Introduction
In recent years, the use of Traditional Chinese
medicine to improve metabolic diseases has gradually become a
research focus. Among numerous natural hypoglycemic active
ingredients, investigators have paid increasing attention to
resveratrol (RSV), which is a polyphenolic plant antitoxin
belonging to the stilbene family, widely found in grapes and other
plants, and has hypoglycemic activity (1). The mechanism of RSV in reducing
insulin resistance (IR) involves the regulation of multiple signal
transduction pathways, such as silent mating type information
regulation 2 homolog-1/peroxisome proliferator-activated receptor γ
coactivator-1α, inhibitor of κB kinase/nuclear factor κВ
inhibitor/nuclear factor κВ, adenosine phosphate kinase (AMPK) and
phosphatidylinositol 3-kinase (PI3K)/protein kinase B
(AKT)/endothelial nitric oxide synthase pathways (2). The in-depth study of these signaling
pathways has clarified some molecular mechanisms of the effects of
RSV (2). In insulin target tissues,
skeletal muscle accounts for 70-80% of the total glucose intake
stimulated by insulin, thus playing a crucial role in regulating
systemic glucose homeostasis (3).
The mammalian target of rapamycin (mTOR) is a
serine/threonine protein kinase with highly conserved structure and
function belonging to the PI3K family. mTOR-mediated signaling
pathways play crucial roles in metabolism, immunity, growth,
development, aging as well as other physiological processes
(4). Research has shown that mTOR
signaling is also involved in the occurrence and development of
numerous metabolic diseases, such as diabetes, hyperlipidemia and
osteoporosis (4). A previous study
found that mTOR functions as a nutrient receptor, and that the mTOR
signal was activated in various tissues of a high-fat diet (HFD) or
obese rodents (5), and was closely
related to the occurrence and development of IR (6,7).
Skeletal muscle anabolism is driven by numerous stimuli such as
growth factors, nutrients (including amino acids and glucose), and
mechanical stress. These stimuli are integrated by the mechanistic
target of mTOR signal transduction cascade (8).
The DNA-damage-inducible transcript factor 4 (DDIT4)
plays a role in insulin signal transduction, DNA damage repair,
nutritional deprivation, oxidative metabolism, endoplasmic
reticulum stress and the hypoxia response (9-14).
Some studies have found that DDIT4 is involved in nutritional
metabolism, and is crucial to insulin-mediated activation of AKT
and mTOR (15-17).
However, at present, there are few reports regarding the actions of
DDIT4 involving the mTOR pathway during IR. Notably, changes in
DDIT4 mRNA and protein have been observed in skeletal muscle under
various physiological conditions (such as nutrient consumption and
resistance exercise) and pathological conditions (including sepsis,
alcoholism, diabetes and obesity) suggesting a role for DDIT4 in
regulating mTOR-dependent skeletal muscle protein metabolism
(18).
In the present study, RSV was administered to
C57BL/6J mice with IR induced by an HFD to observe the effect of
RSV on insulin and mTOR signaling pathways as well as DDIT4, and to
explore the mechanism of the RSV-mediated reduction of IR, to
further understand the specific functions of RSV in glucose and
lipid metabolism.
Materials and methods
Animal experiments
A total of 33 healthy 6-week-old male C57BL/6J mice
weighing ~22 g were purchased from the Hebei Invivo Biotechnology
Co., Ltd. (license no. SCXK 2023-002) and raised in the barrier
system of the animal room at the Clinical Research Center of Hebei
General Hospital (Shijiazhuang, China). The humidity was controlled
at 40-60% with a room temperature at 20-25˚C with a standard 12-h
light-dark cycle. A week after adaptive feeding, mice were randomly
divided into the following two groups: Control group (CON, n=11)
and HFD group (n=22). The CON group was given an ordinary diet
(D12450J: 70% carbohydrate, 20% protein, 10% fat, 3.85 kcal/g), and
the HFD group was given an HFD (D12492J: 20% carbohydrate, 20%
protein, 60% fat, 5.24 kcal/g) (19). All feeds were purchased from Beijing
Huafukang Biotechnology Co., Ltd. After 8 weeks of an HFD, mice
were fasted overnight (12 h) and then underwent an intraperitoneal
glucose tolerance test (IPGTT) (19). The tail tip blood glucose was
measured at point 0 using Roche rapid glucose meter test strips,
followed by intraperitoneal injection of 50% glucose injection at 2
g/kg (20-22).
