Introduction
Pulmonary fibrosis (PF) is a serious interstitial
lung disease characterized by an abnormal accumulation of the
extracellular matrix and destruction of the architecture of the
lungs (1). PF is classified into
the most common idiopathic PF and other types caused by factors
such as genetic mutations or exposure to toxic chemicals or
radiation (2,3). The onset and progression of PF is
mediated by the activation of pulmonary epithelial cells, the
release of cytokines and growth factors, the proliferation and
activation of myofibroblasts, and the abnormal deposition and
transformation of the extracellular matrix (3). Current treatments for PF include
antifibrotic agents, corticosteroids, and immunosuppressive drugs,
but the prognosis for PF remains poor (4,5). The
development of preventive and therapeutic strategies for PF is
therefore an urgent priority.
Plants are rich in polyphenols, which are
non-nutritive secondary metabolites; thus, humans consume
polyphenols in their diet (6). The
intake of these polyphenols has been reported to prevent various
diseases such as diabetes and cardiovascular disease (7,8). Green
tea (Camellia sinensis, Theaceae) is commonly consumed
worldwide and has several dietary benefits such as antioxidative,
anti-inflammatory, antiobesity, and antifibrotic effects (9-12).
(−)-Epigallocatechin-3-O-gallate (EGCG) is the most abundant
and active compound in green tea and has been proven to have
anticancer, anti-inflammatory, vascular-protective, and
antifibrotic effects (13-17).
Previous studies have reported that EGCG suppresses the
accumulation of pulmonary hydroxyproline and bleomycin-induced
pulmonary fibrosis (18,19), although the mechanism of the
antifibrotic effect of EGCG remains unclear.
microRNAs (miRNAs or miRs) are a class of small,
noncoding RNAs that bind directly to mRNA and regulate gene
expression (18), thereby playing
key roles in biological processes such as cell proliferation,
metabolism, inflammation, and fibrosis (19-22).
Furthermore, miRNAs have been shown to be involved in cell-to-cell
communication by transferring from donor cells to recipient cells
via extracellular vesicles (EVs) such as exosomes (23). In PF, miRNA let-7d, an exosomal
miRNA derived from vascular endothelial cells (VECs), has been
reported to regulate fibrosis by modulating the TGF-β signaling
pathway (24). In previous studies,
it has been reported that plant components such as polyphenols and
miRNAs are involved in miRNA-mediated gene regulation in mice and
humans (25-27).
For example, polyphenol extracts from Hibiscus sabdariffa
were shown to regulate the expression of miR-103, miR-107, and
miR-122 and suppress fatty liver disease in hyperlipidemic mice
(25). Plant miR-171 modulated G
protein subunit α 12 signaling, including mechanistic target of
rapamycin, in human embryonic kidney, 293 cells (26). In a clinical study, grape extract
containing resveratrol increased the expression of miR-21,
miR-181b, miR-663, and miR-30c and decreased inflammatory cytokine
levels (27).
In the present study, it was hypothesized that EGCG
exerts its antifibrotic effect via miRNAs contained in EVs from
VECs, and EVs derived from human umbilical vein endothelial cells
(HUVECs) were used to test this hypothesis. In addition, miRNAs
with antifibrotic effects among the EGCG-altered miRNAs were
identified and their influence on the expression of
fibrosis-related genes was evaluated.
Materials and methods
Chemicals and materials
EGCG was purchased from Sigma-Aldrich; Merck KGaA,
and recombinant human TGF-β1 was obtained from Bio-Techne.
OptimaTM MAX-XP Ultracentrifuge, MLS-50 rotor, and
Ultra-Clear centrifuge tubes were purchased from Beckman Coulter,
Inc. miRNA mimic Negative Control, mimic #1 (cat. no. SMC-2003) and
miR-6757-3p mimic (5'-AACACUGGCCUUGCUAUCCCCA-3'; cat. no.
SMM-003-MI0022602) were obtained from Bioneer Corporation.
miR-6757-3p (cat. no. 339306; GeneGlobe ID YP02107562) and U6
primer (cat. no. 339306; GeneGlobe ID YP00203907) were purchased
from Qiagen GmbH.
Cell culture
HUVECs (product no. KE-4109) were purchased from
Kurabo Bio-Medical Department; Kurabo Industries, Ltd. and
maintained with endothelial growth medium 2 (EGM-2; Lonza Group,
Ltd.) containing 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck
KGaA). Human fetal lung fibroblasts (HFL-1; JCRB no. IFO50074) were
obtained from the Japanese Collection of Research Bioresources Cell
Bank and cultured in Dulbecco's modified Eagle's medium (DMEM;
FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% FBS.
These cells were incubated at 37˚C with 5% CO2 in a
humidified chamber.
Isolation of EVs
(ultracentrifugation)
HUVECs were preincubated in 10% FBS/EGM-2 for 24 h
and then incubated in 1% bovine serum albumin (BSA)/Medium 199
(M199) obtained respectively, from Roche Diagnostics and Gibco;
Thermo Fisher Scientific, Inc. After 2 h, HUVECs were treated with
5 µM EGCG. The dose of EGCG was determined according to previous
studies (28,29). Following a 24-h incubation with
M199, the supernatant was collected. Debris and dead cells in the
collected supernatant of the culture were removed by centrifugation
at 2,000 x g for 10 min at 4˚C, and then filtrated through a 0.2-µm
filter (Sartorius Stedim Biotech GmbH). Next, the supernatant of
the culture was ultracentrifuged at 210,000 x g for 38 min at 4˚C.
