Scaffold attachment factor A (SAF-A), or
heterogeneous ribonucleoprotein U (hnRNP U), belonging to the hnRNP
subfamily, is an abundant component of nuclear matrix and hnRNP
particles (1).
Since human SAF-A (Uniprot code: Q00839) contains
825 amino acids, its predicted molecular weight is ~90 kDa. But,
due to modifications such as phosphorylation, SAF-A is usually
observed to be ~120 kDa. SAF-A is widely expressed in various
organs or tissues, such as bone marrow, lymphoid tissues, brain,
heart, lung and kidney (https://www.proteinatlas.org/ENSG00000153187-HNRNPU).
Due to its functional domains, SAF-A binds to both DNA (such as
scaffold-attached region DNA) and RNA (such as chromatin-associated
RNAs) and plays essential roles in several cellular processes such
as chromatin structure regulation (3), transcription (4) and mitosis (5). The present review summarized the
structure, multiple functions and clinical significance of
SAF-A.
SAF-A contains three domains: N-terminal
DNA-binding, C-terminal RNA binding and middle domain (6-8)
(Fig. 1). Notably, the DNA-binding
domain SAF/Acinus/PIAS (SAP) is important for interaction with
matrix association region (MAR) in chromatin (6). Containing a cluster of
arginine/glycine-rich (RGG) repeats, the C-terminus of SAF-A is
necessary for binding to inactive X chromosome regions and
chromatin-associated RNAs (8). The
middle domain of SAF-A, including an SPla and RYanodine receptor
and a nucleotide triphosphate hydrolase region (9,10),
mediates its interaction with different proteins, such as forkhead
box N3(11), Wilms' tumor
suppressor gene 1(9) and severe
fever with thrombocytopenia syndrome virus nucleocapsid protein
(12).
In the eukaryotic cell nucleus, chromatin forms 3-D
structures at multiple levels, including domains, loops, A and B
compartments (relating to active and inactive chromatin,
respectively), topologically associating domains (TADs) and
territories (13-16).
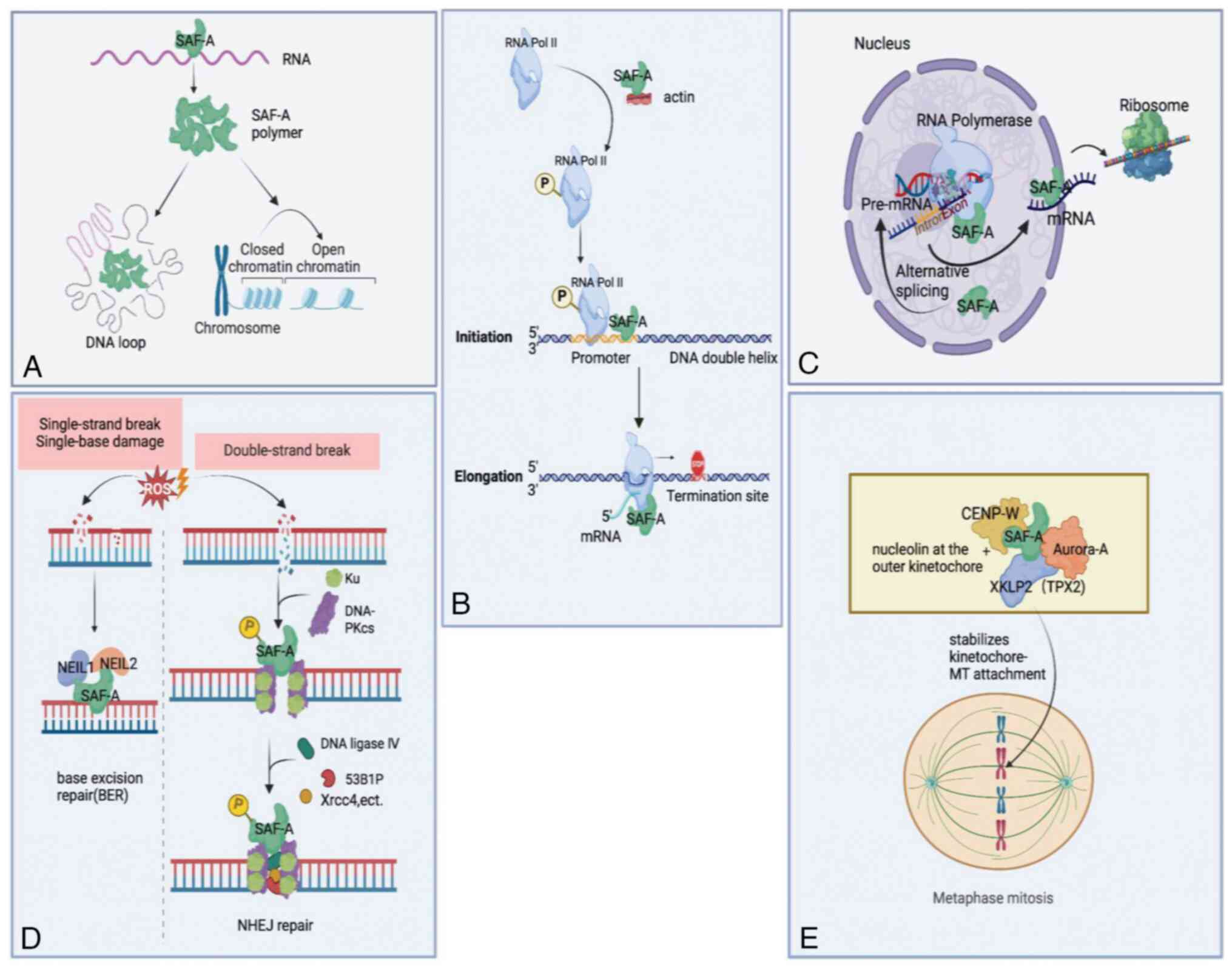
Further studies demonstrated that SAF-A regulates
chromatin structure via RNA. Jiao et al (19) found that SAF-A binds to histone
acetyltransferases p300 in cancer cells (19). Facilitated by heparanase (HPSE)
enhancer RNA, the SAF-A-p300 complex is enriched on the
super-enhancer, consequently leading to chromatin looping between
the HPSE promoter and the super-enhancer. Consistently
interacting with chromatin-associated RNAs, SAF-A forms
oligomerization that induces de-compaction of large-scale human
interphase chromatin structure (8)
(Fig. 2A). In this way, SAF-A
maintains genomic stability (8).