Then, the tail tip blood glucose values were measured at 15, 30, 60
and 120 min using Roche rapid glucose meter test strips after
glucose treatment. The amount of blood used for measuring blood
glucose with a blood glucose meter was approximately 20 µl per
mouse at each time point (20-22).
The area under the glucose curve (AUC) was calculated (23); IPGTT-AUC=0.125 x BG0 + 0.25 x BG15 +
0.375 x BG30 + 0.75 x BG60 + 0.5 x BG120 (BGx=blood glucose values
at different time points) (23).
Eleven mice were randomly selected from the HFD group to form the
high-fat feeding with RSV intervention group (HFD + RSV group), and
were administered 100 mg/kg body weight RSV by gavage for 6 weeks
(24), while other mice in the HFD
group and the CON group were admnistered 0.9% sodium chloride
solution containing 0.1% DMSO by gavage.
After 6 weeks of treatment, IPGTT was performed
again to detect the blood glucose value of each mouse at each time
point, and then the calculated AUC was used to evaluate the degree
of IR. All three groups of mice were intraperitoneally injected
with 1.5 units of insulin (Merck KGaA)/40 g body weight, and then
anesthetized by intraperitoneal injection of 2% pentobarbital
sodium at a dose of 40 mg/kg. After 20 min, blood samples collected
by cardiac puncture were centrifuged at 1,500 x g for 10 min at
4˚C. The serum was collected and stored at -80˚C for the subsequent
determination of serological indicators. Cervical dislocation
caused rapid unconsciousness and death with minimal distress to the
mice, and was a humane way to euthanize the mice. Then, skeletal
muscle samples were quickly removed and stored in liquid nitrogen
for subsequent research. The present study was approved (approval
no. 2023-LW-055) by the Ethics Committee of Hebei General Hospital
(Shijiazhuang, China).
Serological indicators
Detection kits for total cholesterol (TC; cat. no.
A111-1-1), triglyceride (TG; cat. no. A110-1-1), low-density
lipoprotein-cholesterol (LDL-C; cat. no. A113-1-1) and high-density
lipoprotein-cholesterol (HDL-C; cat. no. A112-1-1) were acquired
from the Nanjing Jiancheng Bioengineering Institute in accordance
with the manufacturer's instructions. Insulin concentrations were
obtained using an ELISA kit from ALPCO (cat. no. 80-INSMSU-E01) in
accordance with the manufacturer's instructions. The Qualitative
Insulin Sensitivity Check Index (QUICKI) was determined according
to the equation: QUICKI=1/[log(I0) + log(G0)] (25), with I0=fasting insulin and
G0=fasting glucose.
Histological assessment of skeletal
muscle tissue
The skeletal muscle tissues of mice were fixed in 4%
paraformaldehyde at 4˚C for 6 h. Then, the samples were dehydrated
using a conventional alcohol gradient, clarified using xylene,
embedded in paraffin and cut into 5-µm slices. The sections were
then stained with an H&E Stain Kit (cat. no. G1005; Wuhan
Servicebio Technology Co., Ltd.) using hematoxylin and eosin
solutions for 5 min staining each at room temperature. A Modified
Oil Red O Stain Kit (cat. no. G1261; Beijing Solarbio Science &
Technology Co. Ltd.) was used to detect lipid droplets in skeletal
muscle sections, which were stained with Modified Oil Red O Stain
Solution (included in the aforementioned kit) for 10 min at 25˚C,
and then stained with Mayer's Hematoxylin Solution (included in the
aforementioned kit) for 1 min at 25˚C. The staining of sections was
completed according to the manufacturer's protocol, and stained
sections were examined under a light microscope (magnification,
x400).
C2C12 cell culture
The C2C12 mouse myoblast cell line (cat. no.
CL-0044; Procell Life Science & Technology Co., Ltd.) was
cultured in culture medium (cat. no. CM-0044 Procell Life Science
& Technology Co., Ltd.) until the cells reached 70-80%
confluence. The differentiation medium (cat. no. PM150210A; Procell
Life Science & Technology Co., Ltd.) and 2% horse serum (cat.
no. 164215; Procell Life Science & Technology Co., Ltd.) were
used to induce differentiation of C2C12 cells. Mycoplasma
contamination was not detected in the cells. Differentiated C2C12
cells were then incubated with 0.5 mM palmitic acid (PA; cat. no.