After the supernatant was aspirated, the EV pellets were washed
with PBS and ultracentrifuged (210,000 x g for 38 min at 4˚C).
Following a second aspiration, the pellets were resuspended with
PBS.
Western blotting
EVs were lysed in lysis buffer containing 50 mM
Tris-HCl (pH 7.5), 50 mM sodium fluoride, 0.15 M NaCl, 1% Triton
X-100, 1 mM EDTA, 30 mM sodium pyrophosphate, 1 mM
phenyl-methanesulfonyl fluoride, and 2 mg/ml aprotinin. Protein
concentrations were determined by bicinchoninic acid (BCA) assay.
Laemmli sample buffer containing 0.1 M Tris-HCl buffer (pH 6.8), 1%
SDS, 0.05% mercaptoethanol, 10% glycerol, and 0.001% bromophenol
blue was added and boiled (95˚C, 5 min). Proteins (5 µg/lane) were
separated by reducing 8% (weight/volume) polyacrylamide gel
electrophoresis and electroblotted on nitrocellulose blotting
membranes (Cytiva). The membranes were blocked for 1 h at room
temperature in 0.1% Tween-20/Tris-buffered saline containing 1%
bovine serum albumin (all from Nacalai Tesque, Inc.) and incubated
overnight at 4˚C in the presence of anti-CD9 antibodies (1:100,000
dilution; cat. no. sc-9148; Santa Cruz Biotechnology, Inc.). After
the membranes were washed and then incubated (25˚C, 1 h) with the
appropriate HRP-conjugated goat anti-rabbit secondary antibody
(1:10,000 dilution; cat. no. 12-348; EMD Millipore), the bands were
visualized using TMA-6 chemiluminescence reagent (Lumigen, Inc.;
Beckman Coulter, Inc.). The band images were obtained using FUSION
Solo S (Vilber-Lourmat).
Microarray analysis
Microarray analysis on a 3D-Gene® Human
miRNA Oligo chip ver. 21 (Toray Industries Inc.) was outsourced to
Toray Industries, Inc. Briefly, RNA samples were extracted from EVs
using miRNeasy mini kit (Qiagen GmbH). The RNA concentration was
measured using NanoDrop1000 (Thermo Fisher Scientific, Inc.) and
RNA fluorescence labeling was performed using a 3D-Gene®
miRNA labeling kit (cat. no. TRT-XE211; Toray Industries Inc.).
3D-Gene® Scanner (Toray Industries Inc.) was used for
scanning. Data analysis was performed as follows: The blank value
(average of the C1 blank) was subtracted from the sample
measurements and set to 0 if the value was <0. The fold change
was calculated to be equal to the average of the EGCGs divided by
the average of the control. miRNAs with two or more samples with a
value of 0 were excluded from the analysis. Binding sites of
microRNA were predicted using TargetScan Release 8.0 (https://www.targetscan.org/vert_72/).
Transfection of miRNA mimics into
HFL-1 cells
HFL-1 cells were cultured in 10% FBS/DMEM for 24 h.
Each miRNA mimic was introduced into HFL-1 cells using
LipofectamineTM RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.) as follows. The solution containing the mimics,
RNAiMAX, and DMEM was mixed mildly by pipetting. After a subsequent
incubation for 10 min at room temperature, the medium surrounding
the HFL-1 cells was replaced with the mimic solution (37˚C, 5 h).
The final concentration of the control or miR-6757-3p mimics was 20
nM. The concentration was determined according to a previous study
(30). Subsequently, 5 h after
transfection, HFL-1 cells were treated with 10% FBS/DMEM with or
without TGF-β (the concentration of TGF-β was 5 ng/ml) for 48 h.
Finally, HFL-1 cells were lysed using the TRI reagent®
(Sigma-Aldrich; Merck KGaA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted from HLF-1 cells using the TRI
Reagent® (Molecular Research Center, Inc.). cDNA
synthesis was performed using a PrimeScript RT Reagent Kit (Takara
Bio, Inc.) or miRCURY LNA RT kit (Qiagen GmbH). cDNA of mRNA was
mixed with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad
Laboratories, Inc.) and primers (Table
I). cDNA of miRNA was mixed with miRCURY LNA SYBR Green Master
Mix (Qiagen GmbH) and miR-6757-3p or U6 primers. Gene expression
was assessed using CFX96™ or CFX384™
Real-Time PCR System and CFX Maestro version 3.1.1517.0823 software
(Bio-Rad Laboratories, Inc.). The thermocycling protocol for mRNA
consisted of an initial cycle at 95˚C for 3 min, followed by 50
cycles at 95˚C for 2 sec and 60˚C for 10 sec, and finally from 65˚C
to 95˚C. The thermocycling protocol for miRNA consisted of an
initial cycle at 95˚C for 10 min, followed by 50 cycles at 95˚C for
10 sec and 60˚C for 1 min, and finally 60˚C for 30 sec after
increasing from 65˚C to 95˚C. mRNA expression was normalized to
ACTB. miRNA expression was normalized to U6. Results are shown as
relative values with the mean of the control or TGF-β + cont. mimic
group set to 1 (except for miRNA data in Fig. 2B).