When human monocyte THP-1 cells are infected by vesicular
stomatitis virus (VSV), SAF-A interacts with viral
infection-induced RNAs, mediating the openness and activation of
antiviral immune genes (20).
Puvvula and Moon (10) treated
cancer cells with cell-penetrating peptides derived from SAF-A, and
found that the SAP-derived peptide rather than the RGG-derived one
promotes chromatin compaction in HCT116 (colorectal), T47D (breast)
and UMUC3 (bladder) cancer cells. Based on these observations, RNA
is proposed to be essential for SAF-A-induced chromatin
de-compaction.
Besides RNA, DNA was also reported to be necessary
for SAF-A to regulate chromatin structure. In the study of Kolpa
et al (21), similar to the
C280 SAF-A deletion mutant (only containing RGG domain), the
expression of ∆RGG (containing DNA-binding domain) and G29A mutants
(in lack of DNA-binding ability) was able to release C0T-1 RNA from
chromatin, resulting in chromatin condensation. Despite this, their
regulatory mechanism is different. ∆RGG SAF-A mutant mainly
replaced endogenous SAF-A from chromatin, while the G29A mutant
still combined with C0T-1 RNA and could not bind to chromatin
(21). Therefore, to some degree,
SAF-A may play a role in bridging C0T-1 RNA and chromatin to
regulate chromatin architecture. Depending on the SAP domain, SAF-A
was also found to combine with MARs and the pericentromere tandem
repeats in chromocenters (6,22).
Therefore, it was hypothesized that SAF-A tethers MARs to the
chromocenter to organize nuclear architecture (1).
In eukaryotic cells, transcription of protein-coding
genes, including initiation, elongation and termination phases,
contains numerous regulatory proteins affecting either the RNA
polymerase II (Pol II) machinery or chromatin structure (23-26).
Previous studies have indicated that SAF-A is
involved in initial transcriptional regulation. Actin in the
nucleus regulates transcription by remodeling chromatin, regulating
transcription factor (TF) location, or binding to RNA polymerase
(27-30).
Through extracellular vesicles derived from embryonic stem cells,
SAF-A is transferred into human coronary artery endothelial cells
to combine with actin. The SAF-A-actin complex leads to enhanced
RNA Pol II phosphorylation and its level on vascular endothelial
growth factor (VEGF) promoter, upregulating VEGF expression
(4) (Fig. 2B). When the SAF-A-actin complex is
disrupted by H19 [a long non-coding (lnc) RNA], the phosphorylation
of the Pol II C-terminal domain (CTD) is inhibited, and
consequently, Pol II-mediated transcription is prohibited (31). Additionally, SAF-A was found to
associate with elements within the promoter regions. In Sertoli
cells, SAF-A has been identified to bind directly to the promoter
regions of Sox8 and Sox9, thereby enhancing their
expression (32) (Fig. 2B). Similarly, IL21-anti-sense RNA 1
interacting with SAF-A binds to the IL21 promoter, which is
essential for regulating IL21 transcription (33). Research on embryonic stem cells also
revealed that not only does SAF-A bind to the Oct4 proximal
promoter, but it also interacts with endogenous Pol II. And
depletion of SAF-A impairs Oct4 expression (34).
In previous studies, SAF-A appeared to have two-way
adjusting effects on transcription elongation mediated by Pol II.
According to the study of Obrdlik et al (35), SAF-A, actin and p300/CBP-associated
factor (PCAF) were associated with the phosphorylated Pol II CTD;
and the actin-SAF-A interaction assisted Pol II transcription
elongation depending on PCAF (Fig.
2B). On the contrary, in another study, SAF-A inhibited Pol II
elongation. Through the middle domain, SAF-A was sufficient to
combine with Pol II and repressed TF IIH-mediated Pol II CTD
phosphorylation, which inhibited Pol II elongation (36) (Fig.
2B).
SAF-A has also been implicated in various aspects of
RNA metabolism, including normal and alternative pre-mRNA splicing
and RNA transporting (Fig. 2C).
