Introduction
Tuberculosis (TB) remains a severe global public
health problem, especially in developing countries (1). How to reduce the TB epidemic is a
major issue hindering economic and social development. Detection of
Mycobacterium tuberculosis (Mtb) DNA using Gene Xpert
MTB/RIF assay is more sensitive and rapid for diagnosing TB and
rifampicin resistance (2). However,
due to its costs, environmental limitations, and difficulties in
its supply, it is difficult to carry out screening in a low-income
rural area (3). Interferon-γ
release assay (IGRA) is commonly used in the diagnostic workup of
Mtb but distinguishes poorly between TB and latent TB
infection (LTBI) (4,5).
In addition, the symptoms of patients with TB are
very similar to those of bacterial community-acquired pneumonia
(CAP). Both TB and CAP are infections of the lower respiratory
tract, but they are often considered as separate entities (6). TB is classically a more indolent
disease presenting cavitating lung lesions observed in patients
with a history of cough for three months or longer accompanied by
weight loss, and, often, is not associated with acute respiratory
compromise (7). By contrast, CAP is
generally associated with a short history of several days, is
rapidly progressive, and is more often associated with respiratory
compromise (8). The diagnosis of
CAP is based on the detection of a new infiltrate on a chest
radiograph or other imaging technique in the presence of recently
acquired respiratory signs and symptoms (9). However, clinical findings do not
reliably predict radiologically confirmed PN, as features of TB may
sometimes be quite similar to those of CAP among patients who
experience symptoms at the early stage (10). In addition, the etiology cannot be
simply differentiated clinically or radiologically and is undefined
in ~50% of patients (8). Because
most patients with TB have no sputum specimens or are Mtb
smear/culture-negative, this makes diagnosis more intractable
(11).
Host markers including secreted molecules in blood
have been reported as novel candidate markers to distinguish TB,
such as interferon-γ inducible protein 10 kDa (12), interleukin-2 (IL-2) (13), IL-6(14), C-reactive protein (CRP) (15) and vascular endothelial growth factor
(14). However, the diagnostic
performances of reported biomarkers cannot be applied in low-income
areas due to technical, instrumentation or cost limitations. To
address this problem, the present study retrospectively analyzed
the differences in routinely monitored laboratory variables in
blood tests between AFB- IGRA+ TB and PN, and
built a risk model to differentiate AFB-
IGRA+ TB from PN, and validated its application by an
external independent cohort.
Patients and methods
Study design and criteria for study
inclusion
The patients provided a full medical history,
participated in regular physical examinations, and underwent
routine investigations, including acquired immunodeficiency
syndrome (AIDS) serology, chest radiography, IGRA, and
microbiological sputum examination, where possible.
The inclusion criteria for PN participants were as
follows: i) Meeting the diagnostic criteria of ‘The People's
Republic of China health industry standard (WS 382-2012; http://www.nhc.gov.cn/wjw/s9494/201209/110b324c465740169a863d57a78c18a6.shtml)’.
The clinical diagnosis can be established by any of (a), (b), (c),
(d) plus (e) and excluding TB, lung tumor, non-infectious
interstitial lung disease, pulmonary edema, pulmonary atelectasis,
pulmonary embolism, pulmonary eosinophilic infiltrates, pulmonary
vasculitis, etc. The elements of (a) to (e) are as follows: a)
Newly developed cough and sputum, or aggravation of existing
respiratory symptoms with purulent sputum, with or without chest
pain; b) fever; c) solid lung signs and/or wet rales; d) peripheral
blood WBC >10x109/l or <4x109/l with or
without leftward nuclear shift; and e) chest X-ray showing new
lamellar or patchy infiltrative shadows or interstitial changes
with or without pleural effusion. ii) Sputum culture with a
bacterial pathogenic basis or effective antimicrobial therapy, such
as marked improvement in symptoms such as cough, yellow sputum and
significant uptake of chest imaging. iii) Exclusion of viral,
atypical pathogens, fungal, and Mtb infections. The
inclusion criteria for IGRA+ TB participants were as
follows: Meeting the diagnostic criteria for TB including positive
sputum, bronchoscopic lavage, brush examination, smear microscopy
of biopsy specimens for antacid bacilli, isolation and culture of
Mtb, or positive nucleic acid test and IGRA for Mtb,
or positive pathology of lung tissue biopsy. The inclusion criteria
for AFB- TB participants were as follows: There were no
sputum or negative bacilli smear and negative Mtb, but there
were chest computed tomographic (CT) scans or chest X-ray evidence
and symptoms responding to TB treatment.
The exclusion criteria were as follows: i) Use of
antimicrobial drugs for >24 h; (2) suffering from diseases that can affect
the total number and classification of blood leukocytes, including
leukemia, chronic inflammatory states, etc.; iii) having a recent
(within 3 months) history of glucocorticoid application or ongoing
hormone use; and iv) clearly diagnosed or highly suspected
pulmonary edema, pulmonary embolism, pulmonary atelectasis,
bronchial asthma, viral PN, fungal PN, atypical pathogenic PN,
pulmonary eosinophilic infiltrates and lung cancer.