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene | Forward | Reverse |
|---|
| ACTB |
5'-TGGCACCCAGCACAATGAA-3' |
5'-CTAAGTCATAGTCCGCCTAGAAGCA-3' |
| α-SMA |
5'-CCGACCGAATGCAGAAGGA-3' |
5'-ACAGAGTATTTGCGCTCCGGA-3' |
|
FIBRONECTIN |
5'-GGAGAATTCAAGTGTGACCCTCA-3' |
5'-TGCCACTGTTCTCCTACGTGG-3' |
| TGFBR1 |
5'-GACAACGTCAGGTTCTGGCTCA-3' |
5'-CCGCCACTTTCCTCTCCAAACT-3' |
Statistical analysis
Microarray data (n=3) were analyzed using Student's
t-tests and Microsoft Excel. The results in Fig. 2, Fig.
3 and Fig. 4 (n=4) are
presented as the mean ± standard error of the mean (SEM). The data
were analyzed by a Student's unpaired t-test (Fig. 2) or one-way ANOVA followed by
Dunnett's multiple comparison test (Figs. 3 and 4, vs. TGF-β + cont. mimic) using GraphPad
Prism 5.01 (GraphPad Software, Inc.). A value of P<0.05 was a
considered to indicate a statistically significant difference.
Results
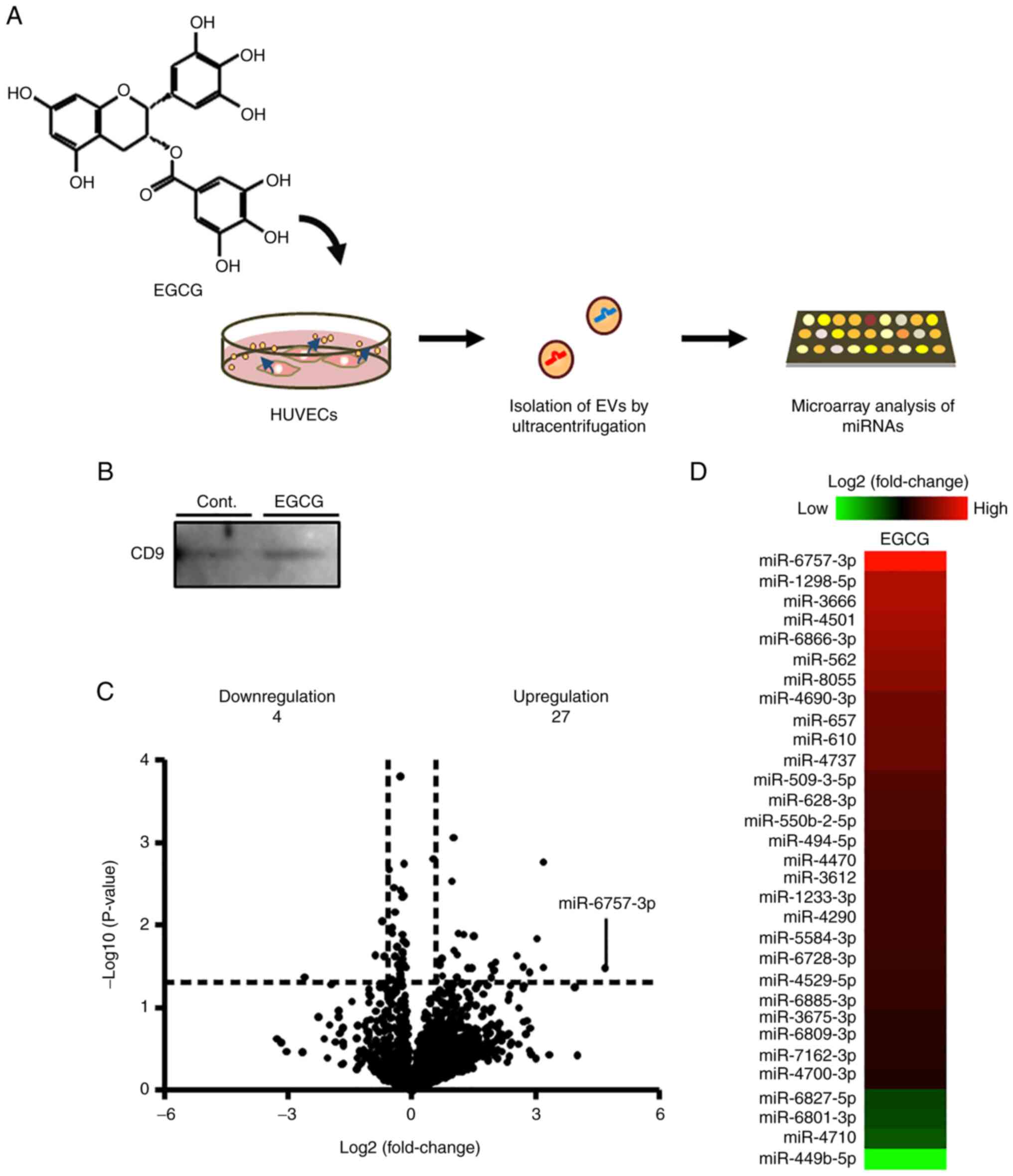
Effects of EGCG on the expression of
miRNAs in EVs derived from HUVECs
Although EGCG exhibits antifibrotic activity, its
mechanism is still unknown. Recently, miRNAs in EVs derived from
VECs were reported to regulate fibrosis (24). In the present study, miRNA
microarray analysis was performed to examine the regulatory effects
of EGCG on the expression of miRNAs in EVs from HUVECs (Fig. 1A). Western blotting revealed the
expression of CD9, an EV marker, which confirmed the successful
isolation of EVs by ultracentrifugation (Fig. 1B). Microarray analysis revealed that
EGCG upregulated 27 miRNAs and downregulated 4 miRNAs in EVs from
HUVECs (Fig. 1C and D; Tables
II and SI).
