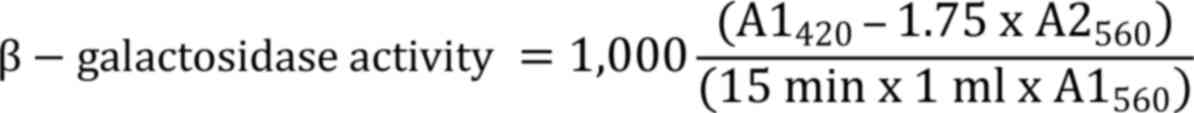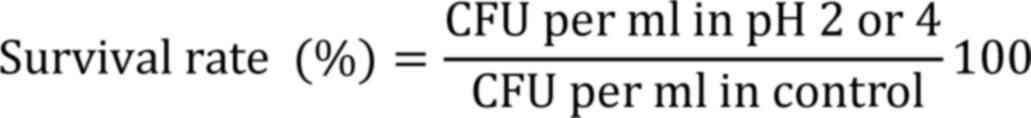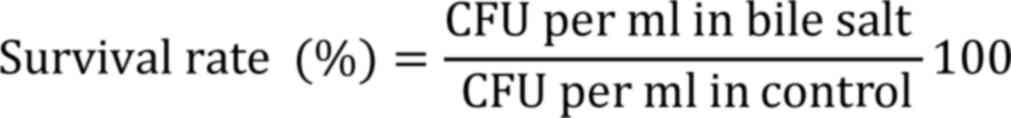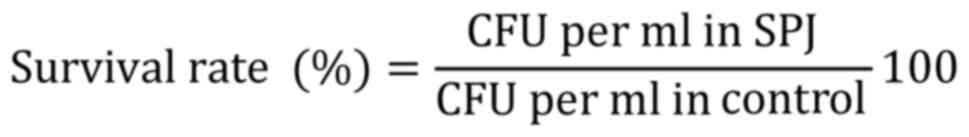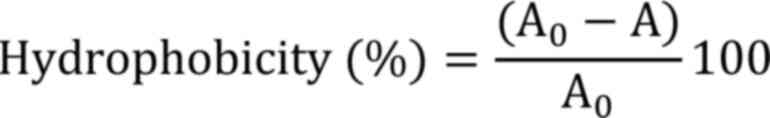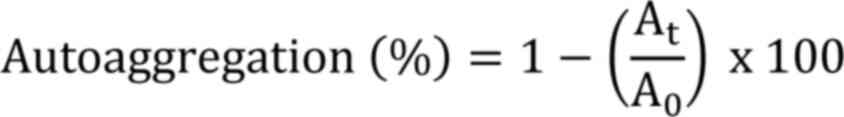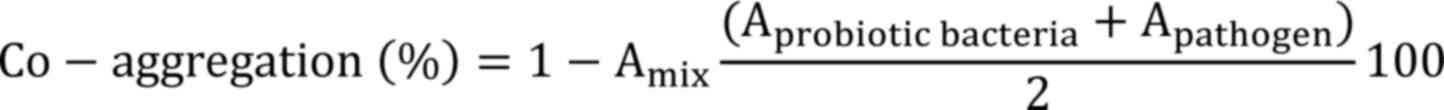Introduction
Lactose intolerance (LI) is a typical condition of
dairy food intolerance, that occurs generally when lactase activity
is decreased in the brush border of human small intestinal mucosa.
LI prevalence shows diversity among regions, human populations,
continents and across the globe (1,2). It
has been observed that 70% of the global human population exhibit
transient lactase activity without LI symptoms, which is influenced
by nutritional and genetic factors (1). Prevalence of lactase non-persistence
condition in Asian and African countries ranges between 80-100%,
however, among Northern European countries the prevalence of LI is
observed to be very low (2,3). Furthermore, hypolactasia in the Asian
continent has rarely been reported, while in the Western world it
is relatively prevalent (1). In the
Indian subcontinent, particularly the northern region, the
frequency of maldigesters was reported to be 48% per 200 subjects
during a breath test, while in the southern region it was observed
to be higher (66%) (4). It has been
documented that, Indo-Aryan migration brought the lactase
persistence (LP) trait to northern India, which was later spread by
intermixing of the native population (2). Thus, it is of interest to study
whether the distribution of the genetic marker responsible for the
LP trait varies between northern and southern Indian populations.
In addition, in Northwest Russia lactase non-persistence ranges
between 16-23% (1-4).
Thus, LI management is a worldwide issue in terms of public health
management.
The most essential enzyme in the dairy sector for
developing low-lactose food stuffs to overcome LI is
β-galactosidase (EC 3.2.1.23) and it is commercially manufactured
from microorganisms such as bacteria, yeast, and fungus. Although
chemically-synthesized enzymes are gaining significance, bacterial
enzymes are preferred as they exhibit high activity and stability
(5). β-galactosidase enzyme is
produced in the small intestine and its deficiency can cause LI.
Diarrhea, stomach discomfort, distention, flatus, and borborgygmi
are typical symptoms of LI, affecting both infants and adults, and
manifest after 30-120 min of lactose absorption (6). Primary deficiency, also known as
hypolactasia, is characterized by partial or complete lack of
β-galactosidase in the small intestine of children of various ages,
while 70% of the population of the world exhibit evidence of usual
symptoms in late adolescence and adulthood. Damage to the small
intestine induced by an overgrowth of enterobacteria in secondary
deficiency results in the destruction of small intestinal cells.
Despite the fact that β-galactosidase is a non-inducible enzyme, it
is found in the jejunum and produced in the microvillus membrane of
the small intestine, where lactose digestion leads to the
production of glucose and galactose monosaccharide, and is absorbed
by enterocytes (7).
Probiotics are live microorganisms, which when
administered in an adequate amount, confer health benefits to the
host (FAO/WHO) (8). The mechanism
of action of beneficial organisms include, competition with
pathogens for adhesion and nutrients, and in addition to the
production of antimicrobial metabolites, enhance host immunity
against pathogens in the gut. The association between human health
and probiotic gut microbiota has been thoroughly studied, with a
particular emphasis on homeostatic and barrier function (9). A wide range of metabolites are
produced from probiotic bacteria, including nonspecific fatty
acids, and highly specific bacteriocins with antimicrobial
properties. Previous studies have shown that probiotic bacteria are
progressively renowned as a means for alleviating intestinal
disorders and treatments have been successful in mouse models for
certain clinical intestinal disorders (10,11).
The most prominent probiotics are Lactiplantibacillus and
Bifidobacteria genera (10,11)
which can be used for prevention and management of numerous
disorders including diarrhea, rotaviral diarrhea, Helicobacter
pylori infection, hyperlipidemia, colitis, acute and chronic
gastroenteritis, irritable bowel syndrome, inflammatory bowel
disease, cirrhosis, pouchitis, vaginosis and maldigestion-related
conditions such as LI, milk protein allergy and soy protein allergy
(6).
In the present study, some of the potential lactic
acid bacterial isolates were used to alleviate LI and analysed for
probiotic potentiality. β-galactosidase-producing lactic acid
bacteria (LAB) were isolated and assessed for acid and bile
tolerance, antibiotic susceptibility, antimicrobial activity,
auto-aggregation and co-aggregation abilities, cell-surface
hydrophobicity, and HT-29 cell adhesion and invasion assays. The
16S rRNA gene consists of highly conserved nucleotide sequences
that can be used to distinguish closely related bacterial species
and to determine the taxonomy and phylogeny of unknown bacteria by
comparing the obtained sequence to known sequences of other
bacteria in the GenBank database (12,13).
Thus, molecular characterization of selected LAB isolates using 16S
rRNA sequence analysis was performed.
Materials and methods
Isolation of LAB from homemade curd
samples
A total of 30 homemade curd samples were collected
from different rural regions of Karnataka state, India and the
samples were stored at 4˚C until further use. With all aseptic
precautions, the samples were homogenized, serially diluted
(tenfold), 0.1 ml of the sample was plated on de Man Rogosa Sharpe
(MRS) agar (Himedia Laboratories Pvt, Ltd.) and incubated for 24 to
48 h at 37˚C. Bacterial colonies developed on MRS media were
serially subcultured by following microdilution technique and pure
cultures were preserved at 4˚C/MRS agar slants.
Screening of β-galactosidase-producing
LAB
A total of 450 LAB isolates were inoculated with MRS
agar medium supplemented with 60 µl X-Gal (20 mg/ml in DMSO;
Himedia Laboratories Pvt, Ltd.) as a chromogenic substrate and 10
µl of iso-propyl-thio-β-D-galactopyranoside (IPTG) (Himedia
Laboratories Pvt, Ltd.) as an inducer for the β-galactosidase.
Following incubation for 48 h at 37˚C, development of blue colonies
indicated β-galactosidase enzyme activity (10,14).
Quantitative assay for
β-galactosidase
β-galactosidase assay of eight isolates was
performed (15,16). Briefly, selected isolates were
centrifuged at 12,000 x g for 5 min at 4˚C (Eppendorf AG 22331;
Eppendorf SE) and washed twice in phosphate-buffered saline (PBS)
and cells were adjusted to 1.0 (560 nm). Furthermore, cells were
permeabilized with 50 µl of toluene/acetone (1:9 v/v) (HiMedia
Laboratories Pvt, Ltd.), vortexed for 7 min and then 100 µl of cell
suspension was added to a tube consisting of 900 µl of phosphate
buffer and 200 µl of O-nitrophenyl-β-D-galactopyranoside
(ONPG; 4 mg/ml) solution (Himedia Laboratories Pvt, Ltd.).
