Introduction
MicroRNAs (miRNAs) are small RNAs of ~21-24
nucleotides (nt) that generally regulate protein expression in
cells and influence cellular processes (1,2).
Particularly, miR-17 has been involved in adenosine deaminase
expression that acts on RNA in melanoma stem cells and regulates
the editing of dedicator of cytokinesis mRNA (3). MiRNAs are intracellularly expressed
molecules, and they regulate gene expression in cells and are
released from cells through various mechanisms (4,5). The
well-known release mechanism is the release of extracellular
vesicles (EVs), such as exosomes that contain miRNAs inside
(6,7). MiRNAs are relatively stable and
somewhat resistant to ribonuclease inside exosomes (8).
There are two major RNA extraction methods (9). One is Guanidinium Thiocyanate
(GuSCN)-Phenol-Chloroform Extraction. In this method, GuSCN is
added and homogenized, and then an acidic solution consisting
mainly of sodium acetate, phenol and chloroform is added and
centrifuged to separate the RNA in the aqueous layer and DNA in the
organic layer. By adding isopropanol to the aqueous layer where the
RNA is eluted, the RNA is precipitated and can be recovered.
Another method is the silica matrix method. In this method, DNA and
RNA are bound to silica-based filters or beads because of their
high affinity for silica. In the case of RNA extraction, DNA and
RNA are bound to silica filters or beads and washed against a
sample of ethanol containing DNase. The advantages of the
GuSCN-Phenol-Chloroform Extraction are that RNA can be extracted
from basically any sample, and the RNA concentration can be
adjusted to some extent by adjusting the amount of water. The
disadvantage is that the RNA recovery rate is lower than that of
the silica membrane filter base, and depends to some extent on the
skill of the technician who performs the extraction. It is also
environmentally unfriendly because of the use of phenol. The
advantages of the silica filter base are that it does not use
phenol, the extraction method is simple, and the recovery rate and
purity are favorable. It is also beneficial for the environment
since most of them do not use phenol. The disadvantages are that
the extractable sample may vary depending on the extraction kit,
and the amount of water used to elute the RNA is fixed, therefore
it is not possible to adjust the concentration by oneself.
Therefore, it is necessary to consider the advantages and
disadvantages and use the RNA extraction method according to the
purpose.
Previous studies have revealed the presence of small
RNAs in blood and various body fluids (10). Serum miRNAs has been a potential
biomarker in the diagnosis and prognosis of various diseases,
including cancer (11-13),
cardiovascular diseases (14) and
neurodegenerative diseases (15).
Researchers frequently use different protocols and kits to perform
serum RNA extractions for miRNA analysis (16). However, each extraction method has
different principles, which may influence the yield and composition
of the extracted serum RNA. Usually, having an RNA extraction kit
that can recover the most RNAs from the serum is essential for
comprehensive RNA analysis, such as microarrays, because it is
easier to conduct analysis with higher concentrations of RNA.
Therefore, the present study compared the RNA yield and composition
of mice serum using five different RNA extraction kits.
Materials and methods
Mice and blood collection
A total of 24 male C57BL/6NJcl mice (8 weeks-old;
body weight, 24.0±0.9 g) were purchased from CLEA Japan. All mice
were provided a solid diet CE-2 (CLEA Japan) and water ad
libitum and were housed in a conventional animal room with
12/12-h light/dark cycle. Mice were housed up to five mice per
cage, and bedding, feed and water were changed weekly. Mice were
observed 2-3 times per day for monitoring, and health or behavior
abnormalities were not observed during the rearing period. Blood
samples from all mice were collected by cardiac blood sampling
under anesthesia with the inhalation anesthetic solution isoflurane
(Pfizer) at the end of the 8-week time points. Small animal
anesthesia machines (Muromachi Kikai) were used to anesthetize the
mice. Isoflurane vaporized to a concentration of 4-5% was inhaled
into the mice and maintained at 2-3% throughout the experiment, and
blood was drawn from the mice's hearts. After anesthesia, ~0.5-1.0
ml of blood was received from the heart, and the mice were promptly
cervically dislocated to minimize distress as a humane endpoint.
The start of anesthesia to the end of blood collection took <10
min per animal. Death was confirmed by respiratory and cardiac
arrest. All mice were euthanized immediately after the experiment.
Serum samples that were separated using BD MicroTainer® SST (Nippon
Becton Dickinson) blood collection tubes were used. Anticoagulants
were not used. Serum was used because RNAs extracted from plasma
contains platelet-derived RNAs (17). The collected blood was centrifuged
at 6,000 x g for 3 min at room temperature to separate the serum.