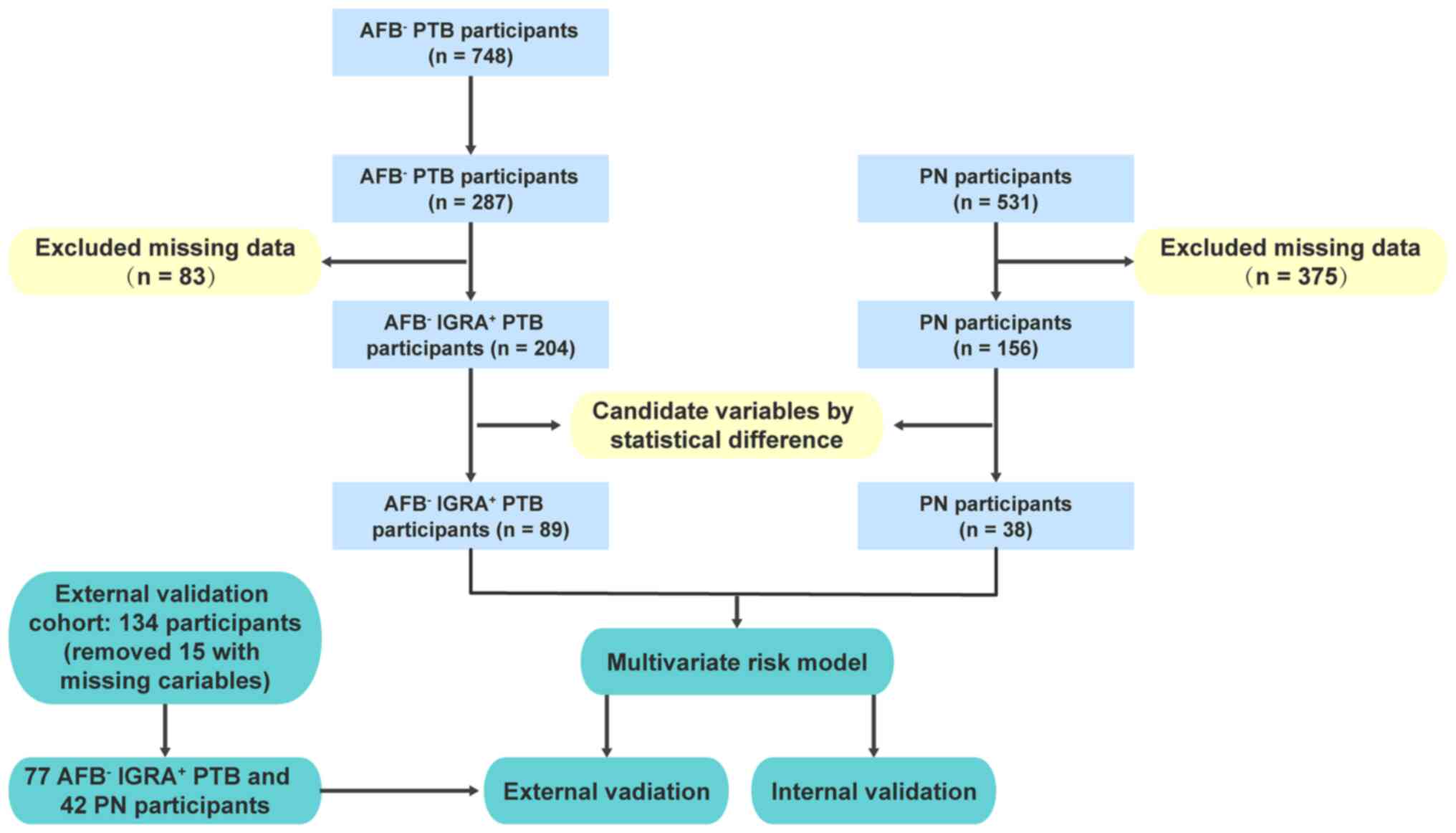
The involved participants were divided into two
groups: The discovery cohort and the external validation cohort.
For the discovery cohort, participants were enrolled at Ganzhou
Fifth Hospital (Ganzhou, China) between August 2018 and August
2020, including 748 AFB- TB participants and 531 PN
participants. As the predefined goal was to assess the ability of
laboratory biomarkers to distinguish IGRA+ patients
presenting with AFB- TB, 287 participants with
IGRA+ TB were subjected for further analysis
(AFB- IGRA+ TB). Participants with >50% of
laboratory data missing were excluded, thus making a total of 89 TB
and 38 PN participants with recorded biomarker values used to
construct the risk model. The external validation cohort of 134
participants in the study was collected from Shenzhen Third
People's Hospital (Shenzhen, China) from June 2018 to June 2019
(Fig. 1). Among them, 15 were
excluded due to missing variables, therefore the external
validation cohort consisted of 77 AFB- IGRA+
TB and 42 PN participants.
Ethical approval and patient consent
to participate
The study protocol was approved by the Ethics
Committee and the Institutional Review Board of Ganzhou Fifth
People's Hospital (registration no. 2020-10) to allow retrospective
access to the records and files of patients. Written informed
consent was waived by the Ethics Committee as this was an
observational and retrospective analysis.