 | Table IIUpregulated and downregulated miRNAs
in extracellular vesicles of human umbilical vein endothelial cells
after treatment with EGCG. |
Table II
Upregulated and downregulated miRNAs
in extracellular vesicles of human umbilical vein endothelial cells
after treatment with EGCG.
| | Upregulation | Downregulation |
|---|
| miRNAa | Fold
changeb |
P-valuec | miRNAa | Fold
changeb |
P-valuec |
|---|
| miR-6757-3p | 25.85 | 0.033 | miR-449b-5p | 0.17 | 0.043 |
| miR-1298-5p | 9.11 | 0.002 | miR-4710 | 0.54 | 0.023 |
| miR-3666 | 9.08 | 0.033 | miR-6801-3p | 0.61 | 0.009 |
| miR-4501 | 8.20 | 0.015 | miR-6827-5p | 0.63 | 0.024 |
| miR-6866-3p | 7.20 | 0.037 | | | |
| miR-562 | 6.52 | 0.032 | | | |
| miR-8055 | 5.86 | 0.023 | | | |
| miR-4690-3p | 4.11 | 0.028 | | | |
| miR-657 | 3.98 | 0.035 | | | |
| miR-610 | 3.82 | 0.030 | | | |
| miR-4737 | 3.79 | 0.043 | | | |
| miR-509-3-5p | 2.82 | 0.013 | | | |
| miR-628-3p | 2.73 | 0.033 | | | |
| miR-550b-2-5p | 2.58 | 0.033 | | | |
| miR-494-5p | 2.39 | 0.013 | | | |
| miR-4470 | 2.32 | 0.050 | | | |
| miR-3612 | 2.19 | 0.013 | | | |
| miR-1233-3p | 2.15 | 0.043 | | | |
| miR-4290 | 2.14 | 0.023 | | | |
| miR-5584-3p | 2.12 | 0.042 | | | |
| miR-6728-3p | 2.02 | 0.020 | | | |
| miR-4529-5p | 2.02 | 0.001 | | | |
| miR-6885-3p | 1.96 | 0.003 | | | |
| miR-3675-3p | 1.67 | 0.041 | | | |
| miR-6809-3p | 1.65 | 0.025 | | | |
| miR-7162-3p | 1.59 | 0.030 | | | |
| miR-4700-3p | 1.50 | 0.047 | | | |
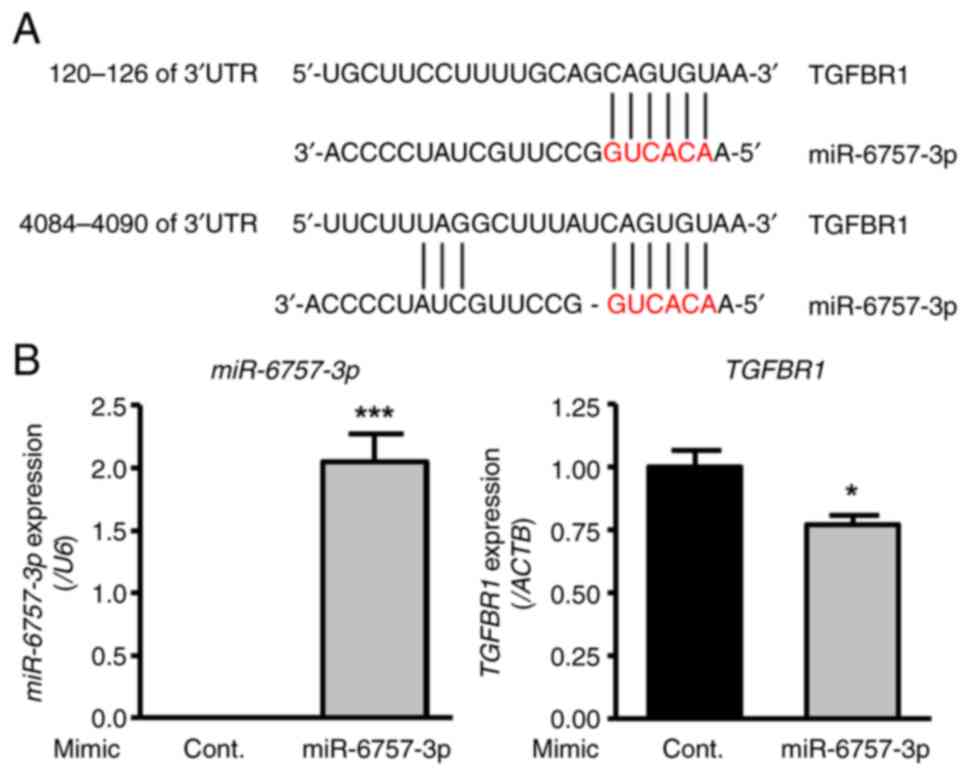
Effect of miR-6757-3p on the
expression of transforming growth factor-β receptor 1 (TGFBR1), a
candidate target gene
miRNAs with antifibrotic effects were investigated.
