1. Introduction
The combined use of genetic tumor testing and
genetically matched targeted medicines has resulted in significant
advances in cancer treatment outcomes over the last few decades
(1). Imatinib has been used for
KIT-mutated gastrointestinal stromal tumor, vemurafenib for BRAF
V600-mutated melanoma (2),
trastuzumab for HER2-amplified breast and gastric cancer (3) and crizotinib for ALK-activated lung
malignancies (4). Modern molecular
biology tools have enhanced the understanding of complex changes
during carcinogenesis in basic research and their parallel use in
clinical practice has improved the identification of malignancies.
To make an accurate diagnosis of a tumor, estimate the prognosis of
a patient and plan tumor therapy, molecular markers and genetic
analyses are essential today (5).
In the era of molecular medicine, with 60 different
molecular analyses, the majority of approaches (57.4%) are using
targeted multigene next-generation sequencing (NGS), accompanied by
whole-exome sequencing (16.4%) and RNA sequencing (13.1%),
array-based comparative genome-wide hybridization (4.9%),
whole-genome sequencing and Sanger sequencing (both 3.3%) and mRNA
sequencing (1.6%). Targeted NGS of the most essential
cancer-associated genes, ideally combined with analysis of
clinically important gene fusions, is the most suited analysis for
detecting actionable changes (e.g., kinase fusion genes) (6). This analysis is readily available and
combines quick sequencing with manageable bioinformatics.
Since most clinicians obtain minimal training in
genetics (7), the abundance of
genetic tests and information poses a serious challenge: In a
recent survey of physicians in a tertiary cancer center, 22%
reported low confidence in their genetic knowledge and 18% expected
to test their patients infrequently (8). The current genetic ‘under-testing’ may
be due to a lack of knowledge; for instance, in the Netherlands,
during the years 2008 to 2014, after crizotinib and EGFR inhibitors
were approved, ~50% of patients with NSCLC had their ALK
rearrangements tested and ~70% had their EGFR mutations tested
(9). It took an additional two
years to bring ALK testing coverage to 80% after it was included in
NSCLC guidelines (10). The
literature search was performed in the PubMed database using the
following key words: Precision therapy and cancer, personalized
medicine and cancer, molecular analysis and cancer therapy and
precision medicine and clinical trials. The last 20 years of
publication were considered. The inclusion criteria were that
precision medicine and cancer were included.
2. Translating complex biomarkers into
molecular diagnostics
The incorporation of mutational profiles and/or gene
expression signatures into a biomarker development strategy is
accompanied by the translation of the clinical outcome-related
biomarker into a robust assay adaptable to widely used analysis
platforms. A review of the scientific literature revealed that most
published biomarkers are insufficient to replace the existing
clinical tests or are only useful for diagnosing advanced disease
phases with low survival rates. Numerous molecular or genetic
biomarkers have indeed been proposed for the diagnosis of various
diseases; however, the majority of these lack the essential
sensitivity and specificity. Although several molecular and
-genetic biomarkers have indeed been proposed for diagnosing
various diseases, they usually lack the essential sensitivity and
specificity. In addition, most predicted metabolomic and proteomic
biomarker results have not yet progressed or proceeded from the
laboratory to clinical trials, since they were stopped at the
initial stage of biomarker identification (11).
Multiple cancer genome databases are currently
available for the interpretation of profiling results, including
canSAR (https://cansar.icr.ac.uk), cBioPortal
(https://www.cbioportal.org/), My Cancer
Genome (https://www.mycancergenome.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC
(https://dcc.icgc.org/) and The Cancer Genome
Atlas (TCGA; https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga),
and various institutes host their own online variant interpretation
tools (such as www.mycancergenome.org, Vanderbilt-Ingram Cancer
Center; pct.mdanderson.org). However, to
translate genomic profiles into treatment recommendations, searches
across numerous databases are usually required, as none of these
repositories appears to be sufficiently useful on their own
(12). Overall, molecular tumor
research is likely to become increasingly significant in the
division of molecular oncology.