Serum was collected from 24 mice. The sera collected were not
pooled and RNA was extracted from each individual. Serum at 100 µl
was dispensed from one mouse in each group, which was considered as
one sample. Four mice were used for each RNA extraction method. The
present study was approved (approval no. AE01-2023-097-1) by the
Hirosaki University Ethics Committee for Animal Experiments
(Hirosaki, Japan), and was conducted under the Hirosaki University
Guidelines for Animal Experiments.
RNA extraction
RNAs were extracted using serum of 100 µl from
8-week-old C57BL/6NJcl male mice and four reagents, including
miRNeasy Serum/Plasma Advanced kit (cat. no. 217204), miRNeasy mini
kit (cat. no. 217004; both from Qiagen KK), TRIzol-LS (cat. no.
10296028), and mirVana™ PARIS™ RNA and Native Protein Purification
Kit (cat. no. AM1556; both from Thermo Fisher Scientific, Inc.),
following the manufacturer's protocol. Additionally, an exoRNeasy
midi kit (cat. no. 77144; Qiagen KK) in two ways was used to
extract RNAs to determine the presence of RNAs in serum EVs. One
method was performed following the manufacturer's protocol to
extract EV RNAs from the serum. The other was utilized to extract
RNAs from the aqueous layer from serum of 200 µl to which QIAzol
Lysis Reagent (cat. no. 79306; Qiagen KK) of 700 µl was added and
separated into two layers to determine the amount of total RNAs in
the serum. The concentration of extracted RNAs was measured by
Qubit™ microRNA Assay Kits (cat. no. Q32880.) and Qubit 4
Fluorometer (cat. no. Q33238; both from Thermo Fisher Scientific,
Inc.) following the manufacturer's protocol. The RNA 6000 Pico 2100
bioanalyzer system (cat. no. 5067-1513) and bioanalyzer small RNA
chip (cat. no. 5067-1548; both from Agilent Technologies, Inc.)
were used to assess the size of extracted RNAs from 8-week-old
C57BL/6NJcl mice.
miRNA microarray
RNAs of 1.8 ng extracted by the aforementioned
extraction method were used for miRNA microarray analysis to
examine serum miRNA expressions, following the manufacturer's
instructions and as previously reported (18). The microRNA Spike In Kit (cat. no.
5190-1934; Agilent Technologies, Inc.) was used to perform quality
checks of the microarray experiments. The RNA samples,
Cyanine-3-labeled fluorescently, were hybridized to SurePrint G3
Mouse 8x60-K miRNA microarray slides (cat. no. G4872A; Agilent
Technologies, Inc.) at 55˚C for 20 h. A SureScan Microarray Scanner
(cat. no. G4900DA; Agilent Technologies, Inc.) was utilized to
detect fluorescence signals using Agilent Feature Extraction 12.0
(Agilent Technologies, Inc.). As a method of evaluating Spike-In,
Agilent Feature Extraction 12.0 was used to verify that the
calculated values of LabelingSpike-InSignal and HybSpike-InSignal
are each >2.5. This indicates that the microarray experiments
are favorable. From all raw data obtained, excluding control
probes, those with signal values 3-fold higher than the error value
were selected by ‘gls Gene Detected’. The selected raw data were
normalized with quantile normalization and displayed
logarithmically. These data were registered with the Gene
Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE246437).
Statistical analysis
The mann-whitney U-test and multiple-test
Steel-Swass method was used to statistically analyze RNA-yield
data. Spearman's rank correlation coefficient was utilized to
assess the correlation coefficient of the microarrays. All tests
were statistically processed with a sample size of four. The
Statcel 4 software (OMS publication, https://oms-publ.main.jp/main/4steps4-hyo1/), was used
for statistical analysis. Correlation coefficients were calculated
and plotted using R (version 4.2.3).
