
functional sequence variation to a different part of the gene,
which is, to a certain extent, linked to an allele of the anonymous
polymorphism explored (25,49). In the present study, it was assumed
that the nucleotide change would likely affect the level of
VDR gene expression, which subsequently influences the
translation rate of the protein or causes certain changes in RNA
stability and translation. This increased the susceptibility of
subjects with the t allele to a low bone density phenotype. The
present study showed that patients with the tt genotype had a
higher PPD (8 mm) than those of the patients with the Tt and TT
genotypes. Therefore, the findings of the present study emphasize
on the assumption that patients with the tt genotype are
susceptible to decreased bone density or decreased immune system
function. Unfortunately, since this was not fully addressed in the
current research, it is recommended to analyze these findings in
future studies.
It is important to note that the present study has
certain limitations such as the small sample size and consisted of
individuals from a single center. Therefore, additional research is
required to identify the specific pathogenesis of this disease and
the functional significance of the polymorphisms of the VDR
gene.
In conclusion, the findings of the present study
suggested that the VDR gene polymorphism was associated with
periodontitis in the Makassar-based population. Indirect evidence
confirms that the severity of periodontal tissue damage may be a
consequence of the presence of the t allele or the tt genotype.
Acknowledgements
We would like to thank Professor Bahruddin Talib
(Department of Prosthodontics, Faculty of Dentistry, Hasanuddin
University, Makassar, Indonesia) and Professor Budu (Department of
Ophthalmology Faculty of Medicine, Hasanuddin University) for their
valuable scientific support, as well as Professor Burhanuddin Daeng
Pasiga (Department of Dental Public Health, Faculty of Dentistry,
Hasanuddin University) for their expert support in data
analysis.
Funding
Funding: The present study was supported by the Ministry of
Research, Technology, and Higher Education of the Republic of
Indonesia (grant no. 1109/D3/PL/2010).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
NH, SO and EM conceived and designed the study, and
wrote the original draft of the manuscript. MR and ASHY analyzed
and interpreted the data, and edited the manuscript. TS and KLO
assisted in the design of the study, curated the data, and reviewed
and edited the manuscript. All authors have read and approved the
final manuscript. NH, SO and MR confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee for Biomedical Research in Humans, Faculty of Medicine,
Hasanuddin University (approval no.
0189/H.04.8.4.5.31/PP36-KOMETIK/2010). All patients in the case and
control groups provided informed consent prior to the initiation of
the study. The experimental procedures were based on the
Declaration of Helsinki.
Patient consent for publication
The patients provided written informed consent for
the publication of any data and/or accompanying images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Heidari Z, Moudi B and Mahmoudzadeh-Sagheb
HJ: Immunomodulatory factors gene polymorphisms in chronic
periodontitis: An overview. BMC Oral Health. 19(29)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Nazemisalman B, Vahabi S, Sabouri E,
Hosseinpour S and Doaju S: Association of vitamin D binding protein
and vitamin D receptor gene polymorphisms in Iranian patients with
chronic periodontitis. Odontol. 107:46–53. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gago EV, Cadarso-Suarez C, Perez-Fernandez
R, Burgos RR, Mugica JD and Iglesias CS: Association between
vitamin D receptor FokI. polymorphism and serum parathyroid hormone
level in patients with chronic renal failure. J Endocrinol Invest.
28:117–121. 2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Khammissa R, Ballyram R, Jadwat Y, Fourie
J, Lemmer J and Feller LJ: Vitamin D deficiency as it relates to
oral immunity and chronic periodontitis. Int J Dent.
2018(7315797)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wan QS, Li L, Yang SK, Liu ZL and Song N:
Role of Vitamin D receptor gene polymorphisms on the susceptibility
to periodontitis: A meta-analysis of a controversial issue. Genet
Test Mol Biomarkers. 23:618–633. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ebersole JL, Lambert J, Bush H, Huja PE
and Basu AJN: Serum nutrient levels and aging effects on
periodontitis. Nutrients. 10(1986)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Suchanecka A, Chmielowiec K, Chmielowiec
J, Trybek G, Masiak J, Michałowska-Sawczyn M, Nowicka R,
Grocholewicz K and Grzywacz A: Vitamin D receptor gene
polymorphisms and cigarette smoking impact on oral health: A
case-control study. Int J Environ Res Public Health.
17(3192)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Toy V and Uslu MO: Do genetic
polymorphisms affect susceptibility to periodontal disease? A
literature review. Niger J Clin Pract. 22:445–453. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nikolopoulos GK, Dimou NL, Hamodrakas SJ
and Bagos PG: Cytokine gene polymorphisms in periodontal disease: A
meta-analysis of 53 studies including 4178 cases and 4590 controls.