Additionally, after a 15-min incubation period at 37˚C, 0.5 ml of 1
M Na2CO3 was added to terminate the reaction,
and absorbance values at 420 and 560 nm (NanoDrop 2000C
UV-Spectrophotometer; Thermo Fisher Scientific, Inc.) were
recorded, with β-galactosidase activity represented in Miller
units:
Where, A1560 denotes the absorbance
before the test and A2560 denotes the absorbance of the
reaction mixture.
Phenotypic and molecular
characterization
β-galactosidase-producing isolates were identified
by colony characteristics viz., size, shape, color and
texture. Furthermore, the isolates were subjected to biochemical
tests and characterized based on Bergey's manual of systematic
bacteriology (17). Furthermore,
carbohydrate fermentation was performed for species level
identification using glucose, fructose, sucrose, galactose,
lactose, maltose, cellobiose, xylose, arabinose, rhamnose,
mannitol, and sorbitol sugars as previously described (18,19).
Identification by 16S rRNA gene
sequence
From the selected bacterial isolates, DNA was
isolated using the CTAB protocol (20) and the extraction was confirmed by
electrophoresis using 0.8% agarose gel. The PCR reaction mixture
contained 400 ng of forward primer and 400 ng reverse primer, 4 µl
10X dNTPs (2.5 mM each), 10 µl of DNA polymerase assay buffer and
3U of 1 µl Taq DNA polymerase enzyme (Sigma-Aldrich; Merck KGaA).
For 16S rRNA gene amplification, prokaryotic universal primers
[Pair 1: ~1,500 bp amplification, forward primer (395),
5'-GGATGAGCCCGCGGCCTA-3' and reverse primer (396),
5'-CGGTGTGTACAAGGCCCGG-3'; Pair 2: ~1,300 bp amplification, forward
primer (63F), 5'-CAGGCCTAACACATGCAAGTC-3' and reverse primer
(1387R), 5'-GGCGGATGTGTACAAGGC-3'] were used and this experiment
was performed at CellKraft Biotech Pvt, Ltd., and designed, using
an ABI thermal cycler (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with the program: Denaturation at 94˚C for 5 min,
followed by 35 cycles at 94˚C for 1 min, annealing at 55˚C for 1
min, and extension at 72˚C for 2 min, and a final extension for 7
min at 72˚C. Amplification was confirmed by electrophoresis of PCR
products using 1% agarose gel and then sequenced by Sanger
sequencing method with an ABI 3130 genetic analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). 16S rRNA sequences
were compared using the Basic Local Alignment Search Tool (BLASTn)
program of the National Center for Biotechnology Information (NCBI)
database. Furthermore, the multiple sequence alignment was aligned
using the MUSCLE program (https://www.drive5.com/muscle/) and the phylogenetic
tree was constructed by the neighbor-joining method using MEGA-X
software (https://www.megasoftware.net/). Partial nucleotide
sequences of 16S rRNA of the identified Lactiplantibacillus
isolates were deposited in the NCBI/GenBank (12,13).
In vitro evaluation for potential
probiotic bacteria
In vitro studies were undertaken to evaluate
the probiotic potentiality of β-galactosidase-producing isolates as
per FAO/WHO (8). All eight isolates
were subjected to simulated gastric juice and bile tolerance tests.
The simulated gastric juice contained 0.3% w/v pepsin and 0.5% w/v
NaCl (Himedia Laboratories Pvt, Ltd.), at pH 2 or 4. A total of 1
ml of cell suspension was centrifuged (12,000 x g, 5 min at 5˚C),
inoculated into 10 ml of gastric juice with pH 2 or 4, incubated at
37˚C for 3 h and cell viability was measured as colony-forming
units (CFU) by plating technique, and the percentage of survival
was calculated as follows:
Bile tolerance was determined by inoculating each
strain (1% v/v) into MRS broth with 0.3% (w/v) of bile salt
(Oxgall; Himedia Laboratories Pvt, Ltd.) and incubated for 3 h at
37˚C. Viability was measured as CFU by plating technique and
compared with the control (without bile salt) (13,21).
Pancreatic enzyme tolerance was calculated according
to a study by Rashmi and Gayathri (13), with slight modifications. Overnight
cultures were centrifuged at 6,000 x g (20 min at 5˚C), inoculated
into simulated pancreatic juice (SPJ; bile 3 g/l and pancreatin 0.1
g/l, sodium phosphate dibasic heptahydrate 50.81 g/l, and NaCl 8.5
g/l in a KH2PO4 buffer at pH 8.0; Himedia
Laboratories Pvt, Ltd.), incubated at 37˚C for 3 h and the
percentage of survival was calculated as follows:
Hemolytic activity
Sheep blood agar (Himedia Laboratories Pvt, Ltd.)
was used for inoculation of selected LAB isolates and incubated for
48 h at 37˚C, and then plates were observed for α, β, or γ
hemolysis (22,23).
Hydrophobicity
For the hydrophobicity assay, two different solvents
viz., non-polar solvent-xylene and polar solvent-chloroform
was used. Centrifuged cells (12,000 x g for 5 min at 5˚C) were
suspended in 50 mM K2HPO4 (pH 6.5) buffer and
adjusted to OD 1.0 at A600 nm and 3 ml of the bacterial
suspensions were mixed with 1 ml of solvent and allowed to stand at
room temperature for 20 min (13,15).
The percentage of bacterial adhesion to the solvent was measured
using the aqueous phase at 600 nm.
Where, A0 and A are the absorbance before
incubation and after incubation, respectively.
Antimicrobial activity against human
pathogenic bacteria
Antimicrobial activity using crude secondary
metabolites of LAB against selected pathogenic bacteria was
performed using the agar well diffusion technique. Cell-free
supernatants (CFS) of each bacterial isolate were prepared and
adjusted to pH 6.5. Certain selected strains of pathogenic bacteria
viz., Escherichia coli (MTCC no. 433), Staphylococcus
aureus (ATCC no. 6538), Pseudomonas aeruginosa (ATCC no.
9027), Salmonella abony (ATCC no. BAA2162) and Listeria
monocytogenes (L. monocytogenes; MTCC no. 1143), were
purchased from the American Type Culture Collection (ATCC) and
Microbial Type Culture Collection (MTCC; IMTech), and 0.1 ml of
pathogens were inoculated onto Mueller Hinton agar media (Himedia
Laboratories Pvt, Ltd.). Furthermore, 7-mm diameter wells were made
in the agar plates and 100 µl of CFS was placed into these wells,
and finally the inhibition zone was measured (mm) after 48 h of
incubation at 37˚C.
Antibiotic sensitivity test
Antibiotic disc diffusion method was performed
according to Kumara et al (23) with modifications. A total of 0.1 ml
of each selected isolate was inoculated onto MRS agar media and
antibiotic disc containing penicillin (P) 10 mcg, erythromycin (E)
15 mcg, ampicillin (AMP) 10 mcg, amikacin (AK) 30 mcg, ofloxacin
(OF) 5 mcg, cefixime (CFM) 5 mcg, ciprofloxacin (CIP) 5 mcg, and
azithromycin (AZM) 15 mcg (Himedia Laboratories Pvt, Ltd.) were
placed and incubated for 24 h at 37˚C. Subsequently, the diameter
of the inhibition zone was measured in mm.
Auto-aggregation and co-aggregation
assays
The ability of bacteria to auto-aggregate and
co-aggregate was assessed according to Armas et al (22) with slight modifications. Stationary
phase cells were centrifuged (5,000 x g for 15 min at 5˚C), pellets
were washed thrice and suspended in PBS and the OD was adjusted to
1 (equivalent to 109 CFU m/l) at 600 nm. The bacterial
suspension (4 ml) was incubated at 37˚C and monitored at a
different time intervals (0 to 5 h) and 0.1 ml of upper suspension
was removed and mixed with 3.9 ml of PBS. The percentage of
absorbance was then measured at 600 nm using the following
formula:
Where, At: Absorbance of the upper layer
mix at a particular time (1 to 5 h).
A0: Absorbance at time zero.
To determine co-aggregation, equal volumes (5 ml;
1:1) of each selected isolate and pathogens [E. coli (MTCC
no. 433) and L. monocytogenes (MTCC no. 1143] were incubated
together at 37˚C for 5 h without disturbance. The absorbance was
then calculated at 600 nm and the percentages of co-aggregation
were determined as follows:
Where, Aprobiotic bacteria: Absorbance of
the Lactiplantibacillus isolates as control,
Apathogen: Absorbance of the pathogen as
a control
Amix: Absorbance of both probiotic
bacteria and the pathogen in a single tube.
Adhesion assay
Bacterial adhesion with human colon cancer cells was
performed (18,24) with some modifications. The human
HT-29 cell line (ATCC no. HTB-38; ATCC) was used, and the cell
culture work was carried out at the Central Research Laboratory,
SDM College of Medical Sciences and Hospital (Dharwad, India).
Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine
serum (both from Himedia Laboratories Pvt, Ltd.) were used to grow
HT-29 cells in 12-well flat-bottom cell culture plates until they
reached 80% confluence. Prior to the experiment, HT-29 cells were
washed gently with PBS twice. Subsequently, the selected eight
isolates were centrifuged (5,000 x g for 15 min at 4˚C) and
suspended in DMEM without antibiotics to provide approximately
109 CFU ml of the bacterial suspension. Additionally,
200 µl of each strain was added to separate wells and incubated for
2 h at 37˚C in 5% CO2 atmosphere. The HT-29 cells were
then washed twice in sterile PBS to remove non-adherent bacteria
before being lysed in 2 ml of 0.1% Triton X-100 in PBS. Cell
lysates were tenfold serially diluted and plated with MRS agar and
incubated for 24 h at 37˚C. The percentage of adherence was
expressed using the formula:
Gram's staining for cell adhesion
assay
The adhesion of all eight isolates to HT-29 cells in
cell culture plates was assessed using the methanol fix technique
for microscopic analysis. Each well received 3 ml of methanol,
which was allowed to stand for 10 min. Furthermore, fixed cells
were stained with Gram's solution (at 28˚C for 5 min) and examined
under oil immersion objective (Olympus Corporation) (24,25).
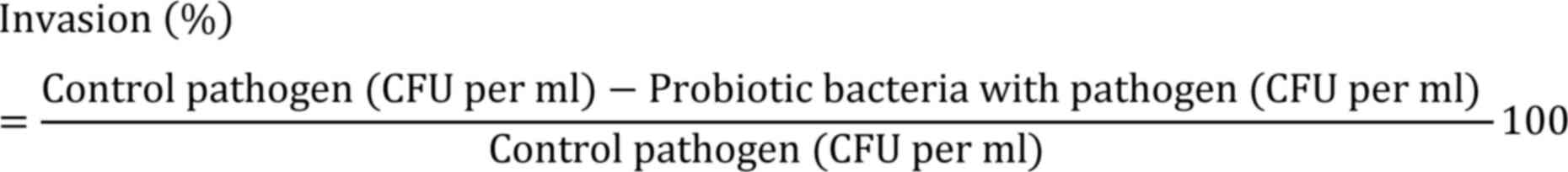
Invasion assay
In vitro methods evaluated the ability of the
eight selected isolates to inhibit the colonization of human
pathogens with intestinal cells (13,26).
HT-29 cells and the eight isolates were inoculated at a
concentration of 109 CFU per well in antibiotic-free
DMEM and incubated for 2 h. L. monocytogenes (MTCC no. 1143
and E. coli (MTCC no. 433) (109 CFU/well) were
inoculated into each well with the antibiotic-free medium and
incubated for 1 h at 37˚C with 5% CO2. Furthermore,
extracellular bacteria in the well were eliminated by transferring
DMEM to 10% FBS with streptomycin 100 U/ml and incubating for 1 h.
Subsequently, 2 ml of 0.1% Triton X-100 in PBS was used to lyse the
treated HT-29 cells. Additionally, to count invading bacteria, 0.1
ml of 10-4, 10-5 and 10-6
dilutions of cell lysates were inoculated with brain heart infusion
agar (Himedia Laboratories Pvt, Ltd.). Invasion assays were
performed on cell lines that had only been exposed to pathogens.
The invasion percentage was determined using the following
formula:
Statistical analysis
All experiments were conducted in triplicate and the
results were reported as the mean ± standard error of the mean
(SEM). Statistical analyses were performed using GraphPad Prism 9
(GraphPad Software, Inc.). Differences between multiple groups were
compared using one-way ANOVA with post hoc Tukey's multiple
comparison tests, Brown-Forsythe test and Bartlett's test and
two-way ANOVA of grouped multiple t-tests. P<0.05 was considered
to indicate a statistically significant difference.
Results
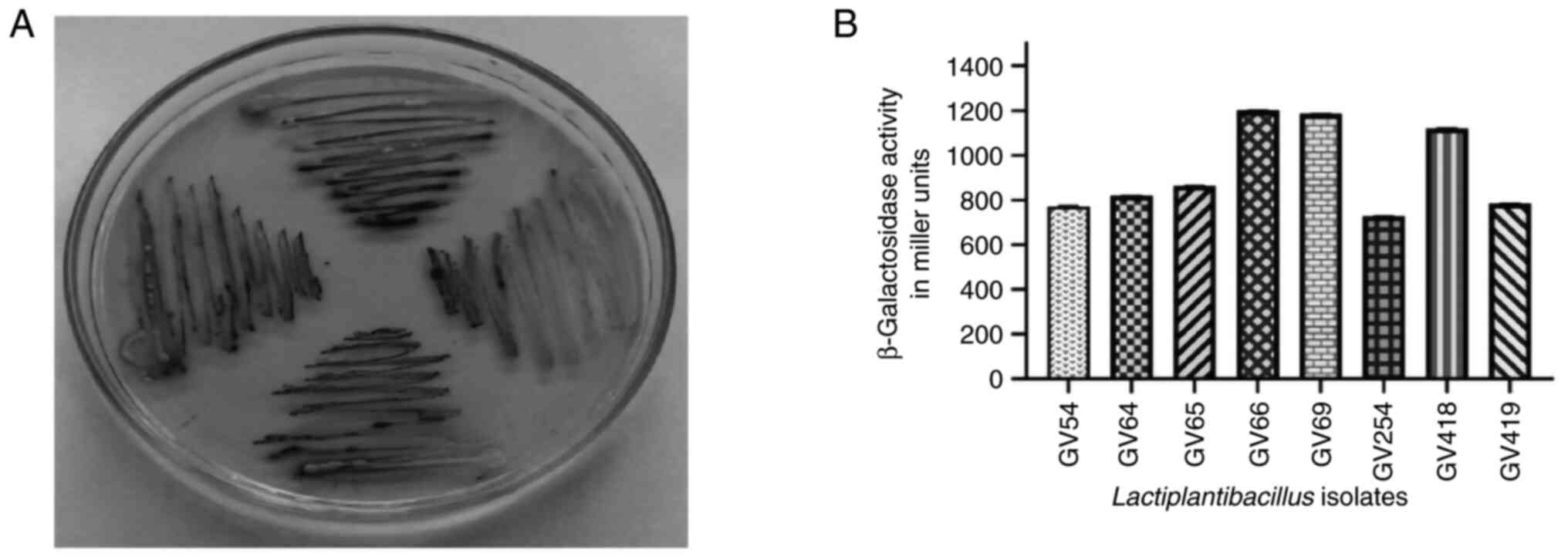
Isolation and screening of
β-galactosidase-producing bacteria
A total of 450 LAB isolates were isolated from
homemade curd samples collected from various regions of Karnataka
(12.97 N 77.50 E), India and all isolates were screened for the
β-galactosidase enzyme by qualitative assay using X-gal plates
(Fig. 1A). Development of
blue-green colored bacterial colonies were selected and the
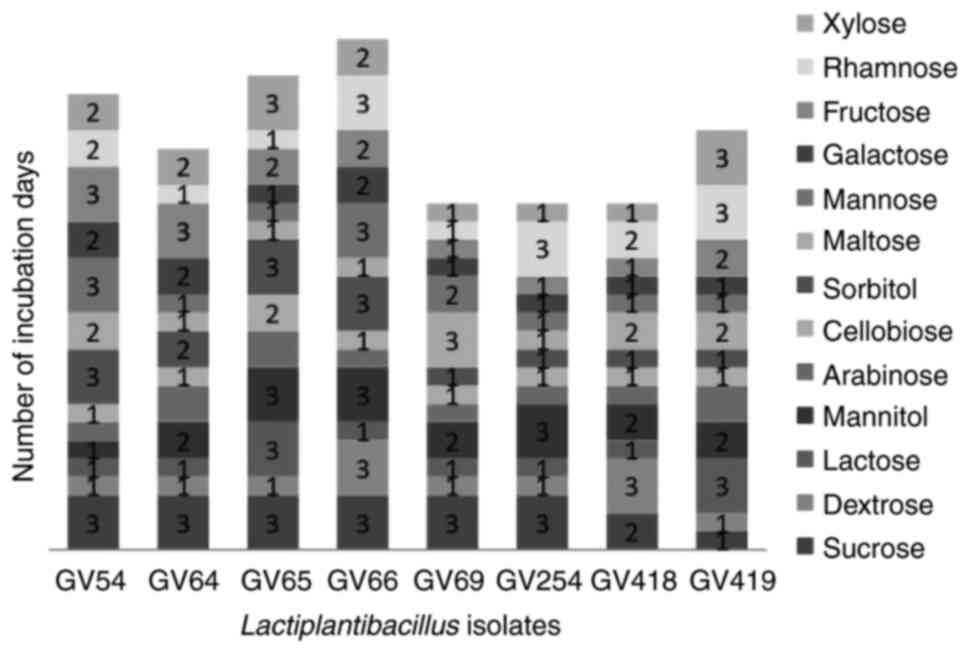
presumptive tests confirmed that selected isolates were
Gram-positive, non-motile, non-spore producing Bacillus and
the carbohydrate fermentation pattern varied among the isolates
(Fig. 2). Based on the quantitative
assay of β-galactosidase activity level ranging from 728.25 to
1,203.32 (U/ml) Miller units (P<0.05), it was revealed that
Lactiplantibacillus sp. GV66 had the highest value of
1,203.32 (U/ml) and L. fermentum GV254 had the lowest value
of 728.25 (U/ml) Miller units.