Results
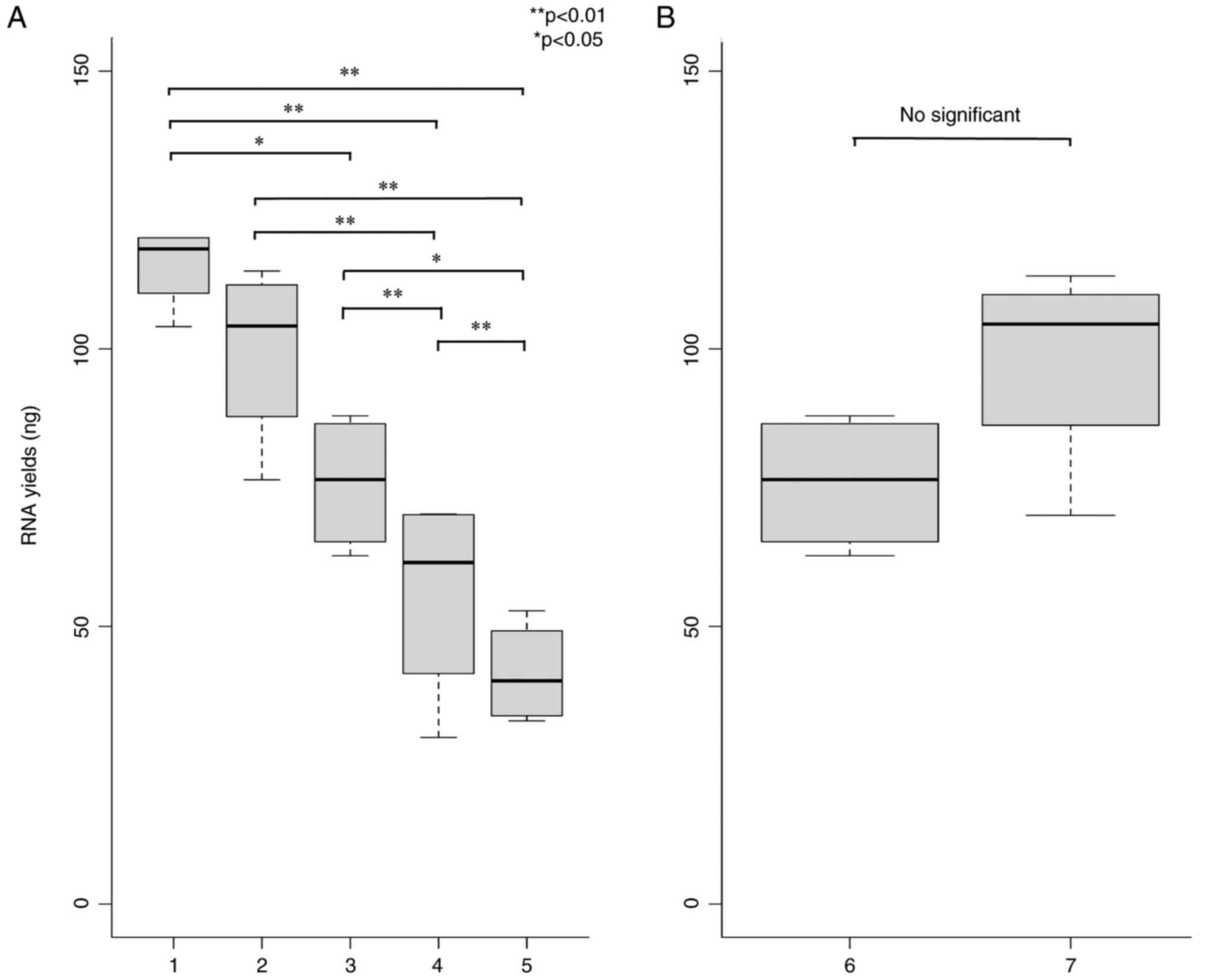
Comparison of the RNA yields from
mouse serum
The RNA yields in mouse serum were compared for each
of the RNA extraction reagents. Qubit™ microRNA Assay Kit was
utilized to measure the RNA concentration, and the yield was
calculated based on the amount of RNase-free water and RNA
concentration. The yields of RNAs in the five RNA extraction
reagents were compared. The results demonstrated significantly
different yields from the miRNeasy Serum/Plasma Advanced kit and
the mirVana™ PARIS™ RNA and Native Protein Purification Kit (from
the miRNeasy mini kit and TRIzol-LS (Fig. 1A). Thus, the miRNeasy Serum/Plasma
Advanced kit or mirVana™ PARIS™ RNA and Native Protein Purification
Kit should be used for the most efficient RNA extraction from mouse
serum. Conversely, the exoRNeasy midi kit was used for two
different extraction methods to assess the proportion of serum RNAs
contained within EVs. The results revealed no statistically
significant difference in yield between the two RNA extraction
methods (Fig. 1B). This indicated
the presence of most serum RNAs in the EVs in serum.