Data collection
The medical records of all participants were
reviewed by experienced TB clinicians, including medical history,
symptoms, clinical signs, microbiological tests, laboratory
findings, chest CT chest X-rays, and treatment measures. A total of
41 laboratory biomarkers were assessed by differential statistics
and odds ratio (OR) calculation for variable selection.
Statistical analyses
For laboratory results, continuous variables were
preprocessed by log2-transformation before analysis. The
laboratory data were verified for skewed distribution using the
Kolmogorov-Smirnov test. In the present study Wilcoxon rank-sum
test was suitable for skewed distribution data (16-18).
P-values were adjusted by false discovery rate (FDR). Variables
between two conditions were defined as statistically significant
when FDR <0.2(19).
A univariate logistic model (glm) was used to
calculate the OR for each laboratory biomarker. The regression
coefficient of the glm was regarded as the log OR. Variables with
an FDR <0.2 or at a statistically significant level (P-value
<0.05) in the glm analysis were candidates for the construction
of a multivariate risk model (lrm) and nomogram, and the final
variables were determined using Akaike's information criterion
(AIC) as a stopping rule. The goodness of fit of the lrm model was
calculated using Hosmer and Lemeshow C statistics test. The
performance of the nomogram was evaluated by the concordance index
(C-index) and assessed by comparing nomogram-predicted vs. actual
observation, and bootstrapping with 1,000 resamples to decrease the
overfit bias was applied for calibration (20,21).
The glm was generated by glmnet package (version 4.1-4) (22) and the nomogram was generated by
DynNom package (version 5.0.2) (23). All analyses and figures were
generated in R version 4.0.3 (https://www.r-project.org/).
Results
Differential laboratory biomarkers
between AFB- IGRA+ TB and PN
The characteristics of the AFB-
IGRA+ TB and PN participants are shown in Table I. No significant differences in age
and sex were found in the first cohort. Males made up the majority
in both cohorts. Clinical types of TB participants included 162
infiltrative pulmonary TB, 19 cavitary pulmonary TB, 8 secondary
pulmonary TB, and 15 tuberculous pleurisy and empyema. Only 44 and
23 participants had received TB-DNA and TB-antibody examination,
yielding 9.09% (4/44) and 13.04% (3/23) positive rates,
respectively.
 | Table IDemographics and baseline
characteristics of AFB- IGRA+ TB and PN
participants. |
Table I
Demographics and baseline
characteristics of AFB- IGRA+ TB and PN
participants.
| | The first
cohort | The second
cohort |
|---|
| Variables | TB | PN | χ2 | P-value | TB | PN |
|---|
| Age | n=204 | n=156 | | | n=77 | n=42 |
|
Mean ±
SD | 46±15 | 47±15 | 0.03 | 0.87 | 29±9 | 41±19 |
|
≤20 | 13 | 12 | - | - | 10 | 7 |
|
21-40 | 56 | 29 | - | - | 57 | 16 |
|
41-60 | 96 | 89 | - | - | 10 | 12 |
|
≥60 | 39 | 26 | - | - | 0 | 7 |
| Sex | | | | | | |
|
Male | 150 | 118 | 3.82 | 0.05 | 52 | 26 |
|
Female | 54 | 38 | 2.78 | 0.10 | 25 | 16 |
| Types | | | | | | |
|
Infiltrative | 162 | - | - | - | 41 | - |
|
Cavitary | 19 | - | - | - | 3 | - |
|
Secondary | 8 | - | - | - | 33 | - |
|
Tuberculous
pleurisy/empyema | 15 | - | - | - | 0 | - |
| Anti-TB
treatment | | | | | | |
|
Yes | 163 | - | - | - | - | - |
|
No | 41 | - | - | - | - | - |
| TB-DNA (FAM) | | | | | | |
|
Positive | 4 | - | - | - | 38 | - |
|
Negative | 40 | - | - | - | 30 | - |
| TB-antibody | | | | | | |
|
Positive | 3 | - | - | - | 20 | - |
|
Negative | 20 | - | - | - | 15 | - |
| Treatment | | | | | | |
|
Initial | 143 | - | - | - | 77 | - |
|
Re-treated | 13 | - | - | - | 0 | - |
The significant variables between the two conditions
were further explored. Using unpaired t-test and setting FDR
<0.2, 13 variables with marked differences were identified
(Table II). Only uric acid (UA)
was elevated (fold change >1.2). Notably, five variables,
including aspartate aminotransferase (AST), total bile acid (TBA),
triglyceride (TG), alanine aminotransferase (ALT), and glutamyl
transpeptidase (GGT), were reduced (fold change <0.83) in TB
compared with PN (Fig. 2A). These
data revealed variable expression differences between the two
conditions, and that some variables may be useful to distinguish
AFB- IGRA+ TB from PN.