It has been identified that SAF-A is responsible for
transporting and stabilizing mRNA via directly binding to it. Zhao
et al (41) found that SAF-A
binds to tumor necrosis factor α (TNF-α), IL-6 and
IL-1β mRNAs and that downregulation of SAF-A expression in
macrophages induces the decreased half-life of these cytokine
mRNAs. Besides, Toll-like receptor signaling in macrophages leads
to SAF-A translocation from nuclear to cytoplasm (41). During this translocation, SAF-A is
proposed to stabilize pro-inflammatory cytokine mRNAs (41). In agreement with the aforementioned
study, SAF-A was found to bind to the 3' untranslated region of
TNF α mRNA to stabilize it and specifically enhance TNF α
expression (42). However, other
studies suggested that SAF-A stabilizes mRNA depending on other
molecules. Pan et al (43)
found that interaction between LIM domains-containing 1 antisense
RNA 1 (LIMD1-AS1) and SAF-A is essential for stabilizing
LIMD1 mRNA in non-small cell lung cancer. Furthermore, Lu
et al (44) reported that
following lipopolysaccharide stimulation in human intestinal
epithelial cells, functional intergenic repeating RNA element
cooperating with SAF-A may regulate the stabilization of vascular
cell adhesion protein 1 and IL12p40 mRNAs through targeting
the AU-rich elements of these mRNAs.
In cells, ionizing radiation (IR) and reactive
oxygen species may result in clustered oxidized bases and DSBs that
are also induced by V(D)J recombination (45-47).
Oxidized base lesions in the human genome are repaired via DNA
glycosylase (DG) repair, also known as base excision repair (BER).
Repair enzymes, including Nei endonuclease VIII-like (NEIL) and
certain non-repair proteins, such as SAF-A, initiate this process
(48,49). In previous studies, SAF-A was
demonstrated to interact directly with NEIL1 and stimulate
NEIL1-mediated repair of oxidative base damage (48,49).
In addition to NEIL1, SAF-A combines with NEIL2, enhancing
NEIL2-initiated transcribed gene-specific repair of oxidized bases
(50) (Fig. 2D).
However, BER of bi-stranded oxidized bases can lead
to additional DSBs, causing loss of DNA fragments. This may be
avoided if DSBs are repaired through non-homologous end joining
(NHEJ) mediated by Ku, DNA-dependent protein kinase catalytic
subunit (DNA-PKcs), X-ray repair cross-complementing protein 4
(XRCC4), DNA ligase IV, and cernunnos-XRCC4-like factor complex,
preceding BER (51-53).
In response to irradiation-induced DSBs, SAF-A is rapidly recruited
to DNA damage sites (54) (Fig. 2D). Besides, DSBs cause
phosphorylated SAF-A at Ser59, which is exclusively dependent on
DNA-PK (55,56) (Fig.
2D). In NHEJ-deficient cells treated with IR or clastogenic
drugs, the extent and duration of SAF-A phosphorylation at Ser59
increase (56), which suggests that
phosphorylated SAF-A may be a biomarker for testing the capacity of
the cells to repair DSBs by NHEJ. A previous study showed that
SAF-A phosphorylation at Ser59 is related to transient NEIL1
release from chromatin and BER prevention. Then, due to
dephosphorylation, SAF-A reactivates BER by relieving DG inhibition
(57). These results suggested that
DSB repair by NHEJ or BER is partly determined by whether SAF-A is
phosphorylated. However, a recent study found that increased SAF-A
stabilizes or recruits NHEJ factors via liquid-liquid phase
separation, including Ku80, 53BP1, DNA-PKcs and the shieldin
complex at DSBs during antibody class-switch recombination
(58) (Fig. 2D). The finding supplies SAF-A may
balance between BER and NHEL through complex mechanisms.
During mitosis, kinetochore-microtubule (MT)
attachment and chromosome congression at the spindle equator is
essential to accurate chromosome segregation (59,60).
Interestingly, SAF-A binds to not only Aurora-A and targeting
protein for Xklp2 (TPX2) at spindle MTs, but also nucleolin at the
outer kinetochore, whereby SAF-A stabilizes kinetochore-MT
attachment (61) (Fig. 2E). Via non-coding RNA, SAF-A
interacts with centromere protein W (CENP-W, an inner centromere
component crucial for forming a functional kinetochore complex).
Notably, SAF-A-CENP-W interaction increases their stability, and
they co-localize at the MT-kinetochore interface during mitosis
(62) (Fig. 2E). Furthermore, SAF-A depletion in
cells leads to delayed mitosis, unsuccessful chromosome alignment,
and spindle assembly (61).
In addition, SAF-A is dynamically phosphorylated
during mitosis, and in human cells, phosphorylation at serine 2
(S2), S3, S4, S59, S66 and S270 is related to mitosis (63-65).
Douglas et al (66) found
SAF-A is phosphorylated at S59 by polo-like kinase 1 instead of
DNA-PK and is dephosphorylated by protein phosphatase 2A. Mutations
of SAF-A S59 in cells lead to aberrant mitoses, including polylobed
nuclei, delayed passage, misaligned and lagging chromosomes
(66). However, the regulatory
mechanism of phosphorylated SAF-A on mitosis remains elusive.
Further studies have demonstrated a correlation
between SAF-A and resistance to chemotherapy. Shi et al
(76) discovered that the knockout
of SAF-A in human T24 cells enhances bladder cancer susceptibility
to cisplatin via inhibition of cell proliferation and impairment of
cellular invasion and migration capabilities. In the research of
Wang et al (77), the
knockdown of SAF-A increases selinexor sensitivities of multiple
myeloma (MM) cells in vitro and in mouse models and MM
patients with relatively low SAF-A expression response to
selinexor.