800508; Merck KGaA) for 24 h, designated the PA group. RSV (cat.
no. R817263-5g; Shanghai Macklin Biochemical Technology Co. Ltd.)
was added to the medium at concentrations of 30 µM at room
temperature to form the PA + RSV group. Some of the PA + RSV group
cells were again divided into two groups: The PA + RSV + MHY1485
group C2C12 cells were treated with 10 µM MHY1485 (cat. no.
HY-B0795; MedChemExpress) to activate mTOR, and the PA + RSV +
DDIT4-siRNA group C2C12 cells were transfected with a small
interfering (si)RNA against DDIT4 (DDIT4-siRNA; Shanghai Genechem
Co. Ltd.), all incubated at 37˚C for 24 h. The cells were collected
for western blotting and cellular TG content determination.
Transfection with siRNAs
Shanghai Genechem Co., Ltd. provided three
DDIT4-siRNAs (DDIT4-siRNA-1: Sense strand,
5'-GGGAAGGAAGUGUUCUCCAGGAAGU-3' and antisense strand,
5'-ACUUCCUGGAGAACACUUCCUUCCC-3'; DDIT4-siRNA-2: Sense strand,
5'-GCAGCUGCUCAUUGAAGAGUGUUGA-3' and antisense strand,
5'-UCAACACUCUUCAAUGAGCAGCUGC-3'; DDIT4-siRNA-3: Sense strand,
5'-GGUGCCCAUGUACUGGAGGAUUCAA-3' and antisense strand,
5'-UUGAAUCCUCCAGUACAUGGGCACC-3'), along with a negative control
siRNA (sense strand, 5'-GGUCUUACGUCAGUCACAAUAUCUG-3' and antisense
strand, 5'-CAGAUAUUGUGACUGACGUAAGACC-3'). To prepare the RNA
oligonucleotide stock solution, 20 pmol of siRNA was mixed with 50
µl of Opti-MEM I reduced serum medium (cat. no. 31985-062; Thermo
Fisher Scientific, Inc.) and stored at -20˚C. Additionally, 1 µl of
Lipofectamine® 2000 (cat. no. 11668019; Invitrogen;
Thermo Fisher Scientific, Inc.) was diluted in 50 µl of Opti-MEM I
medium. For experiments, the RNA oligonucleotide stock solution and
the diluted Lipofectamine 2000 solution were mixed to prepare
transfection complexes at room temperature. Cells were seeded into
24-well plates once they reached ~40% confluence at 37˚C.
Transfection was performed when the cells reached ~60% confluence,
with 100 µl siRNA transfection complexes at a concentration of 50
nM added to each well and incubated at 37˚C for 24 h. Western
blotting was performed 24 h after transfection to assess the
efficiency of the different siRNAs and determine the most effective
one for future experiments. The optimal siRNA was determined to be
DDIT4-siRNA-1.
TG assay
Cellular TG content was measured using a TG
detection kit (cat. no. ab65336; Abcam Plc) according to the
manufacturer's instructions.
Glucose content in culture medium
The glucose oxidase assay kit (cat. no. E1010-500;
Beijing Applygen Gene Technology Co., Ltd.) was used to measure the
remaining glucose content in culture medium as per the
manufacturer's instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from mouse skeletal muscle tissues was
extracted using Trizol reagent [ref. no. DP424; Tiangen Biotech
(Beijing) Co. Ltd.] and was reverse-transcribed into cDNA using a
Fastking RT Kit [ref. no. KR116; Tiangen Biotech (Beijing) Co.
Ltd.] for qPCR. RNA was tested for purity and concentration using
NanoDrop® 2000 (Thermo Fisher Scientific, Inc.).
Amplification was performed using the SuperReal PreMix Plus (SYBR
Green) [ref. no. GFP205; Tiangen Biotech (Beijing) Co. Ltd.] in an
Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher
Scientific, Inc.). The cycling conditions were as follows: 3 min of
pre-denaturation at 95˚C, then 5 sec at 95˚C, and 32 sec at 60˚C,
with 41 cycles in total. The melting point curve was established at
60-95˚C. The expression levels of PI3K, insulin receptor substrate
(IRS)-1, AKT, glucose transporter type 4 (GLUT4), mTOR, p70
ribosomal protein S6 kinase (p70S6K/S6K1) and DDIT4 mRNA in
skeletal muscle tissue were evaluated. β-actin was used as the
internal reference control for genes. Relative gene expression
levels were quantified by the 2-ΔΔCq method (26). The specific primers involved in the
study are listed in Table I.
 | Table IPrimers used for real-time
quantitative polymerase chain reaction. |
Table I
Primers used for real-time
quantitative polymerase chain reaction.