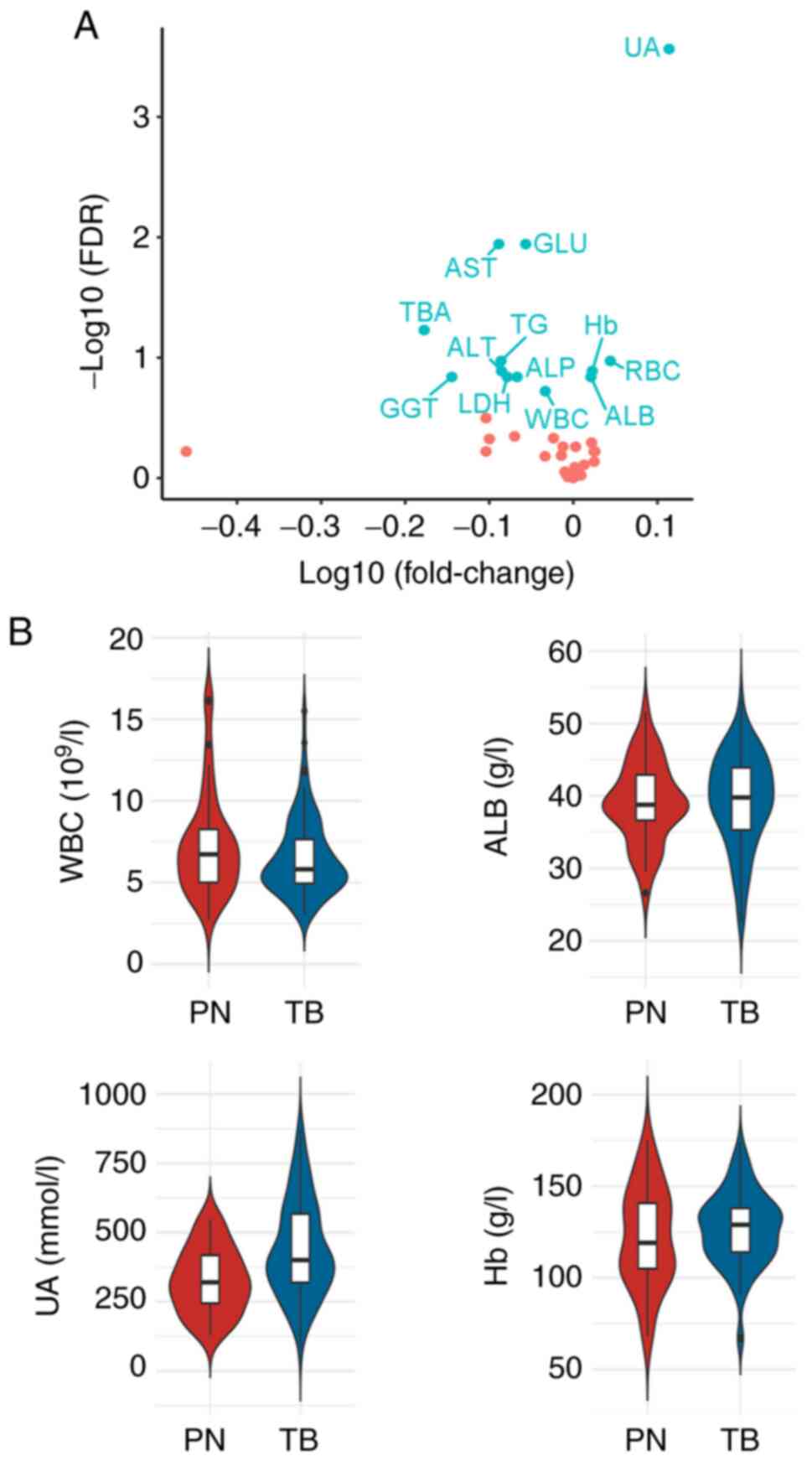 | Figure 2Statistical differential comparison
between AFB- IGRA+ TB and PN. (A) Volcano
plot showing the distribution of all laboratory variables between
the TB and PN. (B) Violin plots showing the values of four
variables between two groups. AFB-, acid-fast bacillus
smear-negative; IGRA+, interferon-γ release
assay-positive; TB, tuberculosis; PN, pneumonia; UA, uric acid;
GLU, glucose; AST, aspartate aminotransferase; TBA, total bile
acid; TG, triglyceride; Hb, hemoglobin; ALT, alanine
aminotransferase; ALP, alkaline phosphatase; RBC, red blood cell;
GGT, glutamyl transpeptidase; LDH, lactate dehydrogenase; WBC,
white blood cell; ALB, albumin. |
 | Table IIStatistical differences and OR values
of each variable in AFB- IGRA+ TB compared
with PN. |
Table II
Statistical differences and OR values
of each variable in AFB- IGRA+ TB compared
with PN.