However, a few studies demonstrated that SAF-A might
suppress tumor cell proliferation and enhance sensitivities to
chemotherapy. Pan et al (43) reported that LIMD1 inhibits
proliferation and promotes apoptosis in non-small cell lung cancer
cells, and SAF-A interacts with lncRNA LIMD1-AS1 to
stabilize LIMD1 mRNA. Puvvula and Moon (10) dealt with various tumor cells with
SAF-A RGG- or SAP-derived peptides. Depending on different
mechanisms, these peptides suppress breast, bladder, colorectal and
prostate cancer cell proliferation. The SAF-A RGG-derived peptides
mainly alter SAF-A splicing and binding targets, while the
SAP-derived regulates global epigenetic marks to induce DNA damage
reaction and cell death (10). Li
et al (78) also identified
that lncRNA SFTA1 could augment cisplatin sensitivity in
treating lung squamous cell carcinoma by upregulating SAF-A.
The contradictory effects of SAF-A on tumors may
result from different tumor types or test methods. Therefore, more
animal and clinical experiments are needed to identify its
functions further.
Previous research revealed that SAF-A plays dual
roles in antiviral response. Through the N-terminal fragment
(aa1-86), SAF-A targets the 3' long terminal repeat of human
immunodeficiency virus type 1 mRNA and blocks viral replication in
cells (79). Furtherly, in both
in vitro and in vivo animal experiments, SAF-A has
been reported to be associated with the initiation of innate
immunity against viruses, including severe fever with
thrombocytopenia syndrome virus, VSV, and herpes simplex virus type
1 by recognizing nuclear or cytoplasmic viral RNA (12,20,80).
Therefore, the diverse roles of SAF-A played in
these experiments may be due to different viruses or hosts, and
more animal experiments are needed to determine the roles of SAF-A
in antiviral immunity.
SAF-A is one of the main components of the nuclear
matrix. Capable of binding both DNA and RNA, SAF-A has crucial
functions in regulating chromatin architecture, transcription, RNA
metabolism, DNA repair and mitosis. Humans or mice with SAF-A
deficiency or mutants display embryonic death (91), lethal cardiomyopathy (37) and neurodevelopmental disorders
(38,90). Therefore, SAF-A may be a valuable
prediction factor or therapeutic target. Besides, SAF-A has been
observed to be related to anti-infection and tumor progression,
though these functions are controversial. Because of this, more
animal experiments and clinical assays are needed.
Not applicable.
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81801551) and the Central
Government Funds of Guiding Local Scientific and Technological
Development for Sichuan (grant no. 2021ZYD0076).
Not applicable.
XC, ML and DZ contributed to the conception of the
study. DZ and LL performed the literature research and wrote the
manuscript. XC and ML revised the manuscript. All authors read and
approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Podgornaya OI: Nuclear organization by
satellite DNA, SAF-A/hnRNPU and matrix attachment regions. Semin
Cell Dev Biol. 128:61–68. 2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fackelmayer FO and Richter A:
hnRNP-U/SAF-A is encoded by two differentially polyadenylated mRNAs
in human cells. Biochim Biophys Acta. 1217:232–234. 1994.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wavelet-Vermuse C, Odnokoz O, Xue Y, Lu X,
Cristofanilli M and Wan Y: CDC20-Mediated hnRNPU ubiquitination
regulates chromatin condensation and anti-cancer drug response.
Cancers (Basel). 14(3732)2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang H, Liu H, Zhao X and Chen X:
Heterogeneous nuclear ribonucleoprotein U-actin complex derived
from extracellular vesicles facilitates proliferation and migration
of human coronary artery endothelial cells by promoting RNA
polymerase II transcription. Bioengineered. 13:11469–11486.
2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sharp JA, Perea-Resa C, Wang W and Blower
MD: Cell division requires RNA eviction from condensing
chromosomes. J Cell Biol. 219(e201910148)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kipp M, Gohring F, Ostendorp T, van Drunen
CM, van Driel R, Przybylski M and Fackelmayer FO: SAF-Box, a
conserved protein domain that specifically recognizes scaffold
attachment region DNA. Mol Cell Biol. 20:7480–7489. 2000.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Helbig R and Fackelmayer FO: Scaffold
attachment factor A (SAF-A) is concentrated in inactive X
chromosome territories through its RGG domain. Chromosoma.
112:173–182. 2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Nozawa RS, Boteva L, Soares DC, Naughton
C, Dun AR, Buckle A, Ramsahoye B, Bruton PC, Saleeb RS, Arnedo M,
et al: SAF-A Regulates Interphase Chromosome Structure through
Oligomerization with Chromatin-Associated RNAs. Cell. 169:1214–1227
e18. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Spraggon L, Dudnakova T, Slight J,
Lustig-Yariv O, Cotterell J, Hastie H and Miles C: hnRNP-U directly
interacts with WT1 and modulates WT1 transcriptional activation.
Oncogene. 26:1484–1491. 2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Puvvula PK and Moon AM: Novel
cell-penetrating peptides derived from scaffold-attachment-factor a
inhibits cancer cell proliferation and survival. Front Oncol.
11(621825)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhu X, Huang B, Zhao F, Lian J, He L,
Zhang Y, Ji L, Zhang J, Yan X, Zeng T, et al: p38-mediated FOXN3
phosphorylation modulates lung inflammation and injury through the
NF-κB signaling pathway. Nucleic Acids Res. 51:2195–2214.