3. Molecular markers for the selection of
cancer therapy
Several medicines have one thing in common: They all
target different parts of cancer hallmarks or features that are
required for successful tumor proliferation and dissemination. The
advancement of molecular-scale technology has been critical in the
discovery of new cancer targets and it is not a coincidence that
better medicines have appeared at the same time that our
understanding of cancer genetics has increased thanks to gene
expression arrays and DNA sequencing. Advanced tumor pathology is
now studied at the molecular level, with immunohistochemical
biomarkers, gene signature classifiers and gene mutations all
providing important information about whether patients may react to
targeted therapy regimes. The broad categories of targeted
medicines utilized in clinical settings are briefly discussed in
the present review, as well as a brief history of
immunohistochemistry, gene expression and DNA sequencing
technologies, before looking at the following three tumor types:
Breast, lung and colorectal malignancies. To review the current
standard therapy for each of these cancer types, prior to focusing
on the pertinent targeted therapies and the pathways they impede,
molecular analyses provide information on the subclinical
manifestation of the disease. Finally, certain strategies that are
critical to the growth of targeted anti-cancer medications may be
planned. Clearly, a deeper knowledge of the mechanisms of action of
drugs and associated biological effects is required, particularly
given that magnetic resonance imaging is also unable to distinguish
tumor cells from edema. Hence, the necessity of being able to
assess a clinical trial drug's effect should be emphasized as a
goal of modern analysis methodologies, such as genomics, proteomics
and even functional imaging analysis (13).
The most significant category is the heterogeneous
population of uncommon tumors, with sarcomas being the most
prevalent, followed by breast, brain, gynecological, lung and
colorectal cancers (14). The use
of NGS technology has proven that there are commonly mutant genes
that are aberrant across various cancer types, which may thus
respond similarly to a specific targeted therapy (15). NGS-based methods that rapidly
generate the mutational profile of a cancer genome in the clinical
setting are now being combined with baseline information about
frequent genomic alterations in cancer generated in the research
setting by sequencing the DNA of thousands of tumors to inform
genome-guided cancer medicine (16).
4. Liquid biopsy and personalized
therapy
In recent years, oncology research has focused on
liquid biopsies, which rely on the detection of cancer-derived
components in patients' biofluids, such as circulating tumor cells
(CTCs) (17,18), circulating tumor DNA (ctDNA), ctRNA
(19) and extracellular vesicles
(EVs) (20), and may reveal
disseminated aggressive clones. Liquid biopsies are not confined to
blood; however, this is what much of the liquid biopsy studies
focus on. Urine, saliva or cervical fluid may be employed, as
genetic information is present in these fluids (21). ctDNAs have emerged as promising
biomarkers, particularly in cancer, and is being widely examined in
translational and clinical research (22). Several efforts are being made to
evaluate the potential of ctDNAs for early cancer screening, and
both qualitative and quantitative cell-free DNA (cfDNA) alterations
have been investigated (23).
Despite extensive study, only a small number of cfDNA-based assays
have been implemented in clinical practice. Conflicting data on
total nuclear cfDNA concentration, for example, make it difficult
to develop and use cfDNA-based tests in the clinic: Plasma cfDNA
concentrations in patients with cancer range from a few ng/ml to
many thousand ng/ml, which overlaps with the range of
concentrations in healthy subjects (24). The finding of cells discharged into
the bloodstream or migrating from tumors is critical and has
resulted from 20 years of intense investigation. CTCs are difficult
to separate and frequently do not represent genetically malignant
cells. Several studies have evaluated the utility of CTCs in
identifying various types of cancer (25,26).
Tanaka et al (27)
demonstrated that CTC enumerations exhibited an unsatisfactory
discriminating capacity between individuals with lung cancer and
those with non-malignant lung lesions [area under the receiver
operating characteristic curve=0.598 (P=0.122)]. Standardization of
pre-analytical parameters and an improved understanding of the
precise origin and structure of cfDNA are additional steps in
implementing its analysis (27).