| Gene | Primer | Sequence
(5'-3') |
|---|
| PI3K | Forward |
5'-CCCATGGGACAACATTCCAA-3' |
| | Reverse |
5'-CATGGCGACAAGCTCGGTA-3' |
| AKT | Forward |
5'-TCAGGATGTGGATCAGCGAGA-3' |
| | Reverse |
5'-CTGCAGGCAGCGGATGATAA-3' |
| IRS-1 | Forward |
5'-GCACCTGGTGGCTCTCTACAC-3' |
| | Reverse |
5'-TCGCTATCCGCGGCAAT-3' |
| GLUT4 | Forward |
5'-GTGACTGGAACACTGGTCCTA-3' |
| | Reverse |
5'-CCAGCCACGTTGCATTGTAG-3' |
| DDIT4 | Forward |
5'-TACTGCCCACCTTTCAGTTG-3' |
| | Reverse |
5'-GTCAGGGACTGGCTGTAACC-3' |
| mTOR | Forward |
5'-GCGGCCTGGAAATGCGGAAGTGG-3' |
| | Reverse |
5'-AAAGCCCCAAGGAGCCCCAACA-3' |
| p70S6K | Forward |
5'-CACTCAGGCCCCCCTACACT-3' |
| | Reverse |
5'-GCCGTCACTGAAAACCAAGTTC-3' |
| β-actin | Forward |
5'-GGTGGGAATGGGTCAGAAGG-3' |
| | Reverse |
5'-AGGTCTCAAACATGATCTGGGT-3' |
Western blotting
Skeletal muscle samples were added to 0.5 ml of
pre-cooled lysate at 4˚C, and then homogenized and stored at 4˚C
overnight. Subsequently, the tissue homogenate was centrifuged at
16,200 x g at 4˚C for 15 min and the supernatants were collected.
C2C12 cells were lysed in RIPA lysis buffer (cat. no. C1053-100;
Applygen Technologies, Inc.) to extract total cellular proteins.
The same amounts of protein (~50 µg/lane) for different group
samples were separated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (5% resolving gel, 10% stacking gel), and then
transferred to polyvinylidene fluoride membranes, which were
blocked in 5% skimmed milk at room temperature for 2 h. The
membranes were then incubated with the primary antibodies at 4˚C
overnight at the following concentrations: Phosphorylated (p)-IRS-1
(rabbit antibody; 1:1,000; cat. no. ab5599; Abcam Ltd.), total
(t)-IRS-1 (rabbit antibody; 1:1,000; cat. no. ab52167; Abcam Ltd.),
p-AKT (rabbit antibody; 1:1,000; cat. no. AA329; Beyotime Institute
of Biotechnology), t-AKT (rabbit antibody; 1:1,000; cat. no. AA326;
Beyotime Institute of Biotechnology), p-PI3K (rabbit antibody;
1:1,000; cat. no. AF5905; Beyotime Institute of Biotechnology),
t-PI3K (rabbit antibody; 1:1,000; cat. no. 20583-1-AP; Proteintech
Group, Inc.), GLUT4 (rabbit antibody; 1:3,000; cat. no. ab65267;
Abcam Ltd.), DDIT4 (rabbit antibody; 1:2,000; cat. no. AF5147;
Beyotime Institute of Biotechnology), p-mTOR (rabbit antibody;
1:4,000; cat. no. ab109268; Abcam Ltd.), t-mTOR (rabbit antibody;
1:4,000; cat. no. ab32028; Abcam Ltd.), p-p70S6K (rabbit antibody;
1:1,000; cat. no. ab59208; Abcam Ltd.), t-p70S6K (rabbit antibody;
1:2,000; cat. no. ab32529; Abcam Ltd.), and β-actin (rabbit
antibody; 1:5,000; cat. no. 20536-1-AP; Proteintech Group Inc.).
After three washes with Tris-buffered saline, the membranes were
incubated with horseradish peroxidase-conjugated secondary
antibodies (anti-rabbit; 1:5,000; cat. no. S0001; Affinity
Biosciences) for 2 h at room temperature. Finally, the washed
membranes were immersed in chemiluminescence solution (cat. no.
34580; Thermo Fisher Scientific, Inc.) for 2 min, and images were
acquired using a Gel Imager System (GDS8000; UVP, Inc.). The Alpha
software processing system (AlphaEaseFC 4.0; ProteinSimple) was
used to measure the optical densities of the target bands.