2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liu BY, Yu XJ and Zhou CM: SAFA initiates
innate immunity against cytoplasmic RNA virus SFTSV infection. PLoS
Pathog. 17(e1010070)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ghirlando R and Felsenfeld G: CTCF: Making
the right connections. Genes Dev. 30:881–891. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fritz AJ, Sehgal N, Pliss A, Xu J and
Berezney R: Chromosome territories and the global regulation of the
genome. Genes Chromosomes Cancer. 58:407–426. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang W, Chandra A, Goldman N, Yoon S,
Ferrari EK, Nguyen SC, Joyce EF and Vahedi G: TCF-1 promotes
chromatin interactions across topologically associating domains in
T cell progenitors. Nat Immunol. 23:1052–1062. 2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Krasikova A, Kulikova T, Rodriguez Ramos
JS and Maslova A: Assignment of the somatic A/B compartments to
chromatin domains in giant transcriptionally active lampbrush
chromosomes. Epigenetics Chromatin. 16(24)2023.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Romig H, Fackelmayer FO, Renz A,
Ramsperger U and Richter A: Characterization of SAF-A, a novel
nuclear DNA binding protein from HeLa cells with high affinity for
nuclear matrix/scaffold attachment DNA elements. EMBO J.
11:3431–3440. 1992.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Fan H, Lv P, Huo X, Wu J, Wang Q, Cheng L,
Liu Y, Tang QQ, Zhang L, Zhang F, et al: The nuclear matrix protein
HNRNPU maintains 3D genome architecture globally in mouse
hepatocytes. Genome Res. 28:192–202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jiao W, Chen Y, Song H, Li D, Mei H, Yang
F, Fang E, Wang X, Huang K, Zheng L and Tong Q: HPSE enhancer RNA
promotes cancer progression through driving chromatin looping and
regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene. 37:2728–2745.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Cao L, Luo Y, Guo X, Liu S, Li S, Li J,
Zhang Z, Zhao Y, Zhang Q, Gao F, et al: SAFA facilitates chromatin
opening of immune genes through interacting with anti-viral host
RNAs. PLoS Pathog. 18(e1010599)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kolpa HJ, Creamer KM, Hall LL and Lawrence
JB: SAF-A mutants disrupt chromatin structure through dominant
negative effects on RNAs associated with chromatin. Mamm Genome.
33:366–381. 2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lobov IB, Tsutsui K, Mitchell AR and
Podgornaya OI: Specificity of SAF-A and lamin B binding in vitro
correlates with the satellite DNA bending state. J Cell Biochem.
83:218–229. 2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Cramer P: Organization and regulation of
gene transcription. Nature. 573:45–54. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Altendorfer E, Mochalova Y and Mayer A:
BRD4: A general regulator of transcription elongation.
Transcription. 13:70–81. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Haberle V and Stark A: Eukaryotic core
promoters and the functional basis of transcription initiation. Nat
Rev Mol Cell Biol. 19:621–637. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gibbons MD, Fang Y, Spicola AP, Linzer N,
Jones SM, Johnson BR, Li L, Xie M and Bungert J: Enhancer-mediated
formation of nuclear transcription initiation domains. Int J Mol
Sci. 23(9290)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Sokolova M, Moore HM, Prajapati B, Dopie
J, Merilainen L, Honkanen M, Matos RC, Poukkula M, Hietakangas V
and Vartiainen MK: Nuclear actin is required for transcription
during drosophila oogenesis. iScience. 9:63–70. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Knoll KR, Eustermann S, Niebauer V,
Oberbeckmann E, Stoehr G, Schall K, Tosi A, Schwarz M, Buchfellner
A, Korber P and Hopfner KP: The nuclear actin-containing Arp8
module is a linker DNA sensor driving INO80 chromatin remodeling.
Nat Struct Mol Biol. 25:823–832. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Almuzzaini B, Sarshad AA, Rahmanto AS,
Hansson ML, Von Euler A, Sangfelt O, Visa N, Farrants AK and
Percipalle P: In β-actin knockouts, epigenetic reprogramming and
rDNA transcription inactivation lead to growth and proliferation
defects. FASEB J. 30:2860–2873. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Al-Sayegh MA, Mahmood SR, Khair SBA, Xie
X, El Gindi M, Kim T, Almansoori A and Percipalle P: beta-actin
contributes to open chromatin for activation of the adipogenic
pioneer factor CEBPA during transcriptional reprograming. Mol Biol
Cell. 31:2511–2521. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bi HS, Yang XY, Yuan JH, Yang F, Xu D, Guo
YJ, Zhang L, Zhou CC, Wang F and Sun SH: H19 inhibits RNA
polymerase II-mediated transcription by disrupting the hnRNP
U-actin complex. Biochim Biophys Acta. 1830:4899–4906.
2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wen Y, Ma X, Wang X, Wang F, Dong J, Wu Y,
Lv C, Liu K, Zhang Y, Zhang Z and Yuan S: hnRNPU in Sertoli cells
cooperates with WT1 and is essential for testicular development by
modulating transcriptional factors Sox8/9. Theranostics.