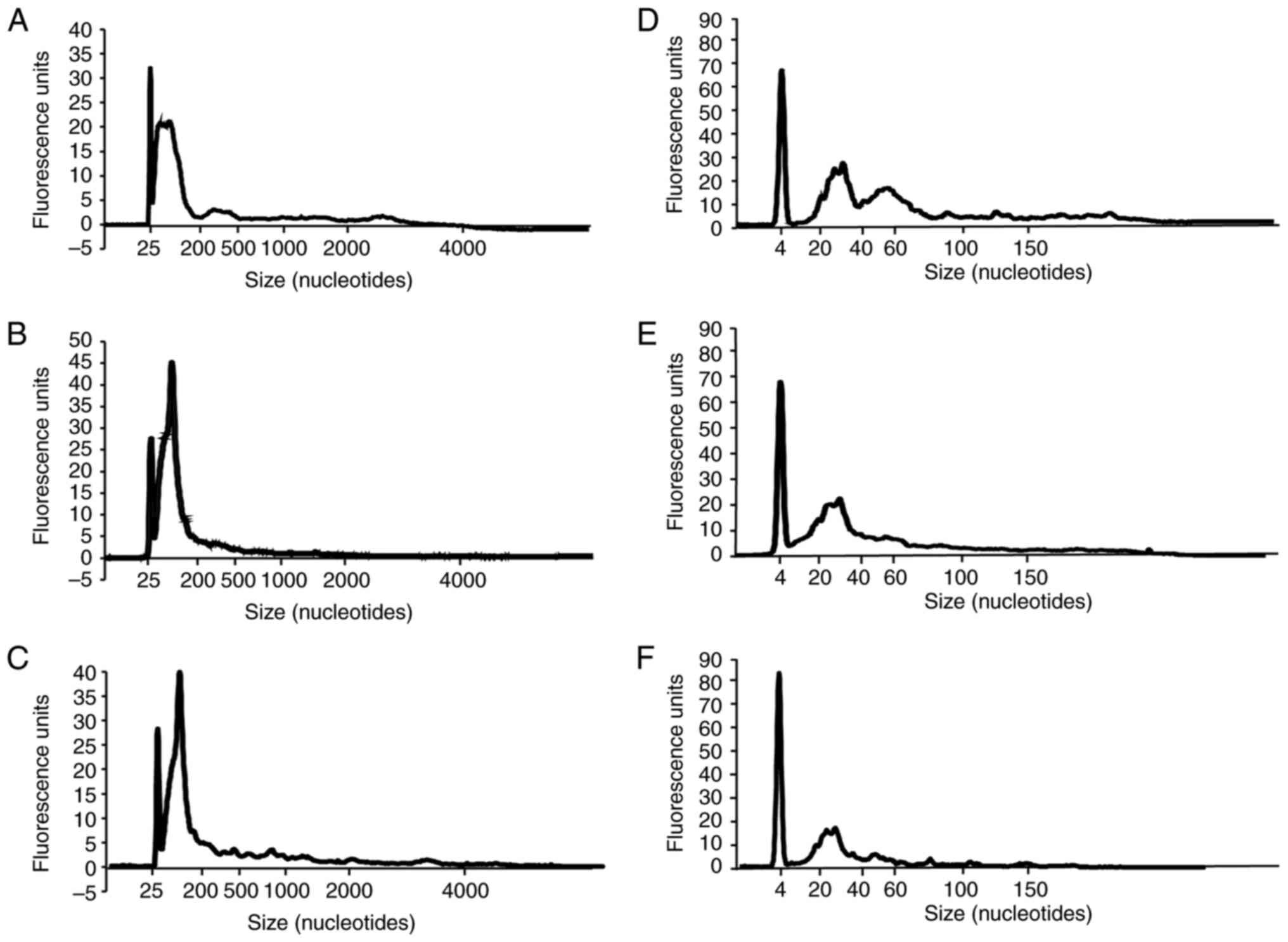
Several small RNAs are present in
serum RNA
The RNA 6000 Pico 2100 bioanalyzer system and the
bioanalyzer small RNA chip in the three RNA extraction reagents
with the hightest RNA yields aforementioned were used to confirm
RNA quality and size in serum. The results of the three RNA
extraction reagents demonstrated that the RNA 6000 Pico bioanalyzer
system confirmed small RNAs of <200 nt (Fig. 2A-C). The bioanalyzer small RNA chip
detected 20-40 nt small RNAs (Fig.
2D-F). Further, serum RNAs extracted by miRNeasy Serum/Plasma
Advanced kit exhibited another peak of ~40-100 nt using a
bioanalyzer small RNA chip (Fig.
2D). This result indicated that most of the RNAs in the serum
are small RNAs. Further, small RNAs from 40-100 nt was also
efficiently extracted from serum using the miRNeasy Serum/Plasma
Advanced kit. This revealed that the extracted RNAs may differ in
composition based on the RNA extraction method.