Statistical analyses
The SPSS v22.0 (IBM, Inc.) was used for data
analysis. The results are presented as the mean ± standard
deviation (SD). Independent sample t-test was used to analyze two
normally distributed sample comparisons. One-way ANOVA was used for
statistical analysis followed by the Bonferroni's multiple
comparison test or Tamhane's multiple comparison test. P<0.05
was considered to indicate a statistically significant difference.
Each experiment was performed in triplicate to ensure reliability
and reproducibility of the results.
Results
Establishment of the C57BL/6J mouse
model with HFD-induced IR
There were no differences in initial body weights
between the CON and HFD groups. From the second week of high-fat
feeding, the body weights of mice in the HFD group were
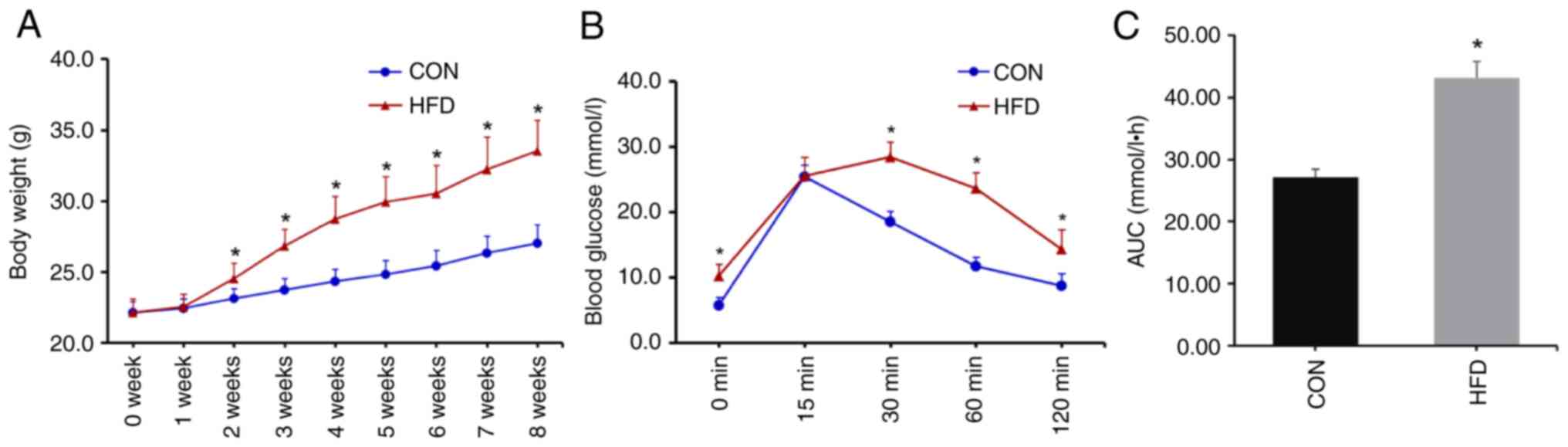
significantly higher than in the CON group (Fig. 1A). The IPGTT was performed after 8
weeks of diet consumption. After fasting for 12 h, there was no
difference in weight loss between the two groups (Table II). Blood glucose levels in the HFD
group were significantly higher at 0, 30, 60 and 120 min than
corresponding levels in the CON group (Fig. 1B). Compared with the CON group, the
AUC was significantly increased in the HFD group (Fig. 1C), indicating that the IR model was
successfully established.
 | Table IIWeight loss in mice after 12 h of
fasting. |
Table II
Weight loss in mice after 12 h of
fasting.
| | First fasting for
12 h | Second fasting for
12 h |
|---|
| Groups | CON group
(n=11) | HFD group
(n=22) | CON group
(n=11) | HFD group
(n=11) | HFD + RSV group
(n=11) |
|---|
| Weight loss
(g) | 2.6±0.2 | 2.7±0.2 | 3.4±0.2 | 3.7±0.3 | 3.6±0.4 |
| t/F | 1.353 | 2.655 |
| P-value | 0.186 | 0.087 |
Effects of RSV administration on body
weight and blood glucose, insulin, as well as lipid levels in mice
with IR
Following high-fat feeding, the body weight of the
HFD group was significantly higher than that in the CON group.