11:10030–10046. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu L, Hu L, Long H, Zheng M, Hu Z, He Y,
Gao X, Du P, Zhao H, Yu D, et al: LncRNA IL21-AS1 interacts with
hnRNPU protein to promote IL21 overexpression and aberrant
differentiation of Tfh cells in systemic lupus erythematosus. Clin
Transl Med. 12(e1117)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Vizlin-Hodzic D, Johansson H, Ryme J,
Simonsson T and Simonsson S: SAF-A has a role in transcriptional
regulation of Oct4 in ES cells through promoter binding. Cell
Reprogram. 13:13–27. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Obrdlik A, Kukalev A, Louvet E, Farrants
AK, Caputo L and Percipalle P: The histone acetyltransferase PCAF
associates with actin and hnRNP U for RNA polymerase II
transcription. Mol Cell Biol. 28:6342–6357. 2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kim MK and Nikodem VM: hnRNP U inhibits
carboxy-terminal domain phosphorylation by TFIIH and represses RNA
polymerase II elongation. Mol Cell Biol. 19:6833–6844.
1999.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ye J, Beetz N, O'Keeffe S, Tapia JC,
Macpherson L, Chen WV, Bassel-Duby R, Olson EN and Maniatis T:
hnRNP U protein is required for normal pre-mRNA splicing and
postnatal heart development and function. Proc Natl Acad Sci USA.
112:E3020–E3029. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sapir T, Kshirsagar A, Gorelik A, Olender
T, Porat Z, Scheffer IE, Goldstein DB, Devinsky O and Reiner O:
Heterogeneous nuclear ribonucleoprotein U (HNRNPU) safeguards the
developing mouse cortex. Nat Commun. 13(4209)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Xiao R, Tang P, Yang B, Huang J, Zhou Y,
Shao C, Li H, Sun H, Zhang Y and Fu XD: Nuclear matrix factor hnRNP
U/SAF-A exerts a global control of alternative splicing by
regulating U2 snRNP maturation. Mol Cell. 45:656–668.
2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Vu NT, Park MA, Shultz JC, Goehe RW,
Hoeferlin LA, Shultz MD, Smith SA, Lynch KW and Chalfant CE: hnRNP
U enhances caspase-9 splicing and is modulated by AKT-dependent
phosphorylation of hnRNP L. J Biol Chem. 288:8575–8584.
2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhao W, Wang L, Zhang M, Wang P, Qi J,
Zhang L and Gao C: Nuclear to cytoplasmic translocation of
heterogeneous nuclear ribonucleoprotein U enhances TLR-induced
proinflammatory cytokine production by stabilizing mRNAs in
macrophages. J Immunol. 188:3179–3187. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yugami M, Kabe Y, Yamaguchi Y, Wada T and
Handa H: hnRNP-U enhances the expression of specific genes by
stabilizing mRNA. FEBS Lett. 581:1–7. 2007.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pan J, Tang Y, Liu S, Li L, Yu B, Lu Y and
Wang Y: LIMD1-AS1 suppressed non-small cell lung cancer progression
through stabilizing LIMD1 mRNA via hnRNP U. Cancer Med.
9:3829–3839. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lu Y, Liu X, Xie M, Liu M, Ye M, Li M,
Chen XM, Li X and Zhou R: The NF-κB-Responsive Long Noncoding RNA
FIRRE Regulates Posttranscriptional Regulation of Inflammatory Gene
Expression through Interacting with hnRNPU. J Immunol.
199:3571–3582. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Bredemeyer AL, Helmink BA, Innes CL,
Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins
JB, Weaver BK, et al: DNA double-strand breaks activate a
multi-functional genetic program in developing lymphocytes. Nature.
456:819–823. 2008.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Cannan WJ, Tsang BP, Wallace SS and
Pederson DS: Nucleosomes suppress the formation of double-strand
DNA breaks during attempted base excision repair of clustered
oxidative damages. J Biol Chem. 289:19881–19893. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Nickoloff JA, Sharma N and Taylor L:
Clustered DNA Double-Strand Breaks: Biological effects and
relevance to cancer radiotherapy. Genes (Basel).
11(99)2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Hegde ML, Banerjee S, Hegde PM, Bellot LJ,
Hazra TK, Boldogh I and Mitra S: Enhancement of NEIL1
protein-initiated oxidized DNA base excision repair by
heterogeneous nuclear ribonucleoprotein U (hnRNP-U) via direct
interaction. J Biol Chem. 287:34202–34211. 2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Dutta A, Yang C, Sengupta S, Mitra S and
Hegde ML: New paradigms in the repair of oxidative damage in human
genome: Mechanisms ensuring repair of mutagenic base lesions during
replication and involvement of accessory proteins. Cell Mol Life
Sci. 72:1679–1698. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Banerjee D, Mandal SM, Das A, Hegde ML,
Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S and Hazra TK:
Preferential repair of oxidized base damage in the transcribed
genes of mammalian cells. J Biol Chem. 286:6006–6016.
2011.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hammel M, Yu Y, Radhakrishnan SK, Chokshi
C, Tsai MS, Matsumoto Y, Kuzdovich M, Remesh SG, Fang S, Tomkinson
AE, et al: An Intrinsically Disordered APLF Links Ku, DNA-PKcs, and
XRCC4-DNA Ligase IV in an extended flexible non-homologous end
joining complex. J Biol Chem. 291:26987–27006. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Weterings E, Verkaik NS, Keijzers G,
Florea BI, Wang SY, Ortega LG, Uematsu N, Chen DJ and van Gent DC:
The Ku80 carboxy terminus stimulates joining and artemis-mediated
processing of DNA ends. Mol Cell Biol. 29:1134–1142.
2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Xing M and Oksenych V: Genetic interaction
between DNA repair factors PAXX, XLF, XRCC4 and DNA-PKcs in human
cells. FEBS Open Bio. 9:1315–1326. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Britton S, Dernoncourt E, Delteil C,
Froment C, Schiltz O, Salles B, Frit P and Calsou P: DNA damage
triggers SAF-A and RNA biogenesis factors exclusion from chromatin
coupled to R-loops removal. Nucleic Acids Res. 42:9047–9062.