Among the miRNAs whose expression increased as a result of EGCG
treatment, miR-6757-3p was focused on because it showed the highest
fold change. TargetScan analysis indicated that miR-6757-3p can
target TGFBR1 (position 120-126 or 4084-4090 of 3'UTR) (Fig. 2A). In addition, TargetScan analysis
of the 26 miRNAs (excluding miR-6757-3p) that were increased in
expression by EGCG treatment revealed that nine miRNAs (miR-3666,
miR-4501, miR-657, miR-509-3-5p, miR-3612, miR-6728-3p,
miR-4529-5p, miR-6885-3p, miR-6809-3p) could target TGFBR1 (data
not shown). The TGFBR1 signaling pathway controls collagen
deposition and fibrosis (31). To
investigate whether miR-6757-3p suppresses TGFBR1 expression,
miR-6757-3p mimics were transfected into HFL-1 cells. As a result,
the transfection of miR-6757-3p mimics upregulated the expression
of miR-6757-3p and downregulated the expression level of TGFBR1
(Fig. 2B). These findings indicated
that miR-6757-3p is able to suppress TGFBR1.
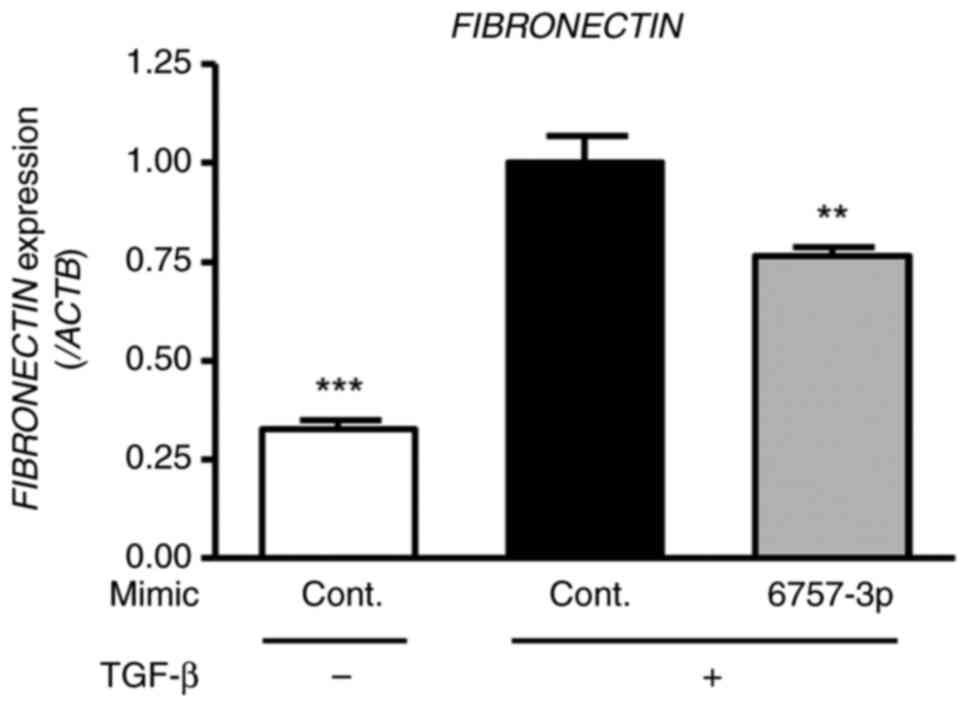
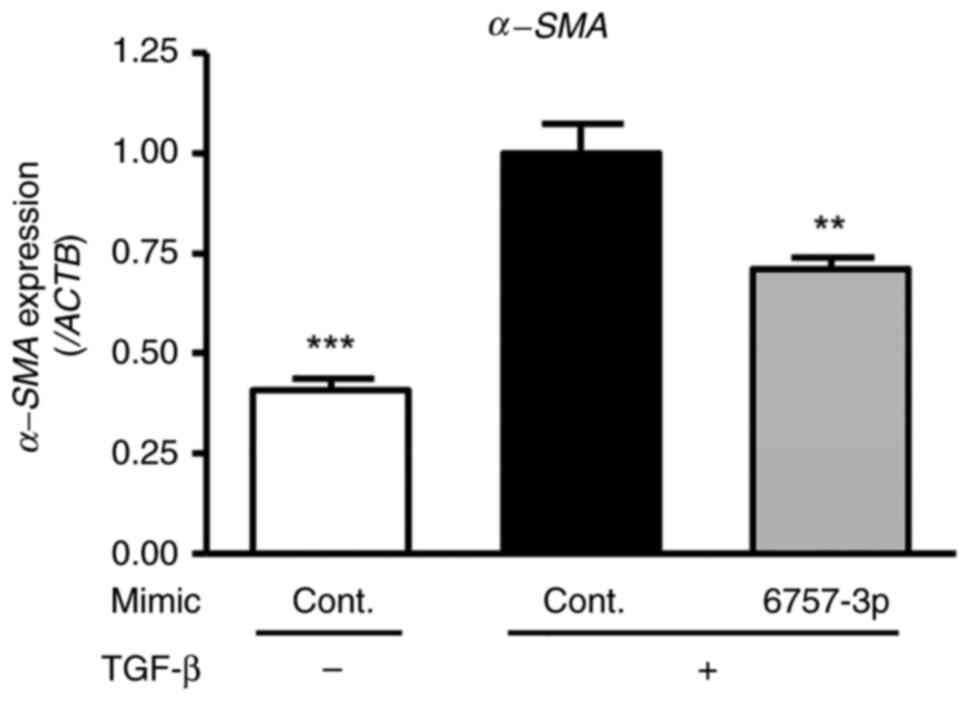
Effects of miR-6757-3p on the
expression of fibrosis-related genes in TGF-β-treated HFL-1
cells
To evaluate the impact of miR-6757-3p on
fibrosis-related gene expression, including fibronectin and
α-smooth muscle actin (α-SMA), HFL-1 cells were transfected with
miR-6757-3p mimics and treated with TGF-β (for a final
concentration of 5 ng/ml) for 48 h. The results revealed that TGF-β
treatment increased fibronectin and α-SMA expression in the HFL-1
cells. Furthermore, there was a significant difference between the
TGF-β + cont. mimic groups and the TGF-β + miR-6757-3p mimic groups
(Figs. 3 and 4). These results indicated that
miR-6757-3p downregulated the expression of fibrosis-related genes
by suppressing the expression of TGFBR1.