After RSV administration for 4 weeks, the body weight of the HFD +
RSV group was lower than that in the HFD group (Fig. 2A).
 | Figure 2Effects of RSV on body weight, and
blood glucose, as well as insulin and lipid levels in mice with
insulin resistance. (A) Body weight of mice with RSV treatment for
6 weeks. (B) The levels of blood glucose at 0, 15, 30, 60 and 120
min of the intraperitoneal glucose tolerance test after RSV
administration for 6 weeks. (C) AUC of glucose. (D) Fasting blood
glucose. (E) Fasting insulin. (F) QUICKI value. (G) TC level. (H)
TG level. (I) HDL-C level. (J) LDL-C level. Data are presented as
the mean ± SD (n=11). One-way ANOVA, followed by Bonferroni's or
Tamhane's multiple comparison post hoc tests, were used for
statistical analysis. *P<0.05 vs. the CON group;
#P<0.05 vs. the HFD group. RSV, resveratrol; AUC,
area under the curve; QUICKI, Qualitative Insulin Sensitivity Check
Index; TC, total cholesterol; TG, triglyceride; HDL-C, high-density
lipoprotein-cholesterol; LDL-C, low-density
lipoprotein-cholesterol; CON, control; HFD, high-fat diet. |
The IPGTT test was performed on three groups of mice
after 6 weeks of treatment with RSV. After fasting for 12 h, there
was no difference in weight loss among the three groups (Table II). The experiment determined that
the maximum percentage of body weight loss that was observed in the
study was 10.6% after fasting for 12 h. Compared with the CON
group, blood glucose levels were higher at 0, 15, 30, 60 and 120
min (Fig. 2B), and the AUC was
increased in the HFD group (Fig.
2C). Compared with the HFD group, blood glucose levels were
lower at 0, 30, 60 and 120 min, with no difference at 15 min
(Fig. 2B), and the AUC was
significantly decreased in the HFD + RSV group (Fig. 2C).
Compared with the CON group, the fasting blood
glucose and insulin levels were significantly higher (Fig. 2D and E), and the QUICKI value was lower in the
HFD group (Fig. 2F). Compared with
the HFD group, fasting blood glucose and insulin levels were
significantly lower (Fig. 2D and