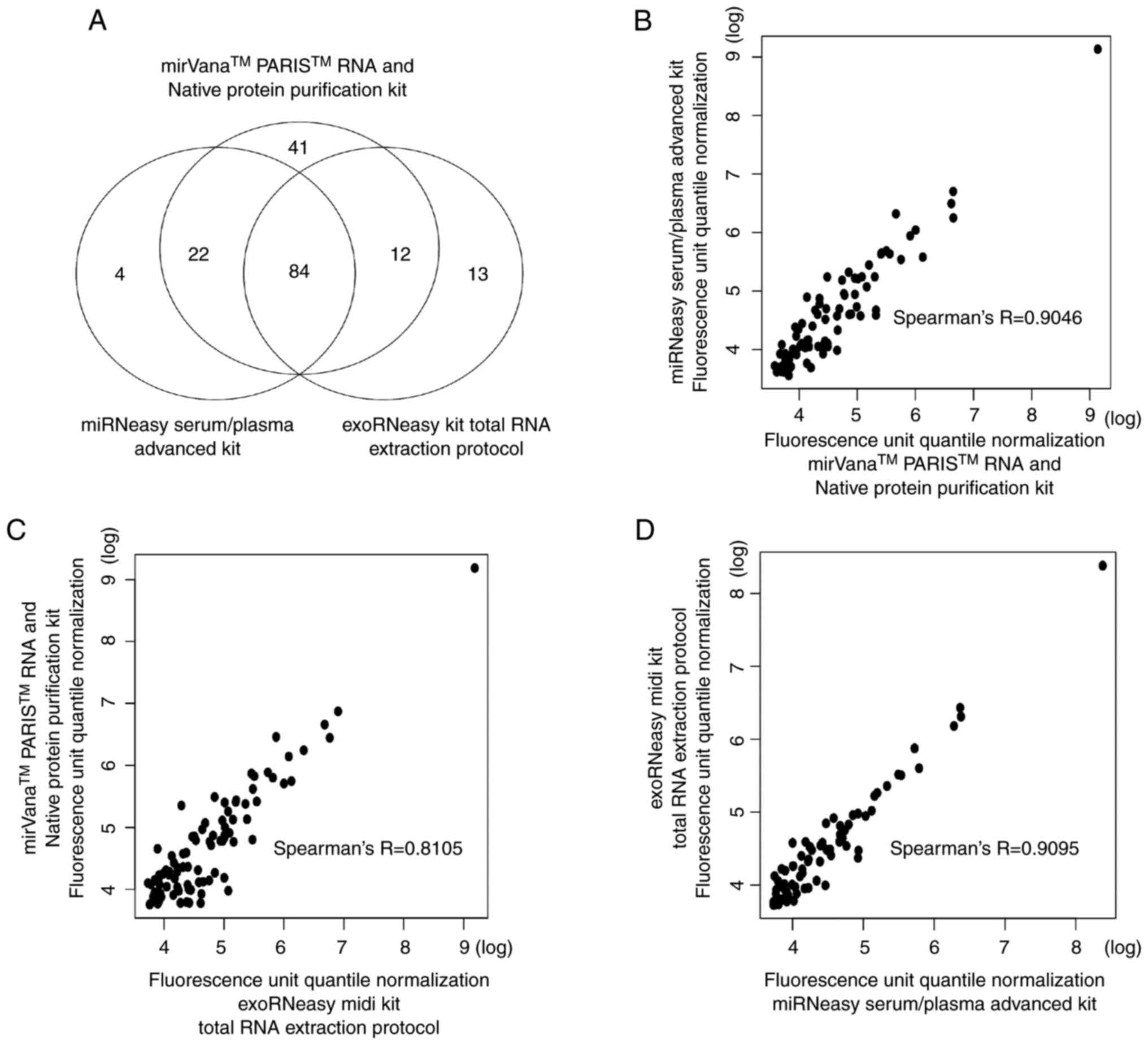
Correlation between miRNA expression
in mouse serum and RNA extraction reagents
The three RNA extraction reagents with the highest
RNA yields were used to perform miRNA microarrays to determine the
differences in serum miRNA expression obtained with each RNA
extraction method and examine the correlations. It was confirmed
that there were no problems with the microarray experiment by
quality check. These three RNA extraction reagents commonly
expressed 84 types of miRNAs (Fig.
3A). However, some types of miRNAs were only detected with
certain RNA extraction reagents. This suggested that different RNA
extraction reagents may cause differences in the types of miRNAs
detected. Furthermore, the correlation between the expression
levels of commonly expressed miRNAs in all combinations of RNA
extraction reagents was examined. There was a high correlation
between the expression of common miRNAs detected by each RNA
extraction reagent (Fig. 3B-D).
These results indicated that the miRNAs commonly expressed by the
three RNA extraction reagents are highly correlated in expression
levels.