J Clin Periodontol. 35:754–767. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Soga Y, Nishimura F, Ohyama H, Maeda H,
Takashiba S and Murayama Y: Tumor necrosis factor-alpha gene
(TNF-alpha)-1031/-863,-857 single-nucleotide polymorphisms (SNPs)
are associated with severe adult periodontitis in Japanese. J Clin
Periodonto. 30:524–531. 2003.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Koutouzis T, Haber D, Shaddox L, Aukhil I
and Wallet SM: Autoreactivity of serum immunoglobulin to
periodontal tissue components: A pilot study. J Periodontol.
80:625–633. 2009.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Holla LI, Fassmann A, Augustin P, Halabala
T, Znojil V and Vanek J: The association of interleukin-4
haplotypes with chronic periodontitis in a Czech population. J
Periodontol. 79:1927–1933. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Donati M, Berglundh T, Hytönen AM,
Hahn-Zoric M, Hanson LÅ and Padyukov L: Association of the- 159
CD14 gene polymorphism and lack of association of the- 308 TNFA and
Q551R IL-4RA polymorphisms with severe chronic periodontitis in
Swedish Caucasians. J Clin Periodontol. 32:474–479. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
van Sorge NM, van der Pol WL and van de
Winkel JG: FcgammaR polymorphisms: Implications for function,
disease susceptibility and immunotherapy. Tiss Antigens.
61:189–202. 2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
LeVan TD, Bloom JW, Bailey TJ, Karp CL,
Halonen M, Martinez FD and Vercelli D: A common single nucleotide
polymorphism in the CD14 promoter decreases the affinity of Sp
protein binding and enhances transcriptional activity. J Immunol.
167:5838–5844. 2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Selvaraj P, Chandra G, Jawahar M, Rani MV,
Rajeshwari DN and Narayanan PR: Regulatory role of vitamin D
receptor gene variants of Bsm I, Apa I, Taq I, and Fok I
polymorphisms on macrophage phagocytosis and lymphoproliferative
response to mycobacterium tuberculosis antigen in pulmonary
tuberculosis. J Clin Immunol. 24:523–532. 2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Gelder CM, Hart KW, Williams OM, Lyons E,
Welsh KI, Campbell IA and Marshall SE: Vitamin D receptor gene
polymorphisms and susceptibility to Mycobacterium malmoense
pulmonary disease. J Infect Dis. 181:2099–2102. 2000.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Banjabi AA, Al-Ghafari AB, Kumosani TA,
Kannan K and Fallatah SM: Genetic influence of vitamin D receptor
gene polymorphisms on osteoporosis risk. Int J Health Sci (Qassim).
14:22–28. 2020.PubMed/NCBI
|
|
19
|
Guo HX, Pan J, Pan HB, Cui SJ and Fang CY:
Correlation of vitamin D receptor gene (ApaI) polymorphism with
periodontitis: A meta-analysis of Chinese population. Food Sci
Nutr. 7:3607–3612. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gunes S, Sumer AP, Keles GC, Kara N,
Koprulu H, Bagci H and Bek Y: Analysis of vitamin D receptor gene
polymorphisms in patients with chronic periodontitis. Indian J Med
Res. 127:58–64. 2008.PubMed/NCBI
|
|
21
|
Laine ML, Loos BG and Crielaard W: Gene
polymorphisms in chronic periodontitis. Int J Dent.
2010(324719)2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Perić M, Cavalier E, Toma S and Lasserre
J: Serum vitamin D levels and chronic periodontitis in adult,
Caucasian population-a systematic review. J Periodontal Res.
53:645–656. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Uwitonze AM, Murererehe J, Ineza MC,
Harelimana EI, Nsabimana U, Uwambaye P, Gatarayiha A and Haq A:
Effects of vitamin D status on oral health. J Steroid Biochem Mol
Biol. 175:190–194. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
World Medical Association. World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194.
2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tachi Y, Shimpuku H, Nosaka Y, Kawamura T,
Shinohara M, Ueda M, Imai H and Ohura K: Vitamin D receptor gene
polymorphism is associated with chronic periodontitis. Life Sci.
73:3313–3321. 2003.PubMed/NCBI View Article : Google Scholar
|
|
26
|
de Brito Júnior RB, Scarel-Caminaga RM,
Trevilatto PC, Souza AP and Barros SP: Polymorphisms in the vitamin
D receptor gene are associated with periodontal disease. J
Periodontol. 75:1090–1095. 2004.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tonetti MS, Greenwell H and Kornman KS:
Staging and grading of periodontitis: Framework and proposal of a
new classification and case definition. J Periodontol. 89 (Suppl
1):S159–S172. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lertpimonchai A, Rattanasiri S, Arj-Ong
Vallibhakara S, Attia J and Thakkinstian A: The association between
oral hygiene and periodontitis: A systematic review and
meta-analysis. Int Dent J. 67:332–343. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Greene JC and Vermillion JR: The
simplified oral hygiene index. J Am Dent Assoc. 68:7–13.
1964.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Boom R, Sol CJ, Salimans MM, Jansen CL,
Wertheim-van Dillen PM and van der Noordaa J: Rapid and simple
method for purification of nucleic acids. J Clin Microbiol.