E), and the QUICKI value was higher
in the HFD + RSV group (Fig. 2F).
These data indicated that RSV may significantly ameliorate the IR
of HFD-fed mice.
In addition, serum concentrations of TG, TC and
LDL-C were significantly increased in the HFD group compared with
the CON group (Fig. 2G, H and J).
Compared with the HFD group, TG and LDL-C were significantly
decreased, and HDL-C was significantly higher in the HFD + RSV
group, with no statistical difference in TC levels between the two
groups (Fig. 2G-J).
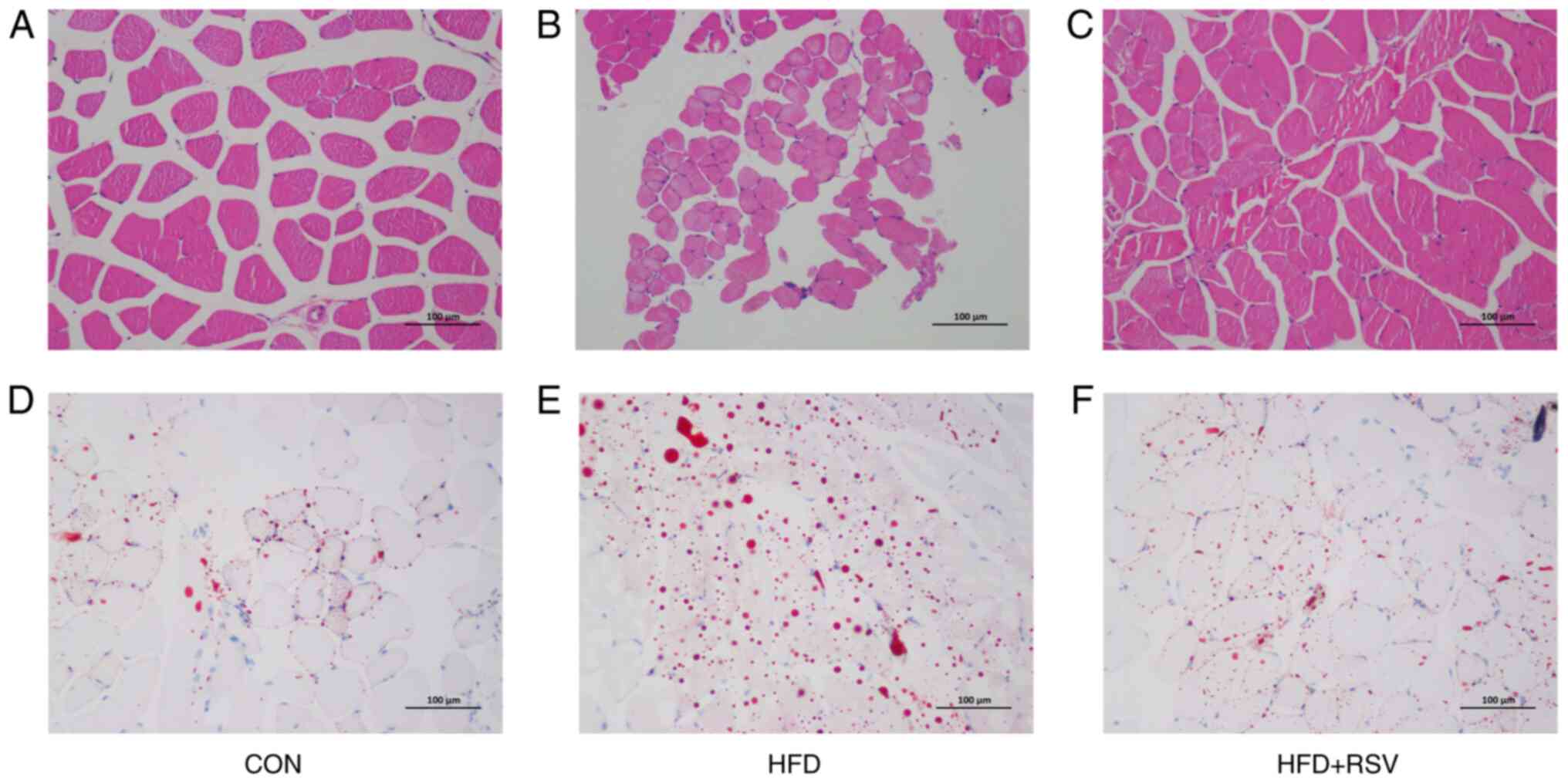
Effects of RSV on histology and lipid
accumulation in skeletal muscle of mice with IR
The H&E staining revealed normal histology of
skeletal muscle cells in the CON group (Fig. 3A). However, in the HDF group, the
cellular structure of skeletal muscle cells was fuzzy and
disordered, with fat drop vacuoles of different sizes in the
cytoplasm, constricting the nuclei, which were closer to the outer
cell membrane (Fig. 3B). In the HFD
+ RSV group, the morphology of skeletal muscle cells resembled
morphological features between those of the CON and HFD groups,
with lipid droplet vacuoles significantly reduced compared with
those in the HFD group (Fig. 3C).
Oil Red O staining showed larger numbers of red fat droplets in
skeletal muscle cells of the HFD group compared with the CON group
(Fig. 3D and E). The number of red fat droplets in
skeletal muscle cells of the HFD + RSV group was decreased compared
with the HFD group, and was between the numbers found in the CON
and HFD groups (Fig. 3D-F).
Effects of RSV on mRNA and protein
expression levels of insulin signaling pathway factors in skeletal
muscle tissue of mice with IR
There were no differences in the mRNA expression
levels of IRS-1, PI3K and AKT in skeletal muscle from the three
groups of mice (Fig. 4A-C). In the
HFD group, the expression level of GLUT4 mRNA was significantly
decreased compared with the CON group (Fig. 4D). Compared with the HFD group, the
expression level of GLUT4 mRNA was upregulated in the HFD + RSV
group (Fig. 4D).