Discussion
In the present study, it was demonstrated that EGCG
altered the expression of 31 miRNAs (a total of 27 miRNAs were
upregulated, and 4 miRNAs were downregulated.) in EVs derived from
HUVECs. These miRNAs may be involved in the physiological effects
of EGCG. The results revealed that miR-6757-3p, which had the
highest fold change ratio after EGCG treatment, decreased
fibrosis-related gene expression in genes such as fibronectin and
α-SMA in pulmonary fibroblasts. These findings indicated that
miR-6757-3p may play a major role in activating the anti-PF effects
of EGCG.
The present study indicated that miR-6757-3p may
target TGFBR1 and downregulate the expression of fibrosis-related
genes. TGFBR1 has been reported to regulate fibrosis through the
TGF-β/Smad signaling pathway (32,33).
In mice constitutively expressing active TGFBR1 in fibroblasts,
promotion of Smad phosphorylation, myofibroblast differentiation,
and fibrosis have been observed (34). Furthermore, a previous study has
revealed that the deletion of TGFBR1 in fibroblasts suppresses
fibrosis (35). TGFBR1 has
attracted attention as a therapeutic target for fibrosis, and the
TGFBR1 kinase inhibitor galunisertib (LY2157299) has reportedly
exhibited liver regeneration-promoting and antifibrotic effects
(36). The TGFBR1 signaling pathway
has also been reported to be involved in myocardial and renal
fibrosis as well as PF (31,37).
Thus, miR-6757-3p could be a new preventive and therapeutic agent
for the treatment of fibrosis in various organs.
In addition, miR-3666, whose expression increased
via EGCG treatment in the present study, was reported to suppress
hepatic steatosis by targeting PPARγ (38). A previous study reported that EGCG
suppresses nonalcoholic fatty liver disease (39). Therefore, miR-3666 in HUVEC-derived
EVs may be associated with the antihepatic steatosis effect of
EGCG.
miRNAs in EVs play key roles in cell-to-cell
communication. It has been demonstrated that EGCG may modulate
intercellular communication via miRNAs in EVs derived from VECs and
regulate various diseases, including PF. In addition, miRNAs
contained in EVs have been reported to be correlated with the
development of diseases and expected to be used as biomarkers
(40). Considering the results
observed using miR-6757-3p, it is hypothesized that it is feasible
to use miR-6757-3p as a marker of the progression and prognosis of
PF. Moreover, the 31 miRNAs whose expression was altered in the
present study could be indicators of EGCG intake. Research into the
effect of certain foods on the expression of miRNAs in EVs is not
extensive to date, but given the results of the present study,
future research is warranted.
The various biological properties of EGCG, including
its anti-inflammatory and anticancer effects, are mediated through
its 67-kDa laminin receptor (67LR) (41,42).
EGCG has also been reported to regulate miRNA expression in
melanoma cells via 67LR (43).
Therefore, it is possible that 67LR is involved in the regulation
of the 31 miRNAs whose expression was altered by EGCG treatment in
the present study. However, further research is needed to
investigate the potential association between 67LR and these 31
miRNAs.
In conclusion, EGCG altered the expression of
various miRNAs including miR-6757-3p, which may target TGFBR1 and
decrease fibrosis-related gene expression, in EVs derived from
VECs. The results of the present study mark an important step
toward a deeper understanding of the relationship between EGCG and
functional miRNAs.
Supplementary Material
Effect of
(–)-epigallocatechin-3-O-gallate on the expression of miRNAs
in extracellular vesicles from human umbilical vein endothelial
cells.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported in part by
Grants-in-Aid for Scientific Research (KAKENHI) from the Japan
Society for the Promotion of Science to HT (grant no. JP20H05683)
and MMu (grant no. JP17H06936).
Availability of data and materials
The microarray data generated in the present study
may be found in the GEO database under accession no. GSE217849 or
at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217849.
The other data used and/or analyzed during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
MMu, YM, MK, YF, and HT were involved in the
conception and design of the study. MMu, YM, MMo, and KM performed
the experiments. YM, MMo, KM, MK, and YF analyzed the data. MMu, YM
and MK wrote the manuscript. MMu, YM, MK, and HT revised the
manuscript. MMu and YM confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The HUVECs purchased from Kurabo Bio-Medical
Department; Kurabo Industries, Ltd. are products that were isolated
from donated human tissue after obtaining permission for their use
in research applications by informed consent or legal
authorization.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wuyts WA, Agostini C, Antoniou KM, Bouros
D, Chambers RC, Cottin V, Egan JJ, Lambrecht BN, Lories R, Parfrey
H, et al: The pathogenesis of pulmonary fibrosis: a moving target.