| Variables | P-value | FDR | Fold change | OR | P-value of OR | 2.5% CI | 97.5% CI |
|---|
| UA | 6.82E-06 | 2.73E-04 | 1.3 | 0.36 | 2.49E-05 | 0.22 | 0.57 |
| GLU | 6.94E-04 | 0.01 | 0.88 | 4.26 | 1.45E-03 | 1.82 | 10.93 |
| AST | 8.54E-04 | 0.01 | 0.82 | 1.78 | 1.47E-03 | 1.26 | 2.56 |
| TBA | 5.90E-03 | 0.06 | 0.66 | 1.59 | 0.01 | 1.14 | 2.3 |
| TG | 1.45E-02 | 0.11 | 0.82 | 2.66 | 0.02 | 1.2 | 6.65 |
| RBC | 0.02 | 0.11 | 1.11 | 0.28 | 0.02 | 0.09 | 0.72 |
| ALT | 0.02 | 0.13 | 0.82 | 1.31 | 0.03 | 1.03 | 1.67 |
| Hb | 0.03 | 0.13 | 1.05 | 0.37 | 0.03 | 0.15 | 0.89 |
| GGT | 0.03 | 0.14 | 0.72 | 1.33 | 0.04 | 1.02 | 1.74 |
| ALP | 0.04 | 0.14 | 0.86 | 1.92 | 0.05 | 1.02 | 3.71 |
| LDH | 0.04 | 0.14 | 0.83 | 1.6 | 0.06 | 1.01 | 2.71 |
| ALB | 0.04 | 0.14 | 1.05 | 0.4 | 0.05 | 0.16 | 0.98 |
| WBC | 0.06 | 0.19 | 0.93 | 1.49 | 0.06 | 0.98 | 2.29 |
To find the odds that TB would progress or not be
given exposure to these laboratory variables, OR was assessed for
each variable by univariate logistic model. Notably, 11 variables
significantly associated with TB progression (P<0.05; Table II) were identified. Among them,
four variables indicated a protective effect in TB progression (OR
<1), including UA, red blood cell (RBC), Hb, and ALB; while
seven variables including GLU, AST, TBA, TG, ALT, GGT, and ALP,
were revealed as risk factors for TB progression (OR >1).
Multivariate risk model to predict TB
progression probability
Combined with the aforementioned results, and using
AIC as a stopping rule, five laboratory variables (age, UA, ALB,
Hb, and WBC) were finally selected to develop a multivariate risk
model with 89 AFB- IGRA+ TB and 38 PN
participants. Notably, the expression distribution of variables in
both groups revealed more unbalance in PN than AFB-
IGRA+ TB (Fig. 2B). The
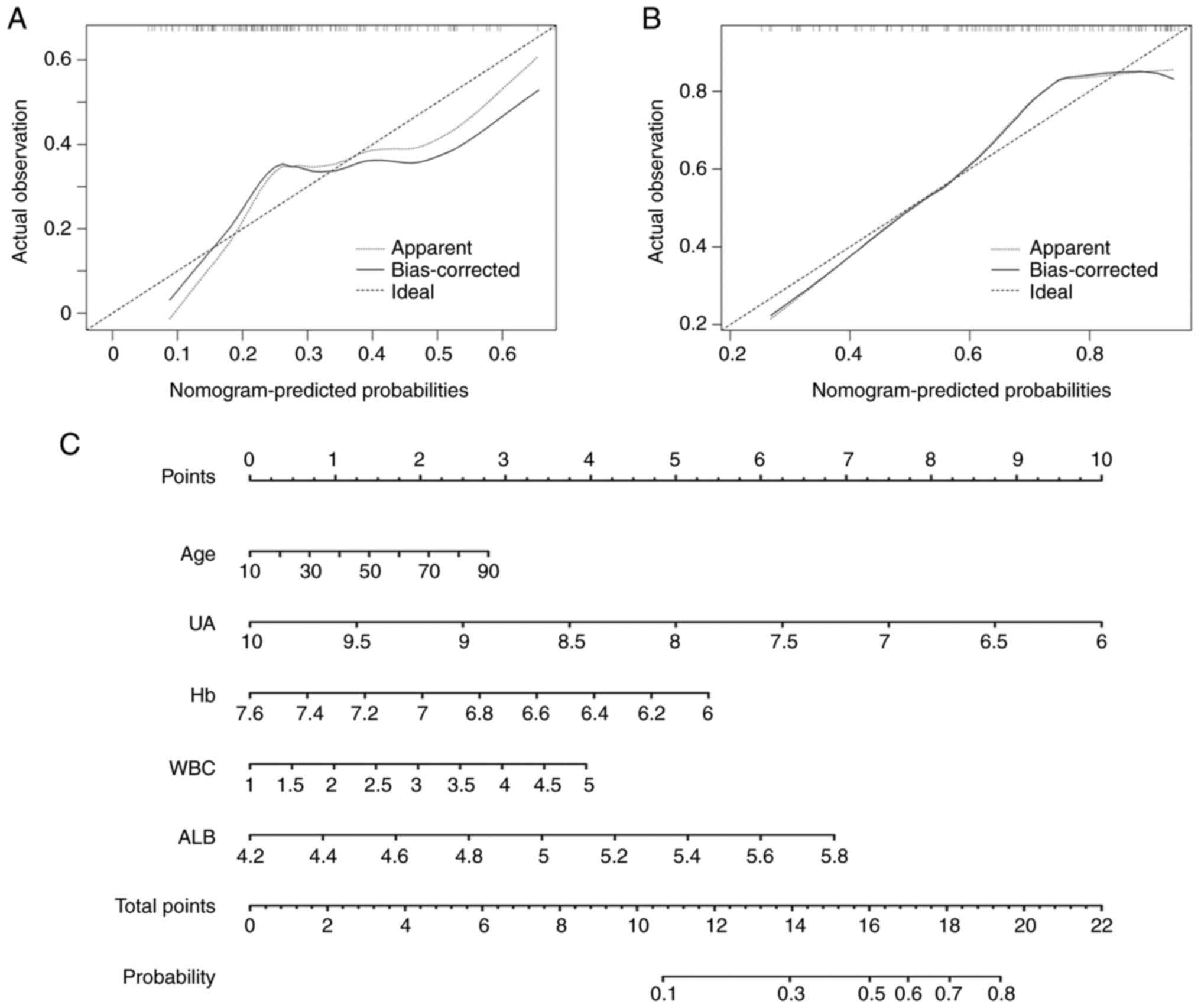
risk model yielded a C-index of 0.7 (95% CI: 0.61, 0.8), with
P=0.01 (chi-square test) (data not shown). The calibration plot
revealed a moderate agreement between the model prediction and the
actual observation (Fig. 3A;
P=0.18, Hosmer-Lemeshow test). Using the nomogram, the values for
each variable were mapped to points on a scale axis ranging from 0
to 10. With a corresponding number of points assigned to given
magnitudes of the variables, the risk probability was calculated by
the corresponding cumulative point score for all the variables
(25). It was revealed that UA had
the most protective effect in TB progression, followed by Hb; while
age, WBC and ALB were shown to be risk factors (Fig. 3C).
Next, an external validation cohort of 134
participants, consisting of 77 AFB- IGRA+ TB
and 42 PN participants were prospectively collected. The C-index of
nomogram for predicting the external cohort was 0.77 (95% CI: 0.68,
0.86) (data not shown). The calibration plot also revealed
consistent results between the prediction by nomogram and actual
observation (Fig. 3B) with P=0.13
(Hosmer-Lemeshow test).
Discussion
In the present study, different profiles were
analyzed between AFB- IGRA+ TB and PN, and
five laboratory variables (age, UA, ALB, Hb and WBC) were selected
to construct a multivariate risk model and nomogram. Internal
validation and a calibration plot showed moderate agreement between
nomogram probability and actual observation, with a C-index of 0.7
(95% CI: 0.61, 0.8). A similar result in an external validation
cohort (C-index: 0.77; 95% CI: 0.68, 0.86) was obtained. These