Discussion
The present study used various RNA extraction
reagents to compare RNA yields, size and small RNAs components from
mouse serum. Serum, not plasma, was used as the specimen. Plasma
generally contains more platelets, and differences in the number of
platelets in individuals may affect the amount and type of miRNAs
in the plasma. It has been reported that platelets also contain
high amounts of miRNAs (19).
Therefore, serum was used to exclude the effect of platelet count
on the amount and type of miRNAs.
RNA extraction methods using miRNeasy Serum/Plasma
Advanced kit and mirVana™ PARIS™ RNA and Native Protein
Purification Kit, following the manufacturer's protocol,
demonstrated the highest RNA yields from 100 µl of mouse serum
(Fig. 1A). Conversely, the yields
of RNAs collected from the RNA extraction methods using the
miRNeasy mini kit and TRIzol-LS were significantly lower than those
of the aforementioned two methods (Fig.
1A). These results revealed that serum RNA extraction methods
using the miRNeasy Serum/Plasma Advanced kit and mirVana™ PARIS™
RNA and Native Protein Purification Kit (efficiently collected
serum RNAs in terms of RNA yields. However, the RNAs must be
concentrated by ethanol precipitation or other methods to obtain
high RNA concentrations, since the mirVana™ PARIS™ RNA and Native
Protein Purification Kit elutes RNAs with 100 µl of RNase-free
water.
Wright et al (20) revealed that miRNeasy Serum/Plasma
Advanced kit is the best extraction kit for blood miRNAs using
sheep serum as sample. This was consistent with the current results
in terms of RNA yields and ease of use. Silica-based or magnetic
beads-based was recommended for RNA extraction of hepatitis C virus
in serum, as used in a recent study (21). The miRNeasy Serum/Plasma Advanced
kit and mirVana™ PARIS™ RNA and Native Protein Purification Kit are
silica-based kits, and TRIzol-LS is a guanidinium phenol-based kit.
Silica-based RNA extraction was considered to be improved for RNA
extraction using samples with low RNA content, such as serum.
There have been several studies on RNA yield from
serum; Tang et al (22) in
their study on RNA extraction from extracellular vesicle-derived
RNA in human serum identified that the exoRNeasy kit had a higher
RNA yield than Trizol-LS. Trakunram et al (23) also reported that RNA extraction of
human serum demonstrated improved RNA yield and purity with the
miRNeasy mini kit compared with Trizol-LS. These results are
consistent with the present study, with improved RNA yield with the
silica filter base than with the phenol base. The present study
also used mouse serum, but similar results were confirmed with
human serum.
RNAs in serum is generally contained inside EVs,
such as exosomes (24-27),
but the extent to which RNAs in serum is present in EVs remains
unclear. Therefore, the RNA yields of the used methods were
compared to extract EV RNAs and total RNAs in serum using the
exoRNeasy midi kit. This result revealed no significant difference
between the amount of serum EV RNAs and that of serum total RNAs,
indicating that most of the serum RNA may be RNAs in EVs (Fig. 1B).
Two types of Agilent Bioanalyzer chips were used for
RNA electrophoresis to determine the size of serum RNAs. The
results revealed the peaks of small RNAs of <200 nt in RNA 6000
Pico 2100 bioanalyzer system, and the peaks of small RNAs of 20-40
nt in the bioanalyzer small RNA chip (Fig. 2). Interestingly, RNAs extracted from
the miRNeasy Serum/Plasma Advanced kit demonstrated a reproducible
bimodal pattern with peaks of ~40-100 nt (Fig. 2D). The peaks of small RNAs at 20-40
nt are mainly miRNAs, whereas the peaks at 40-100 nt are small RNAs
that do not match the size of miRNAs. In the present study,
sufficiently heat-treated serum RNAs were extracted, and the peak
was not caused by miRNA duplication. RNAs that match this size may
consist of precursor miRNAs (28),
transfer RNAs (29,30) and small nucleolar RNAs (snoRNAs)
(31,32). Fitz et al (33) demonstrated that snoRNAs are
encapsulated within EVs and exist extracellularly, and that snoRNAs
in the EVs in serum can be a diagnostic biomarker for Alzheimer's
disease (33). The miRNeasy
Serum/Plasma Advanced kit that efficiently collected 40-100 nt of
small RNAs was unclear, but serum RNAs by miRNeasy Serum/Plasma
Advanced kit may be suitable for extracting small RNAs other than
miRNAs. The novelty of the present study was that it is the first,
to the best of the authors' knowledge, to evaluate the miRNeasy
Serum/Plasma Advanced kit for RNA extraction from serum. In
addition, a bimodal pattern was observed in the miRNeasy
Serum/Plasma Advanced Kit. This is also a novel result, as it had
not been previously reported.