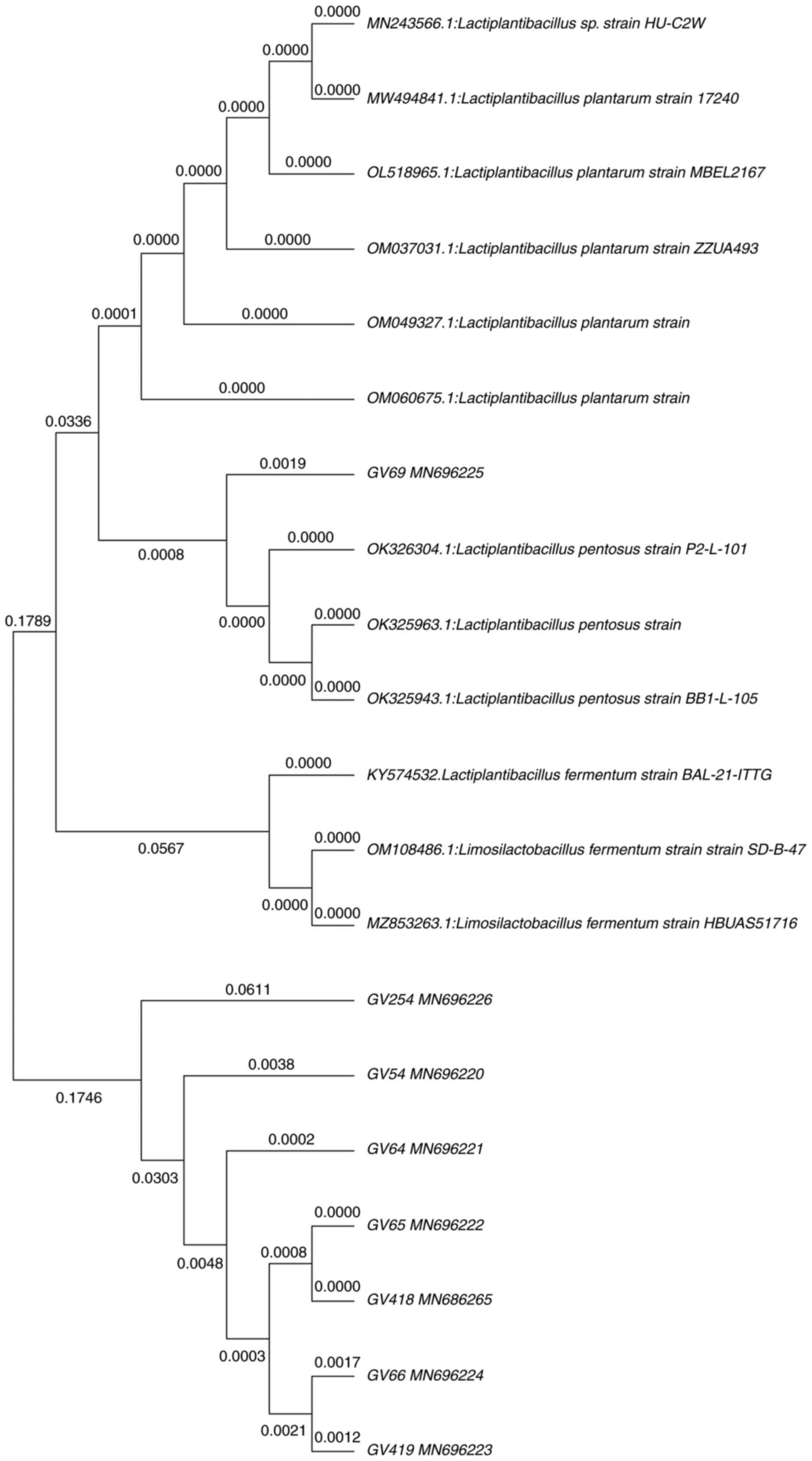
Identification of β-galactosidase
probiotic isolates by 16S rRNA gene sequence
Molecular characterization employing 16S rRNA gene
sequence analysis was performed for eight most potential
β-galactosidase-producing isolates. To estimate an approximate
phylogenetic association, the acquired nucleotide sequences were
compared with existing nucleotide gene sequences from GenBank using
the BLAST tool. Furthermore, nucleotide sequences were aligned
using MUSCLE, and a phylogenetic tree was constructed using the
neighbor-joining method in the MEGA-X software. All eight isolates
belonged to phylum Firmicutes, showing the highest similarity with
the genera Lactiplantibacillus. Fig. 3 illustrates the phylogenetic tree,
in which GV54, GV64, GV69, GV418 showed 99% and GV419 showed 98%
similarity with L. plantarum. However, GV66 showed 98%
similarity with Lactiplantibacillus sp., and GV65 showed 99%
similarity with L. pentosus, whereas GV254 showed 100%
similarity with L. fermentum. Furthermore, nucleotide
sequences were deposited in the GenBank database.
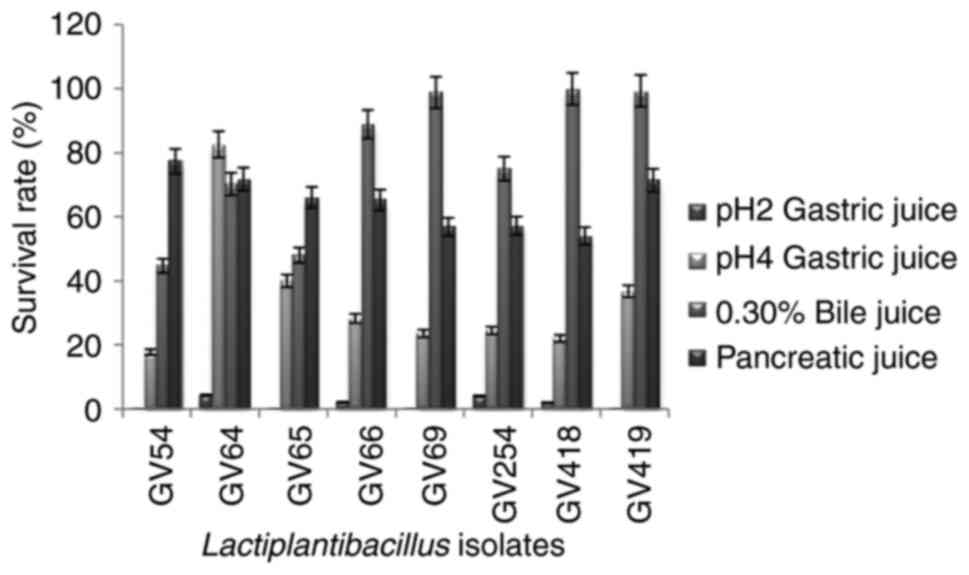
In vitro evaluation of potential
probiotic bacteria
All eight identified β-galactosidase-producing
isolates were subjected to probiotic characterization, in which the
isolates exhibited considerably less tolerance to gastric juice at
pH 2, while at pH 4 the survival rate was increased, and among them
L. plantarum GV64 exhibited the highest tolerance (82.6%)
and L. plantarum GV54 exhibited the lowest tolerance
(17.77%). For bile tolerance L. plantarum GV418 showed the
highest tolerance (99.93%) and L. plantarum GV54 showed the
lowest tolerance (44.68%) to bile juice after 4 h of incubation.
The percentage of tolerance of simulated gastric juice and bile
juice is illustrated in Fig. 4. In
the pancreatic enzyme tolerance test, the survival rate ranged from
54 to 77.33%, and among them L. plantarum GV54 exhibited the
highest tolerance (77.33%) and L. plantarum GV418 exhibited
the lowest tolerance (54%) after 4 h of incubation (Fig. 4). The survival percentage of the
selected eight isolates in gastric, bile and pancreatic juices
confirmed the resistance to upper gastrointestinal conditions. All
eight isolates exhibited γ hemolytic activity and were demonstrated
as non-pathogenic.