Eur Respir J. 41:1207–1218. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Thannickal VJ, Toews GB, White ES, Lynch
JP III and Martinez FJ: Mechanisms of pulmonary fibrosis. Annu Rev
Med. 55:395–417. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Richeldi L, Collard HR and Jones MG:
Idiopathic pulmonary fibrosis. Lancet. 389:1941–1952.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bouros D and Antoniou KM: Current and
future therapeutic approaches in idiopathic pulmonary fibrosis. Eur
Respir J. 26:693–703. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Luppi F, Cerri S, Beghè B, Fabbri LM and
Richeldi L: Corticosteroid and immunomodulatory agents in
idiopathic pulmonary fibrosis. Respir Med. 98:1035–1044.
2004.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zamora-Ros R, Achaintre D, Rothwell JA,
Rinaldi S, Assi N, Ferrari P, Leitzmann M, Boutron-Ruault MC,
Fagherazzi G, Auffret A, et al: Urinary excretions of 34 dietary
polyphenols and their associations with lifestyle factors in the
EPIC cohort study. Sci Rep. 6(26905)2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
van Dam RM, Naidoo N and Landberg R:
Dietary flavonoids and the development of type 2 diabetes and
cardiovascular diseases: Review of recent findings. Curr Opin
Lipidol. 24:25–33. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang X, Ouyang YY, Liu J and Zhao G:
Flavonoid intake and risk of CVD: A systematic review and
meta-analysis of prospective cohort studies. Br J Nutr. 111:1–11.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xing L, Zhang H, Qi R, Tsao R and Mine Y:
Recent advances in the understanding of the health benefits and
molecular mechanisms associated with green tea polyphenols. J Agric
Food Chem. 67:1029–1043. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Azambuja JH, Mancuso RI, Via FID, Torello
CO and Saad STO: Protective effect of green tea and
epigallocatechin-3-gallate in a LPS-induced systemic inflammation
model. J Nutr Biochem. 101(108920)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sae-tan S, Grove KA and Lambert JD: Weight
control and prevention of metabolic syndrome by green tea.
Pharmacol Res. 64:146–154. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tsai CF, Hsu YW, Ting HC, Huang CF and Yen
CC: The in vivo antioxidant and antifibrotic properties of green
tea (Camellia sinensis, Theaceae). Food Chem. 136:1337–1344.
2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wei H, Ge Q, Zhang LY, Xie J, Gan RH, Lu
YG and Zheng DL: EGCG inhibits growth of tumoral lesions on lip and
tongue of K-Ras transgenic mice through the Notch pathway. J Nutr
Biochem. 99(108843)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang M, Zhong H, Zhang X, Huang X, Wang J,
Li Z, Chen M and Xiao Z: EGCG promotes PRKCA expression to
alleviate LPS-induced acute lung injury and inflammatory response.
Sci Rep. 11(11014)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Meng J, Chen Y, Wang J, Qiu J, Chang C, Bi
F, Wu X and Liu W: EGCG protects vascular endothelial cells from
oxidative stress-induced damage by targeting the
autophagy-dependent PI3K-AKT-mTOR pathway. Ann Transl Med.
8(200)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Sriram N, Kalayarasan S and Sudhandiran G:
Epigallocatechin-3-gallate exhibits anti-fibrotic effect by
attenuating bleomycin-induced glycoconjugates, lysosomal hydrolases
and ultrastructural changes in rat model pulmonary fibrosis. Chem
Biol Interact. 180:271–280. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sriram N, Kalayarasan S and Sudhandiran G:
Enhancement of antioxidant defense system by
epigallocatechin-3-gallate during bleomycin induced experimental
pulmonary fibrosis. Biol Pharm Bull. 31:1306–1311. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cai Y, Yu X, Hu S and Yu J: A brief review
on the mechanisms of miRNA regulation. Genomics Proteomics
Bioinformatics. 7:147–154. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shenoy A and Blelloch RH: Regulation of
microRNA function in somatic stem cell proliferation and
differentiation. Nat Rev Mol Cell Biol. 15:565–576. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Jordan SD, Krüger M, Willmes DM, Redemann
N, Wunderlich FT, Brönneke HS, Merkwirth C, Kashkar H, Olkkonen VM,
Böttger T, et al: Obesity-induced overexpression of miRNA-143
inhibits insulin-stimulated AKT activation and impairs glucose
metabolism. Nat Cell Biol. 13:434–446. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Nejad C, Stunden HJ and Gantier MP: A
guide to miRNAs in inflammation and innate immune responses. FEBS
J. 285:3695–3716. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
O'Reilly S: MicroRNAs in fibrosis:
Opportunities and challenges. Arthritis Res Ther.
18(11)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bayraktar R, Van Roosbroeck K and Calin
GA: Cell-to-cell communication: microRNAs as hormones. Mol Oncol.
11:1673–1686. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xie H, Gao YM, Zhang YC, Jia MW, Peng F,
Meng QH and Wang YC: Low let-7d exosomes from pulmonary vascular
endothelial cells drive lung pericyte fibrosis through the
TGFβRI/FoxM1/Smad/β-catenin pathway. J Cell Mol Med.
24:13913–13926. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Joven J, Espinel E, Rull A, Aragonès G,
Rodríguez-Gallego E, Camps J, Micol V, Herranz-López M, Menéndez
JA, Borrás I, et al: Plant-derived polyphenols regulate expression
of miRNA paralogs miR-103/107 and miR-122 and prevent diet-induced
fatty liver disease in hyperlipidemic mice. Biochim Biophys Acta.