findings indicated that five laboratory variables may be used to
predict TB disease probability when a clinical sample is
AFB- IGRA+.
It has been reported that patients with TB tend to
exhibit increased levels of CRP, erythrocyte sedimentation rate
(ESR), and UA, and low levels of Hb (25). An increased UA level was observed in
28.2% of men and 37.5% of women prior to chemotherapy, and more
often during the first 2 months of treatment both in men and women,
which suffered from multiple drug-resistant pulmonary TB (26). In the present study, serum UA was
revealed to be at a significantly higher level in TB (FDR
<0.001), with an OR value of 0.36 (P=2.5E-05) compared with PN
(Table II), indicating that it may
be a specific protective factor in patients with TB. Reduced plasma
ALB concentrations have been reported in TB (27) and may be used as a diagnostic and
prognostic marker in pretreated HIV and TB patients. WBC was
revealed to be significantly increased in patients with TB compared
with healthy controls, and the WBC significantly decreased during
TB treatment (28,29). In the present study, WBC was
statistically significant and a significant risk factor (OR=1.49),
but with no higher counts in fold change compared with PN.
To predict the risk of TB for each AFB-
IGRA+ patient, a nomogram was used to provide a more
accurate profile. With five variables, the nomogram had good
predictive accuracy with a C-index of 0.7. External validation was
essential to confirm it can be applied to patients outside of the
cohort. Thus, a second participant cohort from another center was
recruited, and then assessed on the nomogram, and the result
obtained was consistent with the actual observation (C-index of
0.77).
The present study still had several limitations:
First, in low-income and rural settings, not all patients received
all routine laboratory tests, leading to numerous missing values in
the first cohort of participants. In order to analyze more
biomarkers, participants with >50% of missing data were
excluded, with 41 laboratory variables and a small number of
participants remaining (89 AFB- IGRA+ TBA and
38 PN), resulting in a small sample size. Second, although internal
and external validation exhibited good performance, further
investigations are required to optimize the nomogram in larger
cohorts and more types of pulmonary TB.
In addition, the association between prior TB and
lung cancer has been undefined. Several large cohort studies
provided evidence supporting an association between prior TB and
risk of lung cancer (30,31). In a systematic review and
meta-analysis published in 2011, a previous diagnosis of TB was
associated with increased lung cancer risk [relative risk (RR)=1.76
(95% CI=1.49 to 2.08)] with little variation by smoking status
(32). Similarly, a pooled analysis
from the International Lung Cancer Consortium found a lung cancer
RR of 1.48 (95% CI=1.17 to 1.87) associated with a history of TB,
controlling for smoking status. However, there was no attempt to
differentiate TB from lung cancer with the risk model of the
present study. In future projects, applicability of this model in
other diseases will be further investigated. Following improvement
of this model, such as increasing its applicability, it is
anticipated that it may help clinicians to reduce the cost and time
to diagnose AFB- IGRA+ TB in low-income,
high-burdened, and resource-constrained rural area settings.