Further, 84 miRNAs were expressed in common with the
three types that demonstrated the highest amount of RNA extraction,
but some miRNAs were not confirmed to be expressed in common with
the three types (Fig. 3A). No
issues were concluded in using RNA extraction kits throughout the
experiment. It has been recently reported that miRNA expression
levels in blood are lower than cell/tissue expression levels
(34). In the present study, RNA
was extracted from 100 µl of serum, and it is expected that the
number of miRNAs commonly detected in the three protocols will
increase as the amount of specimen used increases. There are two
possible reasons why the types of miRNAs do not completely match in
all three extraction kits: One reason is that there are individual
differences in the samples. The other may be a bias in the type of
small RNAs extracted due to the characteristics of the RNA
extraction method as demonstrated in Fig. 2.
A high correlation exists in miRNA expression levels
among the RNA extraction kits examined, and any RNA extraction kit
may be used when examining miRNA expressions with microarrays
(Fig. 3B-D). However, the
extraction efficiency of small RNAs other than miRNAs may differ
based on the RNA extraction method. Next-generation sequencing or
other methods are reqired to clarify the components of these 40-100
nt small RNAs in the future. In addition, the sample size in the
present study was small and will need to be reexamined with a
sufficient sample size in the future.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported in part by The JSPS
KAKENHI (grant no. 21H04844) and JST SPRING (grant no. JPMJSP2152)
in Japan.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request The datasets generated and/or analyzed during the current
study are available in the Gene Expression Omnibus repository
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE246437).
Authors' contributions
KY and MC were major contributors to performing the
experiments and writing the manuscript. All authors read and
approved the final manuscript. KY and MC confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
All experiments were performed in accordance with
The Guideline for Animal Experimentation of Hirosaki University.
The present study was approved (approval number: AE01-2023-097-1)
by the Animal Research Committee of Hirosaki University (Hirosaki,
Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Das K and Rao LVM: The role of microRNAs
in inflammation. Int J Mol Sci. 23(15479)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Siddika T and Heinemann IU: Bringing
microRNAs to light: Methods for microRNA quantification and
visualization in live cells. Front Bioeng Biotechnol.
8(619583)2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zhang Y, Yang X, Cui Y and Zhang X:
Suppression of RNA editing by miR-17 inhibits the stemness of
melanoma stem cells. Mol Ther Nucleic Acids. 27:439–455.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liu YJ and Wang C: A review of the
regulatory mechanisms of extracellular vesicles-mediated
intercellular communication. Cell Commun Signal.
21(77)2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Catalano M and O'Driscoll L: Inhibiting
extracellular vesicles formation and release: A review of EV
inhibitors. J Extracell Vesicles. 9(1703244)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Xu D, Di K, Fan B, Wu J, Gu X, Sun Y, Khan
A, Li P and Li Z: MicroRNAs in extracellular vesicles: Sorting
mechanisms, diagnostic value, isolation, and detection technology.
Front Bioeng Biotechnol. 10(948959)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Vaka R, Parent S, Risha Y, Khan S,
Courtman D, Stewart DJ and Davis DR: Extracellular vesicle microRNA
and protein cargo profiling in three clinical-grade stem cell
products reveals key functional pathways. Mol Ther Nucleic Acids.
32:80–93. 2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu HS and Xiao HS: MicroRNAs as potential
biomarkers for gastric cancer. World J Gastroenterol.
20:12007–12017. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ali N, Rampazzo RCP, Costa ADT and Krieger
MA: Current nucleic acid extraction methods and their implications
to point-of-care diagnostics. Biomed Res Int.
2017(9306564)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang P, Wu W, Chen Q and Chen M:
Non-coding RNAs and their integrated networks. J Integr Bioinform.
16(20190027)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li X, Chen W, Li R, Chen X, Huang G, Lu C,
Wen Z, Peng X, Liu K, Zhang C, et al: Bladder cancer diagnosis with
a four-miRNA panel in serum. Future Oncol. 18:3311–3322.
2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Peng X, Wang J, Zhang C, Liu K, Zhao L,
Chen X, Huang G and Lai Y: A three-miRNA panel in serum as a
noninvasive biomarker for colorectal cancer detection. Int J Biol
Markers. 35:74–82. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kim DH, Park H, Choi YJ, Im K, Lee CW, Kim
DS, Pack CG, Kim HY, Choi CM, Lee JC, et al: Identification of
exosomal microRNA panel as diagnostic and prognostic biomarker for
small cell lung cancer. Biomark Res. 11(80)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Han S, Fang J, Yu L, Li B, Hu Y, Chen R,
Li C, Zhao C, Li J, Wang Y, et al: Serum-derived exosomal
has-let-7b-5p as a biomarker for predicting the severity of
coronary stenosis in patients with coronary heart disease and
hyperglycemia. Mol Med Rep. 28(203)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu WL, Lin HW, Lin MR, Yu Y, Liu HH, Dai
YL, Chen LW, Jia WW, He XJ, Li XL, et al: Emerging blood
exosome-based biomarkers for preclinical and clinical Alzheimer's
disease: A meta-analysis and systematic review. Neural Regen Res.