1820:894–899. 2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gismondi A, Nanni V, Monteleone V, Colao
C, Di Marco G and Canini A: Plant miR171 modulates mTOR pathway in
HEK293 cells by targeting GNA12. Mol Biol Rep. 48:435–449.
2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cione E, La Torre C, Cannataro R, Caroleo
MC, Plastina P and Gallelli L: Quercetin, epigallocatechin gallate,
curcumin, and resveratrol: From dietary sources to human MicroRNA
modulation. Molecules. 25(63)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ou HC, Song TY, Yeh YC, Huang CY, Yang SF,
Chiu TH, Tsai KL, Chen KL, Wu YJ, Tsai CS, et al: EGCG protects
against oxidized LDL-induced endothelial dysfunction by inhibiting
LOX-1-mediated signaling. J Appl Physiol (1985). 108:1745–1756.
2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kanlaya R, Peerapen P, Nilnumkhum A,
Plumworasawat S, Sueksakit K and Thongboonkerd V:
Epigallocatechin-3-gallate prevents TGF-β1-induced
epithelial-mesenchymal transition and fibrotic changes of renal
cells via GSK-3β/β-catenin/Snail1 and Nrf2 pathways. J Nutr
Biochem. 76(108266)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Marugame Y, Takeshita N, Yamada S,
Yoshitomi R, Kumazoe M, Fujimura Y and Tachibana H: Sesame lignans
upregulate glutathione S-transferase expression and downregulate
microRNA-669c-3p. Biosci Microbiota Food Health. 41:66–72.
2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tan Z, Jiang X, Zhou W, Deng B, Cai M,
Deng S, Xu Y, Ding W, Chen G, Chen R, et al: Taohong siwu decoction
attenuates myocardial fibrosis by inhibiting fibrosis proliferation
and collagen deposition via TGFBR1 signaling pathway. J
Ethnopharmacol. 270(113838)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xu Z, He B, Jiang Y, Zhang M, Tian Y, Zhou
N, Zhou Y, Chen M, Tang M, Gao J and Peng F: Igf2bp2 knockdown
improves CCl4-induced liver fibrosis and TGF-β-activated
mouse hepatic stellate cells by regulating Tgfbr1. Int
Immunopharmacol. 110(108987)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Schnaper HW, Hayashida T and Poncelet AC:
It's a Smad world: Regulation of TGF-beta signaling in the kidney.
J Am Soc Nephrol. 13:1126–1128. 2002.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Sonnylal S, Denton CP, Zheng B, Keene DR,
He R, Adams HP, Vanpelt CS, Geng YJ, Deng JM, Behringer RR and de
Crombrugghe B: Postnatal induction of transforming growth factor
beta signaling in fibroblasts of mice recapitulates clinical,
histologic, and biochemical features of scleroderma. Arthritis
Rheum. 56:334–344. 2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Khalil H, Kanisicak O, Prasad V, Correll
RN, Fu X, Schips T, Vagnozzi RJ, Liu R, Huynh T, Lee SJ, et al:
Fibroblast-specific TGF-β-Smad2/3 signaling underlies cardiac
fibrosis. J Clin Invest. 127:3770–3783. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Masuda A, Nakamura T, Abe M, Iwamoto H,
Sakaue T, Tanaka T, Suzuki H, Koga H and Torimura T: Promotion of
liver regeneration and anti-fibrotic effects of the TGF-β receptor
kinase inhibitor galunisertib in CCl4-treated mice. Int J Mol Med.
46:427–438. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li J, Yue S, Fang J, Zeng J, Chen S, Tian
J, Nie S, Liu X and Ding H: MicroRNA-10a/b inhibit
TGF-β/Smad-induced renal fibrosis by targeting TGF-β receptor 1 in
diabetic kidney disease. Mol Ther Nucleic Acids. 28:488–499.
2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Mittal S, Inamdar S, Acharya J, Pekhale K,
Kalamkar S, Boppana R and Ghaskadbi S: miR-3666 inhibits
development of hepatic steatosis by negatively regulating PPARγ.
Biochim Biophys Acta Mol Cell Biol Lipids.
1865(158777)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Naito Y, Ushiroda C, Mizushima K, Inoue R,
Yasukawa Z, Abe A and Takagi T: Epigallocatechin-3-gallate (EGCG)
attenuates non-alcoholic fatty liver disease via modulating the
interaction between gut microbiota and bile acids. J Clin Biochem
Nutr. 67:2–9. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Salehi M and Sharifi M: Exosomal miRNAs as
novel cancer biomarkers: Challenges and opportunities. J Cell
Physiol. 233:6370–6380. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Byun EB, Kim WS, Sung NY and Byun EH:
Epigallocatechin-3-gallate regulates anti-inflammatory action
through 67-kDa laminin receptor-mediated tollip signaling induction
in lipopolysaccharide-stimulated human intestinal epithelial cells.
Cell Physiol Biochem. 46:2072–2081. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Tachibana H, Koga K, Fujimura Y and Yamada
K: A receptor for green tea polyphenol EGCG. Nat Struct Mol Biol.
11:380–381. 2004.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yamada S, Tsukamoto S, Huang Y, Makio A,
Kumazoe M, Yamashita S and Tachibana H:
Epigallocatechin-3-O-gallate up-regulates microRNA-let-7b
expression by activating 67-kDa laminin receptor signaling in
melanoma cells. Sci Rep. 6(19225)2016.PubMed/NCBI View Article : Google Scholar
|