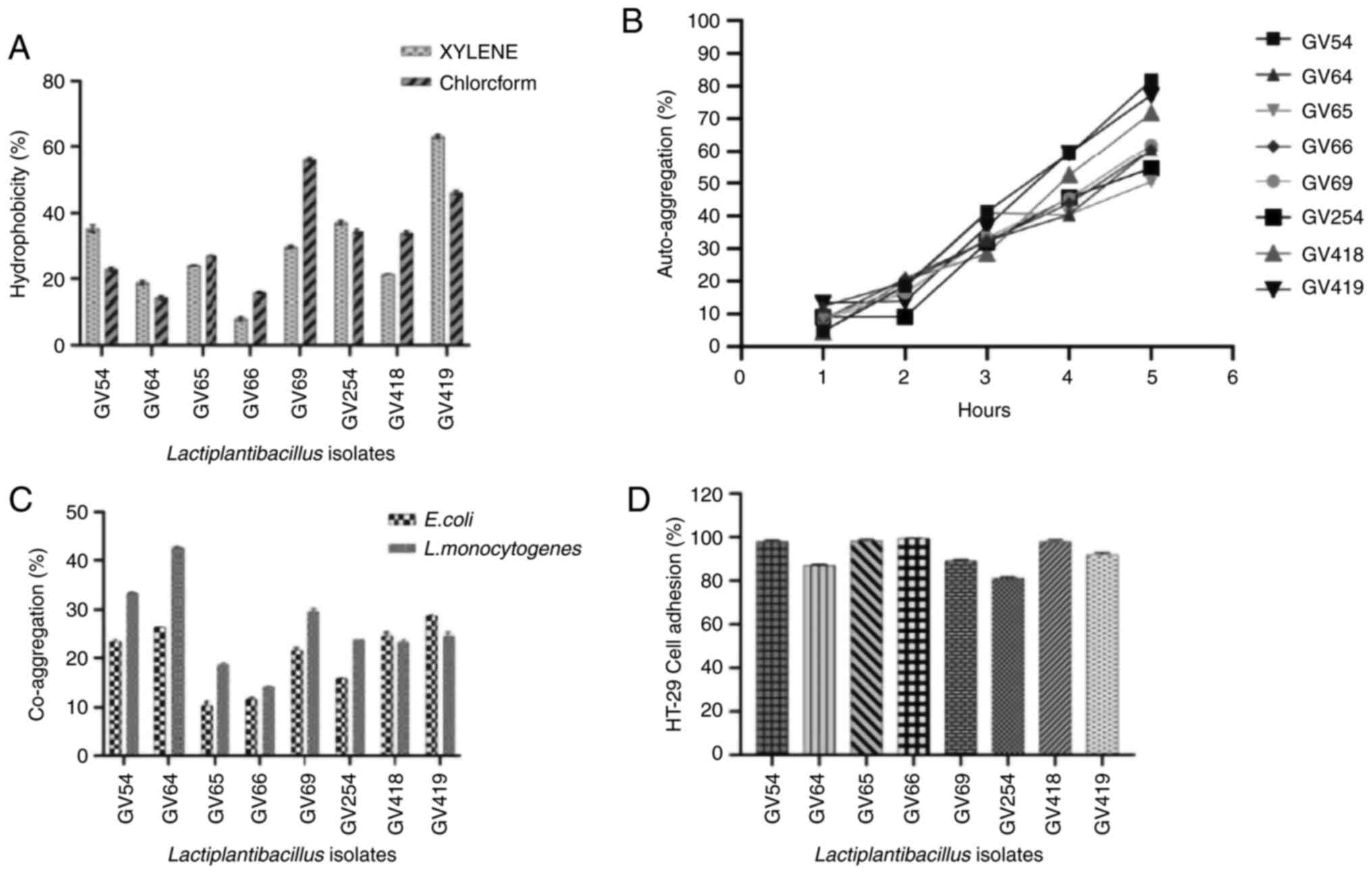
Hydrophobicity
The results of assessment of hydrophobicity
indicated that the eight isolates were hydrophobic, as revealed in
Fig. 5A. L. plantarum GV419
(63%) and L. plantarum GV69 (56%) with maximum affinity,
whereas Lactiplantibacillus sp. GV66 (9%) and L.
plantarum GV64 (15%) with minimal affinity to xylene and
chloroform, respectively.
Antimicrobial activity
The findings of the agar well diffusion method
indicated that the eight isolates have an antagonistic impact on
human pathogens. All isolates exhibited a zone of inhibition
ranging from 4.66±0.57 to 27.00±0.00 mm, whereas L.
plantarum GV64 was resistant to L. monocytogenes (MTCC
no. 1143) and Lactiplantibacillus sp. GV66 and L.
plantarum GV69 were resistant to Pseudomonas aeruginosa
(ATCC no. 9027) (Table I).
 | Table IAntimicrobial activity against human
pathogenic bacteria. |
Table I
Antimicrobial activity against human
pathogenic bacteria.
| | Human pathogenic
bacteria with zone of inhibition in diameter (mm) |
|---|
| Isolates | Escherichia
coli (MTCC no. 433) |
Staphylococcus aureus (ATCC
no. 6538) | Pseudomonas
aeruginosa (ATCC no. 9027) | Salmonella
abony (ATCC no. BAA2162) | Listeria
monocytogenes (MTCC no. 1143) |
|---|
| GV54 | 12.33±0.57 | 12.66±0.57 | 9.33±0.57 | 16.00±1.0 | 10.33±0.57 |
| GV64 | 4.66±0.57 | 11.00±1.00 | 7.33±0.57 | 10.33±0.57 | ND |
| GV65 | 12.66±0.57 | 11.33±0.57 | 12.33±0.57 | 12.33±0.57 | 13.33±0.57 |
| GV66 | 17.66±0.57 | 9.33±0.57 | ND | 14.66±0.57 | 9.33±0.57 |
| GV69 | 27.00±0.00 | 14.66±0.57 | ND | 12.33±0.57 | 18.66±0.57 |
| GV254 | 23.33±0.57 | 11.33±0.57 | 13.66±0.57 | 12.33±0.57 | 9.33±0.57 |
| GV418 | 23.66±0.57 | 7.66±0.57 | 15.00±0.00 | 13.66±0.57 | 10.66±0.57 |
| GV419 | 23.00±0.00 | 9.66±0.57 | 12.33±0.57 | 15.6667 | 9.00±1.00 |
Antibiotic sensitivity
Antibiotic discs were used to assess antibiotic
sensitivity/resistance of the eight isolates, and the assessed
antibiotics suppressed the growth of L. fermentum GV254 and
L. plantarum GV418. On the other hand, L. plantarum
GV54 was cefixime-resistant; L. plantarum GV64 was resistant
to ofloxacin, cefixime, and ciprofloxacin; L. pentosus GV65
was amikacin-resistant; Lactiplantibacillus sp. GV66 was
ofloxacin-resistant; L. plantarum GV69 was resistant to
ampicillin, ofloxacin, and cefixime; and L. plantarum GV419
was resistant to ofloxacin and cefixime [measured in terms of
diameter (mm); Table II].
 | Table IIAssessment of antibiotics for
Lactiplantibacillus isolates with probiotic
potentiality. |
Table II
Assessment of antibiotics for
Lactiplantibacillus isolates with probiotic
potentiality.
| | Zone of inhibition
in diameter (mm) |
|---|
| Isolates | AMP | AK | OF | P | CFM | CIP | E | AZM |
|---|
| GV54 | 10.33±0.57 | 11.66±1.52 | 9.66±0.57 | 10.66±0.57 | ND | 12.66±0.57 | 29.33±0.15 | 24.66±0.57 |
| GV64 | 11.00±0.00 | 12.33±0.57 | ND | 13.66±0.57 | ND | ND | 30.00±00 | 26.33±1.15 |
| GV65 | 9.33±0.57 | ND | 10.33±0.57 | 12.66±0.57 | 9.66±0.57 | 12.33±0.57 | 30.00±00 | 24.33±0.57 |
| GV66 | 6.00±0.00 | 15.66±0.57 | ND | 12.00±0.00 | 10.33±0.57 | 10.33±0.57 | 25.66±0.57 | 20.00±00 |
| GV69 | ND | 14.66±0.57 | ND | 10.66±0.57 | ND | 10.33±0.57 | 29.66±0.57 | 24.66±0.57 |
| GV254 | 12.33±0.57 | 14.66±0.57 | 13.00±0.00 | 10.00±0.00 | 10.00±0.00 | 14.66±0.57 | 27.66±0.57 | 23.33±0.57 |
| GV418 | 12.33±0.57 | 14.00±0.00 | 8.66±0.57 | 7.66±0.57 | 13.33±0.57 | 9.00±0.00 | 30.00±00 | 23.00±00 |
| GV419 | 8.00±0.00 | 14.66±0.57 | ND | 7.33±0.57 | ND | 8.33±0.57 | 24.66±0.57 | 17.66±0.57 |
Auto-aggregation and co-aggregation
assays
The percentage of auto-aggregation was measured
after every hour of incubation, and L. plantarum GV54
exhibited the highest rate of auto-aggregation (81%), while L.
plantarum GV69 and L. plantarum GV419 showed moderate
auto-aggregation (61 and 71%, respectively). L. pentosus
GV65 exhibited minimal auto-aggregation 50% (Fig. 5B). Furthermore, the eight isolates
exhibited co-aggregative properties with both the pathogens, E.
coli (MTCC no. 433) and L. monocytogenes (MTCC no.