17:2381–2390. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mori MA, Ludwig RG, Garcia-Martin R,
Brandão BB and Kahn CR: Extracellular miRNAs: From biomarkers to
mediators of physiology and disease. Cell Metab. 30:656–673.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Clancy L and Freedman JE: Blood-derived
extracellular RNA and platelet pathobiology: Adding pieces to a
complex circulating puzzle. Circ Res. 118:374–376. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chiba M, Uehara H, Niiyama I, Kuwata H and
Monzen S: Changes in miRNA expressions in the injured small
intestine of mice following high-dose radiation exposure. Mol Med
Rep. 21:2452–2458. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Willeit P, Zampetaki A, Dudek K, Kaudewitz
D, King A, Kirkby NS, Crosby-Nwaobi R, Prokopi M, Drozdov I,
Langley SR, et al: Circulating microRNAs as novel biomarkers for
platelet activation. Circ Res. 112:595–600. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wright K, de Silva K, Purdie AC and Plain
KM: Comparison of methods for miRNA isolation and quantification
from ovine plasma. Sci Rep. 10(825)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hongjaisee S, Jabjainai Y, Sakset S,
Preechasuth K, Ngo-Giang-Huong N and Khamduang W: Comparison of
simple RNA extraction methods for molecular diagnosis of hepatitis
C virus in plasma. Diagnostics (Basel). 12(1599)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tang YT, Huang YY, Zheng L, Qin SH, Xu XP,
An TX, Xu Y, Wu YS, Hu XM, Ping BH and Wang Q: Comparison of
isolation methods of exosomes and exosomal RNA from cell culture
medium and serum. Int J Mol Med. 40:834–844. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Trakunram K, Champoochana N, Chaniad P,
Thongsuksai P and Raungrut P: MicroRNA isolation by Trizol-based
method and its stability in stored serum and cDNA derivatives.
Asian Pac J Cancer Prev. 20:1641–1647. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chettimada S, Lorenz DR, Misra V, Wolinsky
SM and Gabuzda D: Small RNA sequencing of extracellular vesicles
identifies circulating miRNAs related to inflammation and oxidative
stress in HIV patients. BMC Immunol. 21(57)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Huang Q, Yang J, Zheng J, Hsueh C, Guo Y
and Zhou L: Characterization of selective exosomal microRNA
expression profile derived from laryngeal squamous cell carcinoma
detected by next generation sequencing. Oncol Rep. 40:2584–2594.
2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li C, Zhou T, Chen J, Li R, Chen H, Luo S,
Chen D, Cai C and Li W: The role of exosomal miRNAs in cancer. J
Transl Med. 20(6)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wang H, Peng R, Wang J, Qin Z and Xue L:
Circulating microRNAs as potential cancer biomarkers: The advantage
and disadvantage. Clin Epigenet. 10(59)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of microRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9(402)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li S, Xu Z and Sheng J: tRNA-derived small
RNA: A novel regulatory small non-coding RNA. Genes (Basel).
9(246)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Liu DSK, Yang QZC, Asim M, Krell J and
Frampton AE: The vlinical significance of transfer RNAs present in
extracellular vesicles. Int J Mol Sci. 23(3692)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huang ZH, Du YP, Wen JT, Lu BF and Zhao Y:
snoRNAs: Functions and mechanisms in biological processes, and
roles in tumor pathophysiology. Cell Death Discov.
8(259)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kufel J and Grzechnik P: Small nucleolar
RNAs tell a different tale. Trends Genet. 35:104–117.
2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fitz NF, Wang J, Kamboh MI, Koldamova R
and Lefterov I: Small nucleolar RNAs in plasma extracellular
vesicles and their discriminatory power as diagnostic biomarkers of
Alzheimer's disease. Neurobiol Dis. 159(105481)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kohama I, Asano N, Matsuzaki J, Yamamoto
Y, Yamamoto T, Takahashi RU, Kobayashi E, Takizawa S, Sakamoto H,
Kato K, et al: Comprehensive serum and tissue microRNA profiling in
dedifferentiated liposarcoma. Oncol Lett. 22(623)2021.PubMed/NCBI View Article : Google Scholar
|