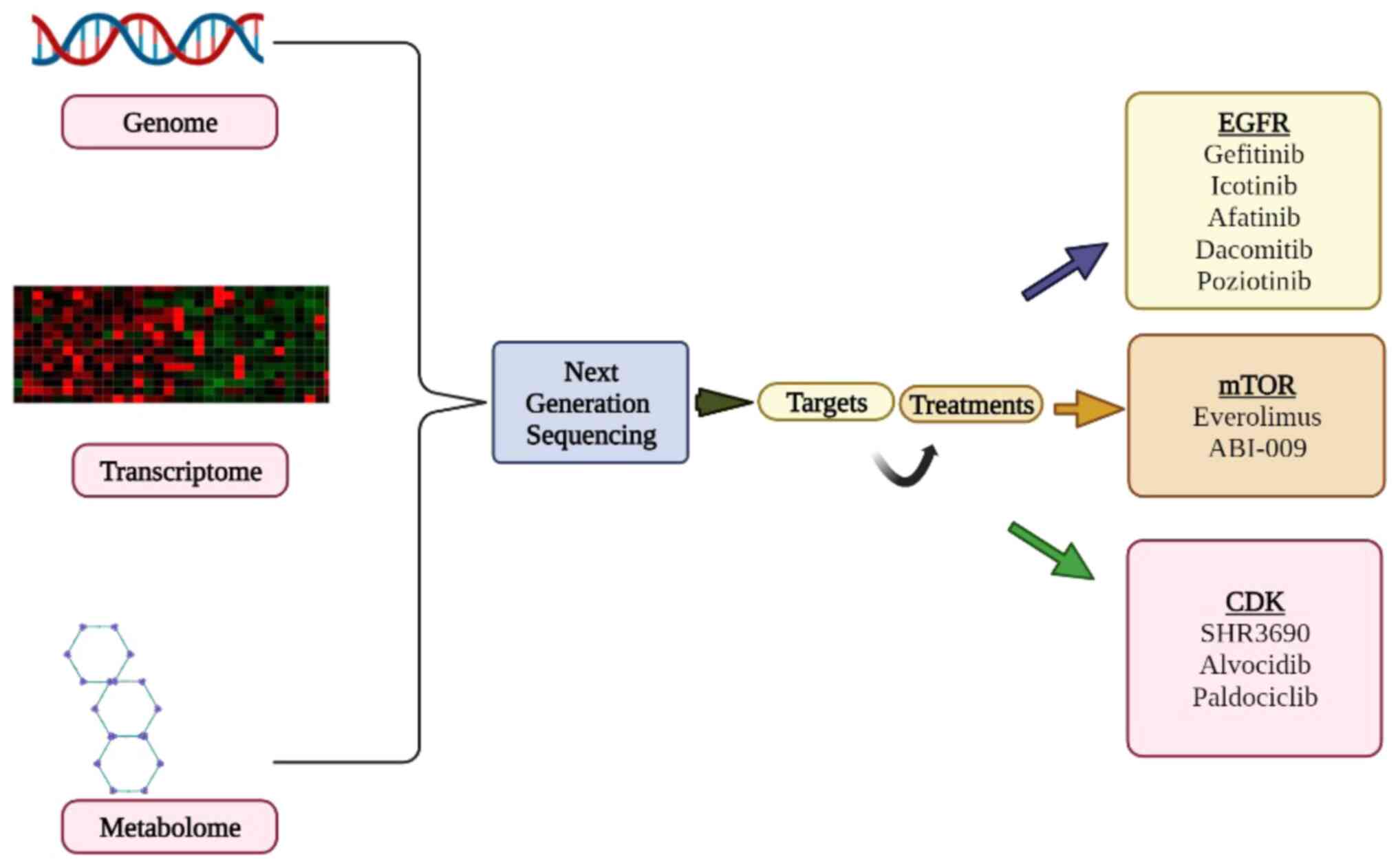
Advances in sophisticated quantitative PCR technologies or NGS
(Fig. 1) will certainly increase
the analytical performance and dependability of future tests
(28).
5. Precision medicine (PM) trials and
treatment algorithm
PM is a method of improving patient outcomes by
combining clinical and molecular patient data to analyze the
genetic causes of a disease (29).
The goal of this technique is to improve patient outcomes while
offering better safety profiles than standard population-based
cancer treatments (30).
The goal of progressive PM clinical studies is to
determine whether tumor molecular profiling has therapeutic utility
and whether treatment selection based on molecular changes delivers
better outcomes than unselective treatment. Treatment algorithms
are used in these trials to allocate patients to specific targeted
medicines based on tumor genetic changes (31).