In conclusion, the present study identified a
five-variable signature to distinguish AFB-
IGRA+ TB from PN patients. A risk model was built to
differentiate AFB- IGRA+ TB from PN, and was
validated in an external independent cohort, which could be applied
in low-income and resource-constrained rural area settings.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Jiangxi Province (grant no. 20202BAB206059) and the
Science and Technology Fund of Guangdong Province (grant no.
sgybey01).
Availability of data and materials
The data that support the findings of this study are
available on request from the corresponding author. The data are
not publicly available due to privacy or ethical restrictions.
Authors' contributions
DX, JZ, FX, QY, KH, WX, HZo and HZh contributed to
the study conception and design. Primary clinical case information,
data collection, and analysis were performed by DX, JZ, QY and FX.
The first draft of the manuscript was written by DX, FX, and HZh.
KH, WX and HZo conducted the literature search, as well as the
screening and quality assessment of the clinical data. DX and HZh
confirm the authenticity of all the raw data. All authors
contributed to this manuscript and have consented to its
submission. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee and the Institutional Review Board of Ganzhou Fifth
People's Hospital (registration no. 2020-10). Written informed
consent was waived by the Ethics Committee as this was an
observational and retrospective analysis study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
WHO: Global Tuberculosis Report 2020.
World Health Organization WHO, Geneva, 2020.
|
|
2
|
Kohli M, Schiller I, Dendukuri N, Yao M,
Dheda K, Denkinger CM, Schumacher SG and Steingart KR: Xpert
MTB/RIF Ultra and Xpert MTB/RIF assays for extrapulmonary
tuberculosis and rifampicin resistance in adults. Cochrane Database
Syst Rev. 1(CD012768)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Trébucq A, Enarson DA, Chiang CY, Van Deun
A, Harries AD, Boillot F, Detjen A, Fujiwara PI, Graham SM,
Monedero I, et al: Xpert® MTB/RIF for national tuberculosis
programmes in low-income countries: When, where and how? Int J
Tuberc Lung Dis. 15:1567–1572. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lim WS: From latent to active TB: Are
IGRAs of any use? Thorax. 71:585–586. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Auguste P, Tsertsvadze A, Pink J, Court R,
McCarthy N, Sutcliffe P and Clarke A: Comparing interferon-gamma
release assays with tuberculin skin test for identifying latent
tuberculosis infection that progresses to active tuberculosis:
Systematic review and meta-analysis. BMC Infect Dis.
17(200)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dheda K, Makambwa E and Esmail A: The
great masquerader: Tuberculosis presenting as community-acquired
pneumonia. Semin Respir Crit Care Med. 41:592–604. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Vessière A, Font H, Gabillard D,
Adonis-Koffi L, Borand L, Chabala C, Khosa C, Mavale S, Moh R,
Mulenga V, et al: Impact of systematic early tuberculosis detection
using Xpert MTB/RIF Ultra in children with severe pneumonia in high
tuberculosis burden countries (TB-Speed pneumonia): A stepped wedge
cluster randomized trial. BMC Pediatr. 21(136)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Grossman RF, Hsueh PR, Gillespie SH and
Blasi F: Community-acquired pneumonia and tuberculosis:
Differential diagnosis and the use of fluoroquinolones. Int J
Infect Dis. 18:14–21. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Williams DJ, Creech CB, Walter EB, Martin
JM, Gerber JS, Newland JG, Howard L, Hofto ME, Staat MA, Oler RE,
et al: Short-vs Standard-course outpatient antibiotic therapy for
community-acquired pneumonia in children: The SCOUT-CAP randomized
clinical trial. JAMA Pediatr. 176:253–261. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hopstaken RM, Muris JW, Knottnerus JA,
Kester AD, Rinkens PE and Dinant GJ: Contributions of symptoms,
signs, erythrocyte sedimentation rate, and C-reactive protein to a
diagnosis of pneumonia in acute lower respiratory tract infection.
Br J Gen Pract. 53:358–364. 2003.PubMed/NCBI
|
|
11
|
Peters JS, McIvor A, Papadopoulos AO,
Masangana T, Gordhan BG, Waja Z, Otwombe K, Letutu M, Kamariza M,
Sterling TR, et al: Differentially culturable tubercle bacteria as
a measure of tuberculosis treatment response. Front Cell Infect
Microbiol. 12(1064148)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Blauenfeldt T, Villar-Hernández R,
García-García E, Latorre I, Holm LL, Muriel-Moreno B, De
Souza-Galvão ML, Millet JP, Sabriá F, Sánchez-Montalva A, et al:
Diagnostic accuracy of interferon gamma-induced protein 10 mRNA
release assay for tuberculosis. J Clin Microbiol. 58:e00848–20.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Qiu X, Wang H, Tang Y, Su X, Ge L, Qu Y
and Mu D: Is interleukin-2 an optimal marker for diagnosing
tuberculosis infection? A systematic review and meta-analysis. Ann
Med. 52:376–385. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ahmad R, Xie L, Pyle M, Suarez MF, Broger
T, Steinberg D, Ame SM, Lucero MG, Szucs MJ, MacMullan M, et al: A
rapid triage test for active pulmonary tuberculosis in adult
patients with persistent cough. Sci Transl Med.