1143), after 5 h of incubation at 37˚C. The rate of co-aggregation
of E. coli and L. monocytogenes ranged between 10 and
28%, and 14 to 42%, respectively. L. monocytogenes exhibited
higher co-aggregation compared to E. coli. Notably, L.
plantarum GV419 exhibited higher co-aggregation (28.67%) of
E. coli compared to L. monocytogenes, while L.
plantarum GV64 (42.71%) exhibited higher co-aggregation of
L. monocytogenes compared to E. coli (Fig. 5C).
Adhesion assay
Adhesion assay of eight potential isolates to human
colorectal adenocarcinoma intestinal epithelial HT-29 cells was
determined and graphically represented in Fig. 5D. Bacterial adherence ability with
Lactiplantibacillus sp. GV66 was 99.43% and L.
fermentum sp. GV254 exhibited considerably less adhesion at
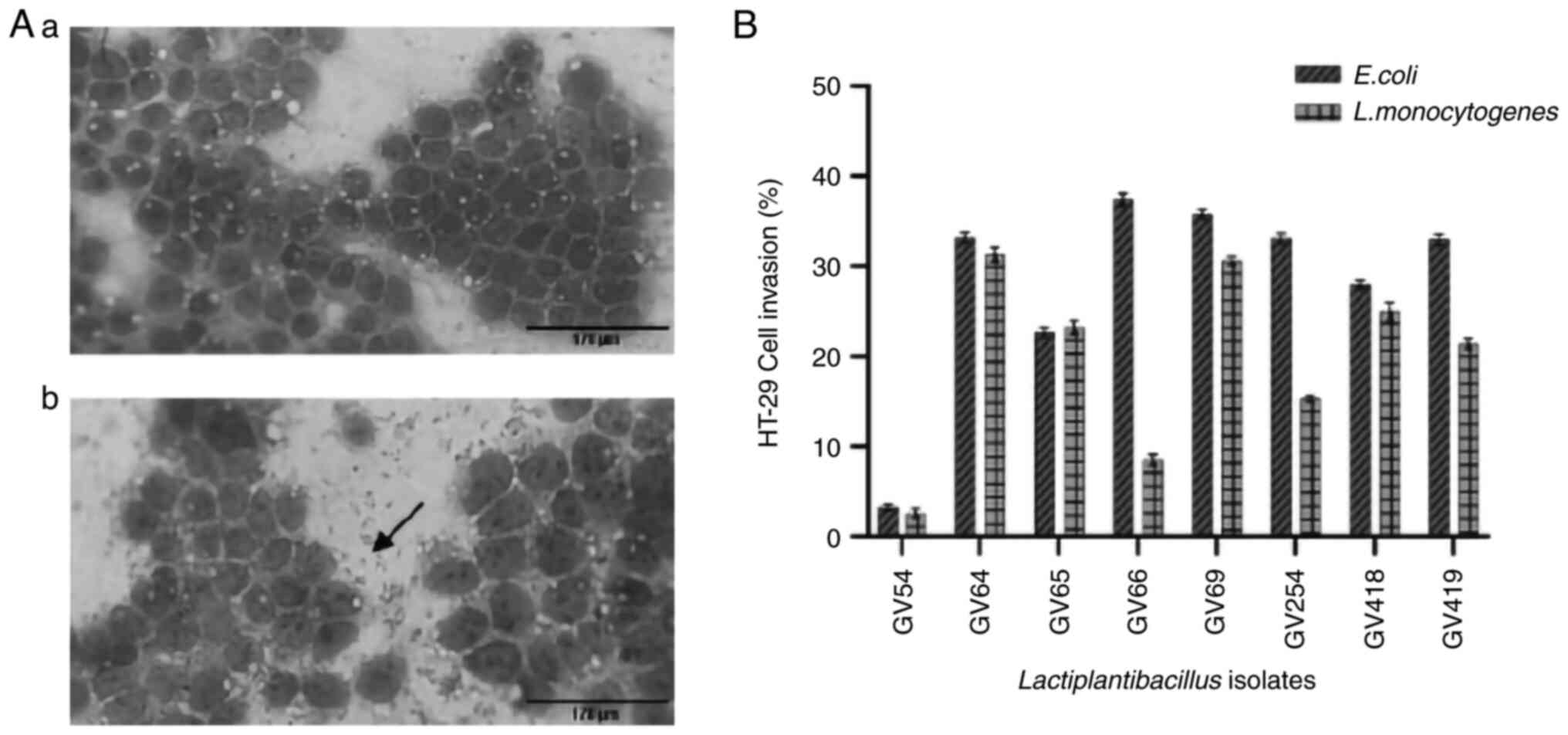
81.14%. The adhesion of all eight isolates was further verified by
direct microscopic observation (Fig.
6A).
HT-29 cell invasion assay
All eight isolates were investigated for the
suppression of E. coli and L. monocytogenes
intracellular invasion, as revealed in Fig. 6B. The inhibition percentage ranged
from 3.26 to 37.41% and 2.56 to 31.31%, respectively. L.
plantarum GV54 had a low percentage of invasion for both
pathogens (3.26 and 2.56%). Lactiplantibacillus sp. GV66 and
L. plantarum GV64 exhibited the highest percentage of
invasion of E. coli (37.41%) and L. monocytogenes
(31.31%), respectively.
Discussion
Fermented foods containing LAB are traditionally
used in daily food intake. Curd, prepared by fermentation of milk
with an inoculum of previously made curd, is used in most
households in India, where it constitutes a significant part of the
daily diet. The LAB that ferment the milk are likely to differ
slightly in each household as there is no standardized starter
culture used to prepare the curd. Although curd is considered to
contain probiotics, there is little documentation in this line. The
present study was undertaken to evaluate the LAB from homemade curd
in southern India for probiotic properties. Probiotic diversity is
very vast, therefore, in the present study, potential LAB isolates
were selected, which are capable of eliminating or reducing LI.
The key findings in the present study successfully
revealed the most promising isolates (GV54, GV64, GV65, GV66, GV69,
GV254, GV418, and GV419) with probiotic characteristics and
β-galactosidase production. LAB that are found in fermented foods
aid lactose digestion by increasing the activity of the
β-galactosidase enzyme with distinct health advantages (19). It has been reported that lactose
from milk digested by β-galactosidase to hydrolyse glucose and
galactose, is absorbed by enterocytes and used as an energy source
(4). LI symptoms can be managed
with dairy products supplemented with β-galactosidase-producing
probiotics, which also inhibit human pathogen adhesion (2). Hence, selected isolates were preferred
for β-galactosidase enzyme activity with X-Gal and ONPG as
substrates (14). While,
Lactococcus casei A13 exhibited negative or zero enzyme
activity, L. delbrueckii subsp. bulgaricus Db1 exhibited
2,053±25 Miller units (15), which
was the highest enzyme activity reported, and served as a positive
control. In the present study, β-galactosidase production was
highest (1,203.32 Miller units) in Lactiplantibacillus sp
GV66 with 58% of enzyme activity compared to the positive control.
Therefore, this isolate is a potential β-galactosidase product and
hence the strain can be further improved for industrial production
of β-galactosidase.
API50 CHL and 16S rRNA sequence analysis may also be
used to identify LAB (27,28). Kumara et al (23) used 16S rRNA gene sequence analysis
to identify four LAB isolates, all of which were demonstrated to be
L. fermentum. The present study identified eight potential
isolates, by molecular characterization using 16S rRNA gene
sequencing, including L. plantarum GV54, GV64, GV69, GV418
and GV419, Lactiplantibacillus sp. GV66, L. pentosus
GV65, L. fermentum GV254, and deposited them in the GenBank
database.
Lactiplantibacillus sp. have been termed
probiotic bacteria due to their ability to survive in gut
conditions such as gastric juice and bile, and exhibit pancreatic
enzyme tolerance, as well as intestinal epithelial cell adhesion
(29). However, the aforementioned
attributes are not completely the same in in vitro testing
as found in the in vivo gut system, but it is still a
valuable tool for rapid screening of possible probiotic strains.
In vivo investigations are more expensive and time-consuming
than in vitro studies for evaluating the probiotic
characteristics of unknown bacteria; hence, in vitro testing
is selected as an alternative (21). Although, an in vivo
validation for the above results is a limitation in this study,
further animal studies are in progress. Bacteria must be able to
survive in intestinal conditions for considerably long periods to
be classified as probiotics (30).
L. fermentum HM3 isolate exhibited better
acid tolerance at pH 3 for 3 h than the reference strain L.
casai Shirota (31). In
addition, Vinderola and Reinheimer (15) reported that S. thermophilus
exhibited tolerance while L. lactis and L. delbrueckii
subsp. Bulgaricus exhibited tolerance at pH 2 and 3. Hsieh
et al (32) reported that
Lactobacillus strains TSP05, TSF331 and TSR332 were
resistant to gastric acid at pH 3.5 in MRS medium for 3 h, and with
regard to the cell viability measured as CFU/ml,
Lactobacillus TSF331 exhibited the highest viability
(4.45x109) in gastric conditions. In the present study,
Lactiplantibacillus strains exhibited better acid tolerance
at pH 4 than pH 2 for 4 h. Among them, L. plantarum GV64
exhibited the highest tolerance (82.6%) and L. plantarum
GV54 showed the lowest tolerance (17.77%). Several studies have
reported that 0.3% (w/v) bile salt concentration in the human gut
also varies according to diet and the level of pancreatic enzyme
secretion (33). Succi et al
(34) also reported that most of
the LAB showed resistance to a 0.5% bile concentration, and Byakika
et al (35) examined whether
the probiotic bacteria were tolerant to 1% bile salt. Hsieh et
al (32) also reported that
0.3% bile was used for a bile tolerance test and
Lactobacillus TSR332 exhibited the highest viability
(4.74x108 CFU/ml) indicating high tolerance to bile
salt. In the present study, a 0.3% bile concentration was used and
eight isolates that exhibited favourable bile tolerance (99.93%)
were selected. Among them, L. plantarum GV418 showed the
highest tolerance (99.93%) and L. plantarum GV54 exhibited
the lowest tolerance (44.68%). Pancreatic enzymes digest various
carbohydrates, proteins, and lipids in the human diet (33). According to Shokryazdan et al
(31), LAB strains were incubated
for 3 h in growth media containing pancreatic enzymes, and the
viability of the L. brevis strain differed. All eight
isolates of the present study, exhibited high pancreatic tolerance
in this investigation, and the degree of tolerance differed among
strains. As aforementioned, the assessed LAB isolates were able to
survive at gastric pH. Notably, to increase 100% viability of
lactic acid bacterial cells, either encapsulation or coating with
inert biomolecules can be performed to improve the likelihood of
survival.