PM in oncology was developed nearly two decades ago
with the introduction of molecular targeted agents (MTAs) and is
now mostly based on the molecular-genetic characteristics of the
patients' malignancies. MTAs affect the function of specific
molecular targets in cell signaling, proliferation, apoptosis,
angiogenesis, metabolism, migration or invasion, whereas cytotoxic
drugs destroy rapidly dividing cells by stimulating the DNA and
cell division machinery. It is now known that the bulk of harmful
genetic modifications are shared by different tumor types (32). MTAs have been indicated to be
beneficial in a variety of tumor types that have a common molecular
abnormality, e.g., trastuzumab and lapatinib for HER2, which is
amplified and overexpressed in 10-15% of breast and gastric
cancers. Molecular changes are frequently observed in a relatively
small fraction of patients with solid tumors. Due to the division
of all classic human cancers into comparatively small subgroups, it
is critical to reach an agreement on the best methods for
establishing the action of experimental therapies in rare molecular
subsets (31).
Clinical trials provide a scientific evaluation of
investigational drugs, technologies or biologics, such as
chemotherapeutic agents, blood products or gene therapies, in human
volunteers. Prior to being granted Food and Drug Administration
(FDA) approval, proposed medicines frequently go through a
rigorously regulated and time-consuming multi-phase clinical trial
process. Significant changes to present clinical trial designs will
be required in the future to move toward a more individualized
approach. An example of a novel accelerated attempt to assess
targeted medicines is an adaptive trial design. Researchers may use
this design to assess study data collected during anticipated
intermediate time periods and change the direction of an
individual's research project or the trial ultimately (33).
Mazo et al (34) provided an in silico analysis,
according to which four well-recognized numerical risk scores
(OncotypeDX, OncoMasTR, EndoPredict and tumor-infiltrating
leucocytes) were significantly associated with neoadjuvant
chemotherapy complete remission to neo-adjuvant chemotherapy in
patients with estrogen receptor-positive and HER-2-negative breast
cancers. The outlook for recurrent acute myeloid leukemia (AML)
remains dismal, but a small number of effective treatments are
available. According to the findings of Uckun et al
(35) phase IB study, older adults
with relapsed AML generally tolerate the integration of the
cytotoxic chemotherapeutic drugs combrestatin A1 diphosphatase
(OXi4503) and cytarabine well. A microvesicles-based study examined
the therapeutic response of glioblastomas to temozolomide and
concurrent radiation therapy (36).
Melo et al (37) indicated
that the serum levels of glypican-1-positive EVs are highly
sensitive and specific for detecting early- and late-stage
pancreatic cancer and correlate with the tumor burden. Uckun et
al (38) described a method to
overcome the blood-brain barrier, as it may restrict the
intra-tumoral availability of therapeutically effective compounds.
They demonstrated that intra-tumoral administration of the RNA
therapeutic OT101 through convection-enhanced delivery inhibits the
immunosuppressive effects of transforming growth factor β2, which
then leads to clinically significant single-agent activity.
Castration-resistant prostate cancer (CRPC) is treated with
docetaxel as the first-line chemotherapy, but resistance is common
due to acquired induction of P-glycoprotein, which is expressed by
the multidrug resistance protein 1 gene. A new taxane is being used
in resistant patients due to its low affinity for P-glycoprotein.
It has been indicated that docetaxel-resistant patients had
significantly higher levels of P-glycoprotein in their serum EVs
than docetaxel-susceptible patients. Therefore, this test has the
prospect to be utilized as a guide for choosing the right taxoid in
patients with CRPC (39). It is
well known that genetic and epigenetic events build up gradually to
form carcinomas. As a result, the current standard of care is
guided by prognostic and predictive biomarkers such as KRAS and
microsatellite instability. Emerging biomarkers and cutting-edge
liquid biopsy platforms, as outlined by Koulis et al
(40), may open the door to novel
combination treatments that target both the tumor microenvironment
and tumor cells. Approximately 5% of NSCLCs have an anaplastic
lymphoma kinase (ALK) mutation. As a result of this discovery,
FDA-approved ALK blockers, such as crizotinib and ceritinib, which
are given to patients who test positive for the ALK mutation, were
created. Another promising application is that of the drug
olaparib, an inhibitor of poly ADP ribose polymerase, for
BRCA-mutant ovarian cancer. The HER2 proto-oncogene, a frequent
target for drugs used in personalized therapy, is overexpressed in
~25% of human breast cancers (41).
However, a sizeable percentage of patients receiving trastuzumab, a
monoclonal antibody against HER2, eventually relapse or experience
progressive disease (42).