28:495–503. 1990.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Williams KR, Wasson SR, Barrett A,
Greenall RF, Jones SR and Bailey EG: Teaching hardy-weinberg
equilibrium using population-level punnett squares: Facilitating
calculation for students with math anxiety. CBE-Life Sciences
Education. 20(ar22)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hamrun N and Hatta M: Polymorphisms of
Vitamin D receptor gene in chronic periodontitis patients
[Indonesia]. JST Kesehatan. 1:165–172. 2011.
|
|
33
|
Hamrun N: Polymorphism of Vitamin D
receptor gene is associated with chronic periodontitis [Indonesia].
Dentika: Dental J. 16:121–125. 2011.
|
|
34
|
Mashhadiabbas F, Neamatzadeh H, Nasiri R,
Foroughi E, Farahnak S, Piroozmand P, Mazaheri M and Zare-Shehneh
M: Association of vitamin D receptor BsmI, TaqI, FokI, and ApaI
polymorphisms with susceptibility of chronic periodontitis: A
systematic review and meta-analysis based on 38 case -control
studies. Dent Res J (Isfahan). 15:155–165. 2018.PubMed/NCBI
|
|
35
|
Ho YP, Lin YC, Yang YH, Chou YH, Ho KY, Wu
YM and Tsai CC: Association of vitamin D receptor gene
polymorphisms and periodontitis in a Taiwanese Han population. J
Dent Sci. 12:360–367. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Loos BG, John RP and Laine ML:
Identification of genetic risk factors for periodontitis and
possible mechanisms of action. J Clin Periodontol. 32:159–169.
2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sun J, Meng H, Cao C, Tachi Y, Shinohara
M, Ueda M, Imai H and Ohura K: Relationship between vitamin D
receptor gene polymorphism and periodontitis. J Periodontal Res.
37:263–267. 2002.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Divaris K, Monda KL, North KE, Olshan AF,
Reynolds LM, Hsueh WC, Lange EM, Moss K, Barros SP, Weyant RJ, et
al: Exploring the genetic basis of chronic periodontitis: A
genome-wide association study. Hum Mol Genet. 22:2312–2314.
2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zmuda JM, Cauley JA and Ferrell R:
Molecular epidemiology of vitamin D receptor gene variants.
Epidemiol Rev. 22:203–207. 2000.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang C, Zhao H, Xiao L, Xie C, Fan W, Sun
S, Xie B and Zhang J: Association between vitamin D receptor gene
polymorphisms and severe chronic periodontitis in a Chinese
population. J Periodontol. 80:603–608. 2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Naito M, Miyaki K, Naito T, Zhang L, Hoshi
K, Hara A, Masaki K, Tohyama S, Muramatsu M, Hamajima N and
Nakayama T: Association between vitamin D receptor gene haplotypes
and chronic periodontitis among Japanese men. Int J Med Sci.
4:216–222. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Uitterlinden AG, Fang Y, Van-Meurs JB,
Pols HA and Van-Leeuwen J: Genetics and biology of vitamin D
receptor polymorphisms. Gene. 338:143–156. 2004.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Marian D, Rusu D, Stratul SI, Calniceanu
H, Sculean A and Anghel A: Association of vitamin D receptor gene
polymorphisms with chronic periodontitis in a population in Western
Romania. Oral Health Prev Dent. 17:157–165. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Jilani M, Mohamed AA, Zeglam HB, Alhudiri
IM, Ramadan AM, Saleh SS, Elkabir M, Amer IB, Ashammakhi N and
Enattah NS: Association between vitamin D receptor gene
polymorphisms and chronic periodontitis among Libyans. Libyan J
Med. 10(26771)2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Borges MA, Figueiredo LC, Brito RB Jr,
Faveri M and Feres M: Microbiological composition associated with
vitamin D receptor gene polymorphism in chronic periodontitis. Braz
Oral Res. 23:203–208. 2009.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Brett P, Zygogianni P, Griffiths G, Tomaz
M, Parkar M, D'Aiuto F and Tonetti M: Functional gene polymorphisms
in aggressive and chronic periodontitis. J Dent Res. 84:1149–1153.
2005.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Kuo LC, Polson AM and Kang T: Associations
between periodontal diseases and systemic diseases: A review of the
inter-relationships and interactions with diabetes, respiratory
diseases, cardiovascular diseases and osteoporosis. Public Health.
122:417–433. 2008.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Amano Y, Komiyama K and Makishima MJ:
Vitamin D and periodontal disease. J Oral Sci. 51:11–20.
2009.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ruiz-Ballesteros AI, Meza-Meza MR,
Vizmanos-Lamotte B, Parra-Rojas I and de la Cruz-Mosso U:
Association of vitamin D metabolism gene polymorphisms with
autoimmunity: Evidence in population genetic studies. Int J Mol
Sci. 21(9626)2020.PubMed/NCBI View Article : Google Scholar
|