The eight isolates in the present study with the
highest hydrophobicity demonstrated maximum adherence to xylene and
chloroform solvents, with L. plantarum GV419 showing 63%
adhesion to xylene and L. plantarum GV69 exhibiting 56%
adhesion to chloroform. The adherence of fecal isolates to the
affinity for n-hexadecane and toluene was examined in another study
(25). The greatest hydrophobicity
of L. brevis CCMA 1284, L. plantarum CCMA 0743, L. plantarum
CCMA 0359 was reported by Fonseca et al (36), and Behbahania et al (37) analyzed L. plantarum strain
L15, which showed 54% adherence to solvents. This suggests that the
hydrophobic and hydrophilic appendages, as well as other
macromolecule components, contribute to the cell surface mosaic's
complexity, resulting in hydrophobicity differences toward
hydrocarbons.
The ability of auto-aggregation and co-aggregation
increased with time, reaching a maximum level at 24 h of incubation
rather than at 5 h (38). The
auto-aggregation ability of the L. plantarum strain was
shown to be the highest in a similar study reported by Fonseca
et al (36), which revealed
that L. paracasei CCMA 0504 and L. paracasei CCMA
0505 exhibited the highest percentage of auto-aggregation. All
eight isolates in the present study exhibited variable values of
auto-aggregation. Notably, L. plantarum GV54 showed the
highest rate of auto-aggregation with 81%, and L. plantarum
GV69 and L. plantarum GV419 exhibited moderate
auto-aggregation with 61 and 71%, respectively. Furthermore, all
isolates exhibited co-aggregative properties, and among them, L.
plantarum GV419 exhibited a high co-aggregation (28.67%) with
E. coli while L. planatrum GV64 (42.71%) with L.
monocytogenes.
The antimicrobial activity against human pathogens
is considered as a main characteristic of probiotic strains that
maintain gut health (21). All
eight isolates were antagonistic against human pathogens in the
present study, with the zone of inhibition ranging from 4.66±0.57
to 27.66±0.57 mm. Asha and Gayathri (18) assessed Lactiplantibacillus
strains from curd samples for their antibacterial activity against
E. coli, V. cholerae, Klebsiella strains, Proteus
strains, and S. dysenteriae. An additional role of the
Lactiplantibacillus strain is to inhibit the colonization of
pathogenic bacteria to human and animal intestines by secreting
several biochemical compounds and enzymes to prevent various
infections. Kumara et al (23) reported that L. fermentum
inhibited colonization of S. ebony, S. aureus, E. coli, P.
aeruginosa, and these bacteria were susceptible to gentamycin,
chloramphenicol, cefoperazone, ampicillin, and resistant to
ciprofloxacin and vancomycin. The susceptibility of
Lactiplantibacillus strains to antibiotics, using the disc
diffusion method, was investigated in the present study and it was
revealed that these strains were susceptible to penicillin,
cefixime, ofloxacin, ciprofloxacin, amikacin, wherein
Lactiplantibacillus sp. GV66 was ofloxacin-resistant, and
L. plantarum GV69 was resistant to ampicillin, ofloxacin and
cefixime.
The assessed strains were susceptible to at least
one of the antibiotics that would prevent the formation of cell
wall and proteins. Two strains of L. paracasei were mildly
sensitive or susceptible to lincomycin, azithromycin, and
penicillin, according to Fonseca et al (36), and L. brevis CCMA1284 strain
was resistant to these three antibiotics. Of note, if a gene
transfer process was involved, then antibiotic resistance would
become a dangerous scenario. Alhough, this process may not be
communicable and would not be a unique criteria of the microbial
genus or species, it would however be a sort of alerting condition.
Furthermore, minimum inhibitory concentration (MIC) values for
antibiotic susceptibility of the Lactobacillus strains were
tested against antibiotics, including ampicillin, gentamicin,
kanamycin, streptomycin, erythromycin, clindamycin, tetracycline,
and chloramphenicol (31) The MIC
values of the reference strain and the Lactobacillus strains
were less than the MIC breakpoint values. Antibiotic resistance
genes have accumulated in probiotics, due to the widespread use of
probiotics in combination with antibiotics to restore gut flora.
There are significant clinical risks if these resistance genes are
transferred from probiotics to pathogens in the colon. The
antibiotic sensitivity of a few antibiotics used in the present
study were found to be within the European Food Safety Authority
(EFSA) breakpoint values for all the examined gluten-hydrolysing
bacteria and were thus deemed safe (13). In concurrence with the
aforementioned evidence, gentamycin, chloramphenicol, cefoperazone,
ampicillin, ciprofloxacin and vancomycin antibiotics were selected
to ensure the safety and efficacy of the assessed probiotic
bacterial isolates.
Probiotic bacteria exhibit cell line attachment and
can colonize with intestinal epithelial cells in order to establish
themselves in the gut (31,36). The HT-29 cell line and the selected
eight isolates were used in the present study for cell adhesion
assay, with Lactiplantibacillus sp. GV66 exhibiting strong
adhesion at 99.43% and L. fermentum GV254 exhibiting
comparatively less adhesion at 81.14%. In other studies,
Lactiplantibacillus strains exhibited strong adherence
abilities to the Caco-2 and HT-29 cell lines (39). Rashmi and Gayathri (13) investigated whether
gluten-hydrolyzing bacteria adhered to the Caco-2 cell line and
inhibited cell invasion by E. coli and L.
monocytogenes. Byakika et al (35) used the goat ileum to assess cell
adhesion, and Behbahani et al (37) used scanning electron microscopy to
identify that the adhesion level of the L. plantarum strain
L15 to Caco-2 cells was 12%. Aissi et al (24) employed HT-29, Caco-2 and INT-407
cells, as well as Bifidobacterial strains, and
microscopically studied them. HT-29 cell invasion assay by E.
coli and L. monocytogenes was performed in the present
study using an in vitro approach. L. plantarum GV54
had a low percentage of invasion by the pathogens E. coli
and L. monocytogenes (3.26 and 2.56%). By contrast,
Lactiplantibacillus sp. GV66 and L. plantarum GV64
exhibited the highest percentage of invasion of E. coli
(37.41%) and L. monocytogenes (31.31%), respectively, thus
revealing their significant potential as probiotic bacteria.
Vinderola and Reinheimer (15) assessed the β-galactosidase activity
in L. delbrueckii substrains bulgaricus, L.
acidophilus, and other Lactiplantibacillus stains
ranging from 1,301 to 2,053 Miller units. Gheytanchi et al
(14) also reported the
β-galactosidase enzyme activity in L. delbrueckii substrains
bulgaricus and L. casei (ranging from 867 to 1,966 U/ml)
isolated from cheese. Lactiplantibacillus strains with
substantial β-galactosidase activity were identified in the present
study; among them Lactiplantibacillus sp. GV66 had the
highest value at 1,203.32 (U/ml) Miller units. All of these
positive traits of Lactiplantibacillus sp. render this
strain ideal for use in probiotic formulations, either alone or in
combination with other advantageous probiotic-bacterial isolates.
Lactiplantibacillus sp. that produces β-galactosidase could
be used as a probiotic supplement to help individuals with LI.
Thus, the use of probiotics may lead to a promising method in
prevention or management of LI. In addition, it is possible to
improve and optimize the enzyme activity and development of milk
products with potential probiotics/enzymes for the management of
LI.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Sequence data that support the findings of the
present study have been deposited in GenBank with the primary
accession no. MN686265: MN696220-MN696226 (https://www.ncbi.nlm.nih.gov/search/all/?term=MN686265:MN696220-MN696226
[accn]). All data used or analyzed during the present study are
included within this article. All other data are available from the
corresponding authors upon reasonable request.