According to a recent study, EVs with the specific markers EGFR,
p-EGFR and genomic DNA were produced in large quantities after
treatment with the EGFR inhibitor cetuximab (43). This study further supported the idea
that ‘targeted agents may induce cancer cells to change the EV
emission profiles reflective of drug-related therapeutic stress’.
Thus, it may be assumed that such EV emission profiles may be
defined and used to evaluate the efficacy of various treatments in
different patients. The concept of acclimation is not entirely new.
In radiation oncology, it has been successfully used and
implemented in recent years (44).
As the knowledge of biomarkers and that particular
EGFR mutations lead to superior outcomes with EGFR tyrosine kinase
inhibitors (45), bio-markers have
emerged as an essential aspect in planning treatment for NSCLC.
Erlotinib, KRAS/BRAF (sorafinib), retinoid-EGFR signaling
(bexarotene and erlotinib) and VEGFR (vantetanib) are among the
targets. The primary objective of the study was the 8-week disease
control rate (DCR), which was defined by Response Evaluation
Criteria in Solid Tumors as a full or partial response or stable
disease. Treatment efficacy was characterized as >80%
probability of obtaining a DCR of >30% in similar patients, with
efficacy of treatment defined as >80% possibility of reaching a
DCR of >30% (46).
6. Conclusions
In conclusion, none of the studies that were chosen
employed a standard scale to classify molecular changes based on
their therapeutic value. The need for genetic testing will grow
along with the rapid advancement of the genetic understanding and
development of medical science. Whole-genome and whole-exome
sequencing technologies are already widely employed for research
objectives, also including providing prognostic or predictive
profiles or screening patients for early clinical trials.
Whole-genome and whole-exome sequencing technologies are projected
to become the standard of care in the near future. Clinicians will
be challenged with increasingly complex genomic information and a
growing number of platforms from which to choose. While large-scale
sequencing is far more instructive in most circumstances, tailored
in-depth sequencing may be better in others.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Data sharing is not applicable.
Authors' contributions
SS: Literature search, conception and writing. LS:
Literature search, writing and preparation of the figure. Both
authors read and approved the final version of the article. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Garraway LA: Genomics-driven oncology:
Framework for an emerging paradigm. J Clin Oncol. 31:1806–1814.
2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Blanke CD, Rankin C, Demetri GD, Ryan CW,
Von Mehren M, Benjamin RS, Raymond AK, Bramwell VH, Baker LH, Maki
RG, et al: Phase III randomized, intergroup trial assessing
imatinib mesylate at two dose levels in patients with unresectable
or metastatic gastrointestinal stromal tumors expressing the kit
receptor tyrosine kinase: S0033. J Clin Oncol. 26:626–632.
2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et
al: Improved survival with vemurafenib in melanoma with BRAF V600E
mutation. N Engl J Med. 364:2507–2516. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa
K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al:
First-line crizotinib versus chemotherapy in ALK-positive lung
cancer. N Engl J Med. 371:2167–2177. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
MacConaill LE: Existing and emerging
technologies for tumor genomic profiling. J Clin Oncol.
31:1815–1824. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Luchini C, Lawlor RT, Milella M and Scarpa
A: Molecular tumor boards in clinical practice. Trends Cancer.
6:738–744. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Schwaederle M, Parker BA, Schwab RB, Fanta
PT, Boles SG, Daniels GA, Bazhenova LA, Subramanian R, Coutinho AC,
Ojeda-Fournier H, et al: Molecular tumor board: The University of
California-San Diego Moores cancer center experience. Oncologist.
19:631–636. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gray SW, Hicks-Courant K, Cronin A,
Rollins BJ and Weeks JC: Physicians' attitudes about multiplex
tumor genomic testing. J Clin Oncol. 32:1317–1323. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vander Velden DL, Van Herpen CML, Van
Laarhoven HWM, Smit EF, Groen HJM, Willems SM, Nederlof PM,
Langenberg MHG, Cuppen E, Sleijfer S, et al: Molecular tumor
boards: Current practice and future needs. Ann Oncol. 8:3070–3075.
2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Theelen WS, Mittempergher L, Willems SM,
Bosma AJ, Peters DD, van der Noort V, Japenga EJ, Peeters T, Koole
K, Šuštić T, et al: FGFR1, 2 and 3 protein overexpression and
molecular aberrations of FGFR3 in early stage non-small cell lung
cancer. J Pathol Clin Res Res. 2:223–233. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Goodsaid FM and Mendrick DL: Translational
medicine and the value of biomarker qualification. Sci Transl Med.