2014.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Berglund FM and Clarke PR: hnRNP-U is a
specific DNA-dependent protein kinase substrate phosphorylated in
response to DNA double-strand breaks. Biochem Biophys Res Commun.
381:59–64. 2009.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Britton S, Froment C, Frit P, Monsarrat B,
Salles B and Calsou P: Cell nonhomologous end joining capacity
controls SAF-A phosphorylation by DNA-PK in response to DNA
double-strand breaks inducers. Cell Cycle. 8:3717–3722.
2009.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hegde ML, Dutta A, Yang C, Mantha AK,
Hegde PM, Pandey A, Sengupta S, Yu Y, Calsou P, Chen D, et al:
Scaffold attachment factor A (SAF-A) and Ku temporally regulate
repair of radiation-induced clustered genome lesions. Oncotarget.
7:54430–54444. 2016.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Refaat AM, Nakata M, Husain A, Kosako H,
Honjo T and Begum NA: HNRNPU facilitates antibody class-switch
recombination through C-NHEJ promotion and R-loop suppression. Cell
Rep. 42(112284)2023.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Tanaka TU: Kinetochore-microtubule
interactions: Steps towards bi-orientation. EMBO J. 29:4070–4082.
2010.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Biggins S and Walczak CF: Captivating
capture: How microtubules attach to kinetochores. Curr Biol.
13:R449–R460. 2003.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ma N, Matsunaga S, Morimoto A, Sakashita
G, Urano T, Uchiyama S and Fukui K: The nuclear scaffold protein
SAF-A is required for kinetochore-microtubule attachment and
contributes to the targeting of Aurora-A to mitotic spindles. J
Cell Sci. 124:394–404. 2011.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Chun Y, Kim R and Lee S: Centromere
Protein (CENP)-W interacts with heterogeneous nuclear
ribonucleoprotein (hnRNP) U and may contribute to
kinetochore-microtubule attachment in mitotic cells. PLoS One.
11(e0149127)2016.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kettenbach AN, Schweppe DK, Faherty BK,
Pechenick D, Pletnev AA and Gerber SA: Quantitative
phosphoproteomics identifies substrates and functional modules of
Aurora and Polo-like kinase activities in mitotic cells. Sci
Signal. 4(rs5)2011.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Malik R, Lenobel R, Santamaria A, Ries A,
Nigg EA and Korner R: Quantitative analysis of the human spindle
phosphoproteome at distinct mitotic stages. J Proteome Res.
8:4553–4563. 2009.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Wang Z, Udeshi ND, Slawson C, Compton PD,
Sakabe K, Cheung WD, Shabanowitz J, Hunt DF and Hart GW: Extensive
crosstalk between O-GlcNAcylation and phosphorylation regulates
cytokinesis. Sci Signal. 3(ra2)2010.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Douglas P, Ye R, Morrice N, Britton S,
Trinkle-Mulcahy L and Lees-Miller SP: Phosphorylation of
SAF-A/hnRNP-U Serine 59 by Polo-Like Kinase 1 Is Required for
Mitosis. Mol Cell Biol. 35:2699–2713. 2015.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Shi W, Wang Q, Bian Y, Fan Y, Zhou Y, Feng
T, Li Z and Cao X: Long noncoding RNA PANDA promotes esophageal
squamous carcinoma cell progress by dissociating from NF-YA but
interact with SAFA. Pathol Res Pract. 215(152604)2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Liang Y, Fan Y, Liu Y and Fan H: HNRNPU
promotes the progression of hepatocellular carcinoma by enhancing
CDK2 transcription. Exp Cell Res. 409(112898)2021.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Song H, Li D, Wang X, Fang E, Yang F, Hu
A, Wang J, Guo Y, Liu Y, Li H, et al: HNF4A-AS1/hnRNPU/CTCF axis as
a therapeutic target for aerobic glycolysis and neuroblastoma
progression. J Hematol Oncol. 13(24)2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Xu CL, Chen Y, Zhu TT, Sun ZJ and Chu JH:
Clinical significance and pathogenesis analysis of heterogeneous
nuclear ribonucleoprotein U in acute myeloid leukemia. Zhonghua Xue
Ye Xue Za Zhi. 43:745–752. 2022.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
71
|
Han BY, Liu Z, Hu X and Ling H: HNRNPU
promotes the progression of triple-negative breast cancer via RNA
transcription and alternative splicing mechanisms. Cell Death Dis.
13(940)2022.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Dong YY, Wang MY, Jing JJ, Wu YJ, Li H,
Yuan Y and Sun LP: Alternative Splicing Factor Heterogeneous
Nuclear Ribonucleoprotein U as a promising biomarker for gastric
cancer risk and prognosis with tumor-promoting properties. Am J
Pathol. 194:13–29. 2024.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Chen T, Zheng W, Chen J, Lin S, Zou Z, Li
X and Tan Z: Systematic analysis of survival-associated alternative
splicing signatures in clear cell renal cell carcinoma. J Cell
Biochem. 121:4074–4084. 2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Zhang W, Zheng Z, Wang K, Mao W, Li X,
Wang G, Zhang Y, Huang J, Zhang N, Wu P, et al: piRNA-1742 promotes
renal cell carcinoma malignancy by regulating USP8 stability
through binding to hnRNPU and thereby inhibiting MUC12
ubiquitination. Exp Mol Med. 55:1258–1271. 2023.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Hu WM, Li M, Ning JZ, Tang YQ, Song TB, Li
LZ, Zou F, Cheng F and Yu WM: FAM171B stabilizes vimentin and
enhances CCL2-mediated TAM infiltration to promote bladder cancer
progression. J Exp Clin Cancer Res. 42(290)2023.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Shi ZD, Hao L, Han XX, Wu ZX, Pang K, Dong
Y, Qin JX, Wang GY, Zhang XM, Xia T, et al: Targeting HNRNPU to
overcome cisplatin resistance in bladder cancer. Mol Cancer.