Authors' contributions
MV executed the planned experimental work, wrote
the manuscript and performed the data analysis. DG conceived and
designed the study, as well as acquired and analysed the data. VK,
CSP and MB drafted the work, and revised it critically for
important intellectual content. DG and VK confirm the authenticity
of all the raw data. All authors read and approved the final
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lomer MC, Parkes GC and Sanderson JD:
Review Article: Lactose intolerance in clinical practice-myths and
realities. Aliment Pharmacol Ther. 27:93–103. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gayathri D and Vasudha M: Lactose
Intolerance with Special Emphasis on Probiotics for Management. EC
Nutrition. 13:325–332. 2018.
|
|
3
|
Ingram CJ, Mulcare CA, Itan Y, Thomas MG
and Swallow DM: Lactose digestion and the evolutionary genetics of
lactase persistence. Human Genet. 124:579–591. 2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Babu J, Kumar S, Babu P, Prasad JH and
Ghoshal UC: Frequency of lactose malabsorption among healthy
Southern and Northern Indian populations by genetic analysis and
lactose hydrogen breath and tolerance tests. Am J Clin Nutr.
91:140–146. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sriphannam W, Lumyong S, Niumsap P, Ashida
H, Yamamoto K and Khanongnuch C: A selected probiotic strain of
Lactobacillus fermentum CM33 isolated from breast-fed infants as a
potential source of β-galactosidase for prebiotic oligosaccharide
synthesis. J Microbiol. 50:119–126. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Harrington LK and Mayberry JF: A
re-appraisal of lactose intolerance. Int J Clin Pract.
62:1541–1546. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Heyman MB: Committee on Nutrition. Lactose
intolerance in infants, children, and adolescents. Pediatrics.
118:1279–1286. 2006.PubMed/NCBI View Article : Google Scholar
|
|
8
|
FAO/WHO: Working Group on Drafting
Guidelines for the Evaluation of Probiotics in Food, London
Ontario, Canada, 2002.
|
|
9
|
Patel RM and Lin PW: Developmental biology
of gut-probiotic interaction. Gut Microbes. 1:186–195.
2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pan Q, Zhu J, Liu L, Cong Y, Hu F, Li J
and Yu X: Functional identification of a putative β-galactosidase
gene in the special lac gene cluster of Lactobacillus acidophilus.
Curr Microbiol. 60:172–178. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Pagnini C, Saeed R, Bamias G, Arseneau KO,
Pizarro TT and Cominelli F: Probiotics promote gut health through
stimulation of epithelial innate immunity. Proc Natl Acad Sci USA.
107:454–459. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Swamy CT, Gayathri D, Devaraja TN,
Bandekar M, D'Souza SE, Meena RM and Ramaiah N: Plant growth
promoting potential and phylogenetic characteristics of a
lichenized nitrogen fixing bacterium, Enterobacter cloacae. J Basic
Microbiol. 56:1369–1379. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rashmi BS and Gayathri D: Molecular
characterization of gluten hydrolyzing Bacillus sp. and their
efficacy and biotherapeutic potential as probiotics using Caco-2
cell line. J Appl Microbiol. 123:759–772. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gheytanchi E, Heshmati F, Shargh BK,
Nowroozi J and Movahedzadeh F: Study on β-galactosidase enzyme
produced by isolated lactobacilli from milk and cheese. Afr J
Microbiol Res. 4:454–458. 2010.
|
|
15
|
Vinderola CG and Reinheimer JA: Lactic
acid starter and probiotic bacteria: A comparative ‘in vitro’ study
of probiotic characteristics and biological barrier resistance.
Food Res Int. 36:895–904. 2003.
|
|
16
|
Li J, Zhang W, Wang C, Yu Q, Dai R and Pei
X: Lactococcus lactis expressing food-grade β-galactosidase
alleviates lactose intolerance symptoms in post-weaning Balb/c
mice. Appl Microbiol Biotechnol. 96:1499–1506. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vos P, Garrity G, Jones D, Krieg NR,
Ludwig W, Rainey FA, et al: Bergey's manual of systematic
bacteriology. The Firmicutes (Vol. 3). SSBM, 2011.
|
|
18
|
Asha and Gayathri D: Antagonistic
potential of Lactobacillus against enteropathogenic bacteria;
purification and characterization of their bacteriocins. Adv J Food
Sci Technol. 4:265–269. 2012.
|
|
19
|
Princely S, Basha NS, Kirubakaran JJ and
Dhanaraju MD: Biochemical characterization, partial purification
and production of an intracellular beta-galactosidase from
Streptococcus thermophilus grown in whey. Euro J Exp Bio.
3:242–251. 2013.
|
|
20
|
William S, Feil H and Copeland A:
Bacterial genomic DNA isolation using CTAB. Sigma. (50
(6876))2012.
|
|
21
|
Shokryazdan P, Sieo CC, Kalavathy R, Liang
JB, Alitheen NB, Faseleh Jahromi M and Ho YW: Probiotic potential
of Lactobacillus strains with antimicrobial activity against some
human pathogenic strains. Biomed Res Int.
2014(927268)2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Armas F, Camperio C and Marianelli C: In
vitro assessment of the probiotic potential of Lactococcus lactis
LMG 7930 against ruminant mastitis-causing pathogens. PLoS One.
12(e0169543)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kumara SS, Bashisht A, Venkateswaran G,
Hariprasad P and Gayathri D: Characterization of novel
Lactobacillus fermentum from curd samples of indigenous cows from
Malnad region, Karnataka, for their aflatoxin B1 binding
and probiotic properties. Probiotics Antimicrob Proteins.
11:1100–1109. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Aissi EA, Lecocq M, Brassart C and
Bouquelet S: Adhesion of some Bifidobacterial strains to human
enterocyte-like cells and binding to mucosal glycoproteins. Microb
Ecol Health Dis. 13:32–39. 2001.
|
|
25
|
Duary RK, Rajput YS, Batish VK and Grover
S: Assessing the adhesion of putative indigenous probiotic
lactobacilli to human colonic epithelial cells. Indian J Med Res.
134:664–671. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Botes M, Loos B, van Reenen CA and Dicks
LM: Adhesion of the probiotic strains Enterococcus mundtii ST4SA
and Lactiplantibacillus plantarum 423 to Caco-2 cells under
conditions simulating the intestinal tract, and in the presence of
antibiotics and anti-inflammatory medicaments. Arch Microbiol.
190:573–584. 2008.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Maleki Kakelar H, Barzegari A, Hanifian S,
Barar J and Omidi Y: Isolation and molecular identification of
Lactobacillus with probiotic potential from abomasums driven
rennet. Food Chem. 272:709–714. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Reuben RC, Roy PC, Sarkar SL, Rubayet Ul,
Alam ASM and Jahid IK: Characterization and evaluation of lactic
acid bacteria from indigenous raw milk for potential probiotic
properties. J Dairy Sci. 103:1223–1237. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bin Masalam MS, Bahieldin A, Alharbi MG,
Al-Masaudi S, Al-Jaouni SK, Harakeh SM and Al-Hindi RR: Isolation,
molecular characterization and probiotic potential of lactic acid
bacteria in Saudi raw and fermented milk. Evid Based Complement
Alternat Med. 2018(7970463)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang CY, Lin PR, Ng CC and Shyu YT:
Probiotic properties of Lactobacillus strains isolated from the
feces of breast-fed infants and Taiwanese pickled cabbage.
Anaerobe. 16:578–585. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shokryazdan P, Faseleh Jahromi M, Liang JB
and Ho YW: Probiotics: From isolation to application. J Am Coll
Nutr. 36:666–676. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hsieh PS, Chen CW, Kuo YW and Ho HH:
Lactobacillus spp. reduces ethanol-induced liver oxidative stress
and inflammation in a mouse model of alcoholic steatohepatitis. Exp
Ther Med. 21(188)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zago M, Fornasari ME, Carminati D, Burns
P, Suàrez V, Vinderola G, Reinheimer J and Giraffa G:
Characterization and probiotic potential of Lactiplantibacillus
plantarum strains isolated from cheeses. Food Microbiol.
28:1033–1040. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Succi M, Tremonte P, Reale A, Sorrentino
E, Grazia L, Pacifico S and Coppola R: Bile salt and acid tolerance
of Lactobacillus rhamnosus strains isolated from Parmigiano
Reggiano cheese. FEMS Microbiol Lett. 244:129–137. 2005.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Byakika S, Mukisa IM, Byaruhanga YB and
Muyanja C: Probiotic potential of lactic acid starter cultures
isolated from a traditional fermented sorghum-millet beverage. Int
J Microbiol. 2020(7825943)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Fonseca HC, de Sousa Melo D, Ramos CL,
Dias DR and Schwan RF: Probiotic properties of lactobacilli and
their ability to inhibit the adhesion of enteropathogenic bacteria
to Caco-2 and HT-29 cells. Probiotics Antimicrob Proteins.
13:102–112. 2021.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Behbahani BA, Noshad M and Falah F:
Inhibition of Escherichia coli adhesion to human intestinal Caco-2
cells by probiotic candidate Lactiplantibacillus plantarum strain
L15. Microb Pathog. 136(103677)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Goh YJ and Klaenhammer TR: Functional
roles of aggregation-promoting factor in stress tolerance and
adherence of Lactiplantibacillus acidophilus NCFM. Appl Environ
Microbiol. 76:5005–5012. 2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Del Piano M, Morelli L, Strozzi GP,
Allesina S, Barba M, Deidda F, Lorenzini P, Ballaré M, Montino F,
Orsello M, et al: Probiotics: From research to consumer. Dig Liver
Dis. 38 (Suppl 2):S248–S255. 2006.PubMed/NCBI View Article : Google Scholar
|