11(eaaz9925)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yoon C, Semitala FC, Atuhumuza E, Katende
J, Mwebe S, Asege L, Armstrong DT, Andama AO, Dowdy DW, Davis JL,
et al: Point-of-care C-reactive protein-based tuberculosis
screening for people living with HIV: A diagnostic accuracy study.
Lancet Infect Dis. 17:1285–1292. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3(Article3)2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Smyth GK: limma: Linear Models for
Microarray Data. In: Bioinformatics and Computational Biology
Solutions Using R and Bioconductor. Gentleman R., Carey V.J., Huber
W., Irizarry R.A., Dudoit S (eds). Springer, New York, NY, pp
397-420 (, 2005).
|
|
18
|
Lim WK and Lim AW: A Comparison Of Usual
t-Test Statistic and Modified t-Test Statistics on Skewed
Distribution Functions. J modern applied statistical methods:
JMASM. 15:67–89. 2016.
|
|
19
|
Capanu M and Seshan VE: False discovery
rates for rare variants from sequenced data. Genet Epidemiol.
39:65–76. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tang LQ, Li CF, Li J, Chen WH, Chen QY,
Yuan LX, Lai XP, He Y, Xu YX, Hu DP, et al: Establishment and
validation of prognostic nomograms for endemic nasopharyngeal
carcinoma. J Natl Cancer Inst. 108(djv291)2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jang JY, Park T, Lee S, Kim Y, Lee SY, Kim
SW, Kim SC, Song KB, Yamamoto M, Hatori T, et al: Proposed nomogram
predicting the individual risk of malignancy in the patients with
branch duct type intraductal papillary mucinous neoplasms of the
pancreas. Ann Surg. 266:1062–1068. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Friedman J, Hastie T and Tibshirani R:
Regularization paths for generalized linear models via coordinate
descent. J Stat Softw. 33:1–22. 2010.PubMed/NCBI
|
|
23
|
Jalali A, Alvarez-Iglesias A, Roshan D and
Newell J: Visualising statistical models using dynamic nomograms.
PLoS One. 14(e0225253)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Balachandran VP, Gonen M, Smith JJ and
DeMatteo RP: Nomograms in oncology: More than meets the eye. Lancet
Oncol. 16:e173–e180. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gil-Santana L, Cruz LAB, Arriaga MB,
Miranda PFC, Fukutani KF, Silveira-Mattos PS, Silva EC, Oliveira
MG, Mesquita EDD, Rauwerdink A, et al: Tuberculosis-associated
anemia is linked to a distinct inflammatory profile that persists
after initiation of antitubercular therapy. Sci Rep.
9(1381)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Аbdullaev RY, Komissarova OG, Chumakova ES
and Odinet VS: Level of uric acid in blood serum of new pulmonary
tuberculosis patients with multiple drug resistance. Tuberculosis
and Lung Diseases. 95:31–36. 2017.
|
|
27
|
Bisaso KR, Owen JS, Ojara FW, Namuwenge
PM, Mugisha A, Mbuagbaw L, Luboobi LS and Mukonzo JK:
Characterizing plasma albumin concentration changes in TB/HIV
patients on anti retroviral and anti-tuberculosis therapy. In
Silico Pharmacol. 2(3)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Rohini K, Surekha Bhat M, Srikumar PS and
Mahesh Kumar A: Assessment of hematological parameters in pulmonary
tuberculosis patients. Indian J Clin Biochem. 31:332–335.
2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Carole C, Kokhreidze E, Tukvadze N, Banu
S, Uddin MKM, Biswas S, Russomando G, Acosta CCD, Arenas R,
Ranaivomanana PP, et al: Association of baseline white blood cell
counts with tuberculosis treatment outcome: A prospective
multicentered cohort study. Int J Infect Dis. 100:199–206.
2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yu YH, Liao CC, Hsu WH, Chen HJ, Liao WC,
Muo CH, Sung FC and Chen CY: Increased lung cancer risk among
patients with pulmonary tuberculosis: A population cohort study. J
Thorac Oncol. 6:32–37. 2011.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wu CY, Hu HY, Pu CY, Huang N, Shen HC, Li
CP and Chou YJ: Pulmonary tuberculosis increases the risk of lung
cancer: A population-based cohort study. Cancer. 117:618–624.
2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Brenner DR, McLaughlin JR and Hung RJ:
Previous lung diseases and lung cancer risk: A systematic review
and meta-analysis. PLoS One. 6(e17479)2011.PubMed/NCBI View Article : Google Scholar
|