2(47ps44)2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ortiz MV, Kobos R, Walsh M, Slotkin EK,
Roberts S, Berger MF, Hameed M, Solit D, Ladanyi M, Shukla N and
Kentsis A: Integrating genomics into clinical pediatric oncology
using the molecular tumor board at the memorial sloan kettering
cancer center. Pediatr Blood Cancer. 63:1368–1374. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tobin NP, Foukakis T, De Petris L and
Bergh J: The importance of molecular markers for diagnosis and
selection of targeted treatments in patients with cancer. J Intern
Med. 278:545–570. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chinot OL, Wick W, Mason W, Henriksson R,
Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea
D, et al: Bevacizumab plus radiotherapy-temozolomide for newly
diagnosed glioblastoma. N Engl J Med. 370:709–722. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mardis ER: The translation of cancer
genomics: Time for a revolution in clinical cancer care. Genome
Med. 6(22)2014.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Andre F, Mardis E, Salm M, Soria JC, Siu
LL and Swanton C: Prioritizing targets for precision cancer
medicine. Ann Oncol. 25:2295–2303. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Aaltonen KE, Novosadová V, Bendahl PO,
Graffman C, Larsson AM and Rydén L: Molecular characterization of
circulating tumor cells from patients with metastatic breast cancer
reflects evolutionary changes in gene expression under the pressure
of systemic therapy. Oncotarget. 8:45544–45565. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hong X, Sullivan RJ, Kalinich M, Kwan TT,
Giobbie-Hurder A, Pan S, LiCausi JA, Milner JD, Nieman LT, Wittner
BS, et al: Molecular signatures of circulating melanoma cells for
monitoring early response to immune checkpoint therapy. Proc Natl
Acad Sci USA. 115:2467–2472. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Egyud M, Tejani M, Pennathur A, Luketich
J, Sridhar P, Yamada E, Ståhlberg A, Filges S, Krzyzanowski P,
Jackson J, et al: Detection of circulating tumor DNA in plasma: A
potential biomarker for esophageal adenocarcinoma. Ann Thorac Surg.
108:343–349. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Francaviglia I, Magliacane G, Lazzari C,
Grassini G, Brunetto E, Dal Cin E, Girlando S, Medicina D, Smart
CE, Bulotta A, et al: Identification and monitoring of somatic
mutations in circulating cell-free tumor DNA in lung cancer
patients. Lung Cancer. 134:225–232. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Tanos R and Thierry AR: Clinical relevance
of liquid biopsy for cancer screening. Transl Cancer Res. 7 (Suppl
2):S105–S129. 2018.
|
|
22
|
Thierry AR, El Messaoudi S, Gahan PB,
Anker P and Stroun M: Origins, structures, and functions of
circulating DNA in oncology. Cancer Metastasis Rev. 35:347–376.
2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fleischhacker M and Schmidt B: Circulating
nucleic acids (CNAs) and cancer-a survey. Biochim Biophys Acta.
1775:181–232. 2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Mouliere F, El Messaoudi S, Pang D,
Dritschilo A and Thierry AR: Multi-marker analysis of circulating
cell-free DNA toward personalized medicine for colorectal cancer.
Mol Oncol. 8:927–941. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang Z, Fan W, Deng Q, Tang S, Wang P, Xu
P, Wang J and Yu M: The prognostic and diagnostic value of
circulating tumor cells in bladder cancer and upper tract
urothelial carcinoma: A meta-analysis of 30 published studies.
Oncotarget. 8:59527–59538. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hofman P: Liquid biopsy for early
detection of lung cancer. Curr Opin Oncol. 29:73–78.
2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tanaka F, Yoneda K, Kondo N, Hashimoto M,
Takuwa T, Matsumoto S, Okumura Y, Rahman S, Tsubota N, Tsujimura T,
et al: Circulating tumor cell as a diagnostic marker in primary
lung cancer. Clin Cancer Res. 15:6980–6986. 2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Krishnamurthy N, Spencer E, Torkamani A
and Nicholson L: Liquid biopsies for cancer: Coming to a patient
near you. J Clin Med. 6(3)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Desmond-Hellmann S: Toward precision
medicine: A new social contract? Sci Transl Med.