21(37)2022.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Wang X, Xu J, Li Q, Zhang Y, Lin Z, Zhai
X, Wang F, Huang J, Gao Q, Wen J, et al: RNA-binding protein hnRNPU
regulates multiple myeloma resistance to selinexor. Cancer Lett.
580(216486)2024.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Li L, Yin JY, He FZ, Huang MS, Zhu T, Gao
YF, Chen YX, Zhou DB, Chen X, Sun LQ, et al: Long noncoding RNA
SFTA1P promoted apoptosis and increased cisplatin chemosensitivity
via regulating the hnRNP-U-GADD45A axis in lung squamous cell
carcinoma. Oncotarget. 8:97476–97489. 2017.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Valente ST and Goff SP: Inhibition of
HIV-1 gene expression by a fragment of hnRNP U. Mol Cell.
23:597–605. 2006.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Cao L, Liu S, Li Y, Yang G, Luo Y, Li S,
Du H, Zhao Y, Wang D, Chen J, et al: The Nuclear Matrix Protein
SAFA Surveils Viral RNA and Facilitates Immunity by Activating
Antiviral Enhancers and Super-enhancers. Cell Host Microbe.
26:369–384 e8. 2019.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Gupta AK, Drazba JA and Banerjee AK:
Specific interaction of heterogeneous nuclear ribonucleoprotein
particle U with the leader RNA sequence of vesicular stomatitis
virus. J Virol. 72:8532–8540. 1998.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Hu X, Wu X, Ding Z, Chen Z and Wu H:
Characterization and functional analysis of chicken dsRNA binding
protein hnRNPU. Dev Comp Immunol. 138(104521)2023.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Zhou H, Yan Y, Gao J, Ma M, Liu Y, Shi X,
Zhang Q and Xu X: Heterogeneous Nuclear Protein U Degraded the
m(6)A Methylated TRAF3 Transcript by YTHDF2 To Promote Porcine
Epidemic Diarrhea Virus Replication. J Virol.
97(e0175122)2023.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Caliebe A, Kroes HY, van der Smagt JJ,
Martin-Subero JI, Tonnies H, van 't Slot R, Nievelstein RA, Muhle
H, Stephani U, Alfke K, et al: Four patients with speech delay,
seizures and variable corpus callosum thickness sharing a 0.440 Mb
deletion in region 1q44 containing the HNRPU gene. Eur J Med Genet.
53:179–185. 2010.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Depienne C, Nava C, Keren B, Heide S,
Rastetter A, Passemard S, Chantot-Bastaraud S, Moutard ML, Agrawal
PB, VanNoy G, et al: Genetic and phenotypic dissection of 1q43q44
microdeletion syndrome and neurodevelopmental phenotypes associated
with mutations in ZBTB18 and HNRNPU. Hum Genet. 136:463–479.
2017.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Epi4K Consortium; Epilepsy Phenome/Genome
Project. Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D,
Eichler EE, Epstein MP, Glauser T, et al: De novo mutations in
epileptic encephalopathies. Nature. 501:217–221. 2013.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Shimada S, Oguni H, Otani Y, Nishikawa A,
Ito S, Eto K, Nakazawa T, Yamamoto-Shimojima K, Takanashi JI,
Nagata S and Yamamoto T: An episode of acute encephalopathy with
biphasic seizures and late reduced diffusion followed by hemiplegia
and intractable epilepsy observed in a patient with a novel
frameshift mutation in HNRNPU. Brain Dev. 40:813–818.
2018.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Durkin A, Albaba S, Fry AE, Morton JE,
Douglas A, Beleza A, Williams D, Volker-Touw CML, Lynch SA, Canham
N, et al: Clinical findings of 21 previously unreported probands
with HNRNPU-related syndrome and comprehensive literature review.
Am J Med Genet A. 182:1637–1654. 2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Wang T, Hoekzema K, Vecchio D, Wu H,
Sulovari A, Coe BP, Gillentine MA, Wilfert AB, Perez-Jurado LA,
Kvarnung M, et al: Large-scale targeted sequencing identifies risk
genes for neurodevelopmental disorders. Nat Commun.
11(4932)2020.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Taylor J, Spiller M, Ranguin K, Vitobello
A, Philippe C, Bruel AL, Cappuccio G, Brunetti-Pierri N, Willems M,
Isidor B, et al: Expanding the phenotype of HNRNPU-related
neurodevelopmental disorder with emphasis on seizure phenotype and
review of literature. Am J Med Genet A. 188:1497–1514.
2022.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Roshon MJ and Ruley HE: Hypomorphic
mutation in hnRNP U results in post-implantation lethality.
Transgenic Res. 14:179–192. 2005.PubMed/NCBI View Article : Google Scholar
|