4(129ed3)2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Heckman-Stoddard BM and Smith JJ:
Precision medicine clinical trials: Defining new treatment
strategies. In seminars in oncology nursing. Vol. 30. WB Saunders,
pp109-116, 2014.
|
|
31
|
Le Tourneau C, Kamal M, Tsimberidou AM,
Bedard P, Pierron G, Callens C, Rouleau E, Vincent-Salomon A,
Servant N, Alt M, et al: Treatment algorithms based on tumor
molecular profiling: The essence of precision medicine trials. J
Natl Cancer Inst. 108(v362)2015.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ciriello G, Miller ML, Aksoy BA,
Senbabaoglu Y, Schultz N and Sander C: Emerging landscape of
oncogenic signatures across human cancers. Nat Genet. 45:1127–1133.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
33
|
Berry DA: Adaptive clinical trials in
oncology. Nat Rev Clin Oncol. 9:199–207. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Mazo C, Barron S, Mooney C and Gallagher
WM: Multi-gene prognostic signatures and prediction of pathological
complete response to neoadjuvant chemotherapy in ER-positive,
HER2-negative breast cancer patients. Cancers (Basel).
12(1133)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Uckun FM, Cogle CR, Lin TL, Qazi S, Trieu
VN, Schiller G and Watts JM: A phase 1B clinical study of
combretastatin A1 diphosphate (OXi4503) and cytarabine (ARA-C) in
combination (OXA) for patients with relapsed or refractory acute
myeloid leukemia. Cancers (Basel). 12(74)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shao H, Chung J, Balaj L, Charest A,
Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R and
Lee H: Protein typing of circulating microvesicles allows real-time
monitoring of glioblastoma therapy. Nat Med. 18:1835–1840.
2012.PubMed/NCBI View
Article : Google Scholar
|
|
37
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Uckun FM, Qazi S, Hwang L and Trieu VN:
Recurrent or refractory high-grade gliomas treated by
convection-enhanced delivery of a TGFβ 2-targeting RNA therapeutic:
A post-hoc analysis with long-term follow-up. Cancers (Basel).
11(1892)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kato T, Mizutani K, Kameyama K, Kawakami
K, Fujita Y, Nakane K, Kanimoto Y, Ehara H, Ito H, Seishima M, et
al: Serum exosomal P-glycoprotein is a potential marker to diagnose
docetaxel resistance and select a taxoid for patients with prostate
cancer. Urol Oncol. 33:385.e15–e20. 2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Koulis C, Yap R, Engel R, Jardé T, Wilkins
S, Solon G, Shapiro JD, Abud H and McMurrick P: Personalized
medicine-current and emerging predictive and prognostic biomarkers
in colorectal cancer. Cancers (Basel). 12(812)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, et al: Identification of the transforming EML4-ALK
fusion gene in non-small-cell lung cancer. Nature. 448:561–566.
2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Rexer BN and Arteaga CL: Intrinsic and
acquired resistance to HER2-targeted therapies in HER2
gene-amplified breast cancer: Mechanisms and clinical implications.
Crit Rev Oncog. 17:1–16. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Montermini L, Meehan B, Gamier D, Lee WJ,
Lee TH, Guha A, Al-Nedawi K and Rak J: Inhibition of oncogenic
epidermal growth factor receptor kinase triggers release of
exosome-like extracellular vesicles and impacts their
phosphoprotein and DNA content. J Biol Chem. 290:24534–24546.
2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Van Dommelen SM, van der Meel R, van
Solinge WW, Coimbra M, Vader P and Schiffelers RM: Cetuximab
treatment alters the content of extracellular vesicles released
from tumor cells. Nanomedicine (Lond). 11:881–890. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Douillard JY, Shepherd FA, Hirsh V, Mok T,
Socinski MA, Gervais R, Liao ML, Bischoff H, Reck M, Sellers MV, et
al: Molecular predictors of outcome with gefitinib and docetaxel in
previously treated non-small-cell lung cancer: Data from the
randomized phase III INTEREST trial. J Clin Oncol. 28:744–752.
2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kim ES, Herbst RS, Wistuba II, Lee JJ,
Blumenschein GR Jr, Tsao A, Stewart DJ, Hicks ME, Erasmus J Jr,
Gupta S, et al: The BATTLE trial: Personalizing therapy for lung
cancer. Cancer Discov. 1:44–53. 2011.PubMed/NCBI View Article : Google Scholar
|