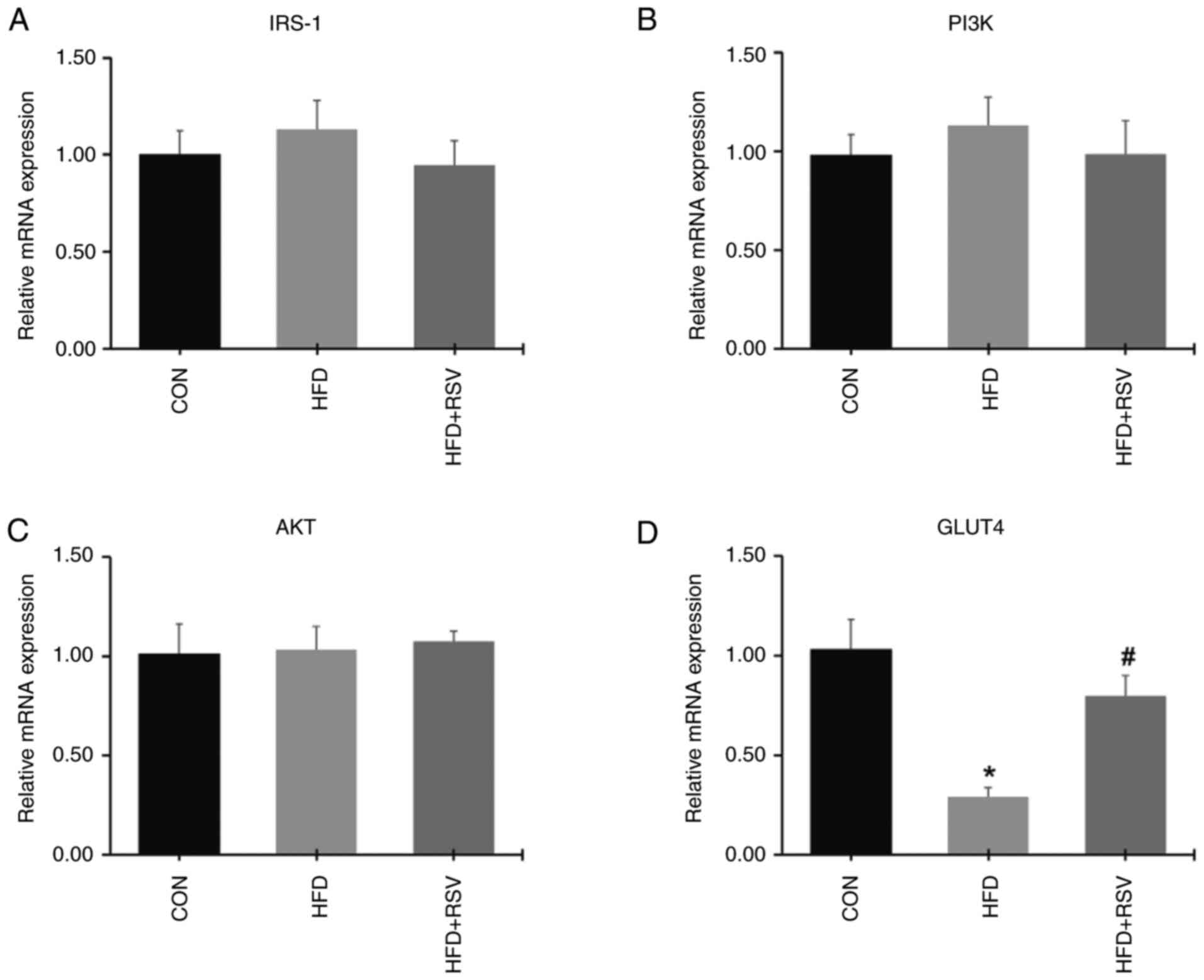 | Figure 4Effect of RSV on the expression
levels of IRS-1, PI3K, AKT and GLUT4 mRNA in skeletal tissue. (A)
IRS-1, (B) PI3K, (C) AKT and (D) GLUT4 mRNA levels. Data are
presented as the mean ± SD (n=11). *P<0.05 vs. the
CON group; #P<0.05 vs. the HFD group. RSV,
resveratrol; IRS-1, insulin receptor substrate-1; PI3K,
phosphatidylinositol 3-kinase; AKT, protein kinase B; GLUT4,
glucose transporter 4; CON, control; HFD, high-fat diet. |
Western blotting showed that there were no
differences in the total protein expression levels of IRS-1, PI3K
and AKT in skeletal muscle from the three groups (Fig. 5A). Compared with the CON group, the
expression levels of p-PI3K, p-AKT and GLUT4 were decreased, while
the expression of p-IRS-1 was increased in the HFD group (Fig. 5A and E). Moreover, the phosphorylated
protein-to-total protein ratios for PI3K and AKT were
downregulated, while that for IRS-1 was upregulated, in the HFD vs.
the CON groups (Fig. 5B-D).
Compared with the HFD group, the protein expression levels of
p-PI3K, p-AKT and GLUT4 were increased, while the expression of
p-IRS-1 was decreased in the HFD + RSV group (Fig. 5A and E). Furthermore, the phosphorylated
protein-to-total protein ratios for PI3K and AKT were upregulated,
while that for IRS-1 was downregulated, in the HFD + RSV group
relative to the HFD group (Fig.
5B-D). These data suggested that administration of RSV in mice
with IR could activate the IRS-1/PI3K/AKT/GLUT4 signaling
pathway.
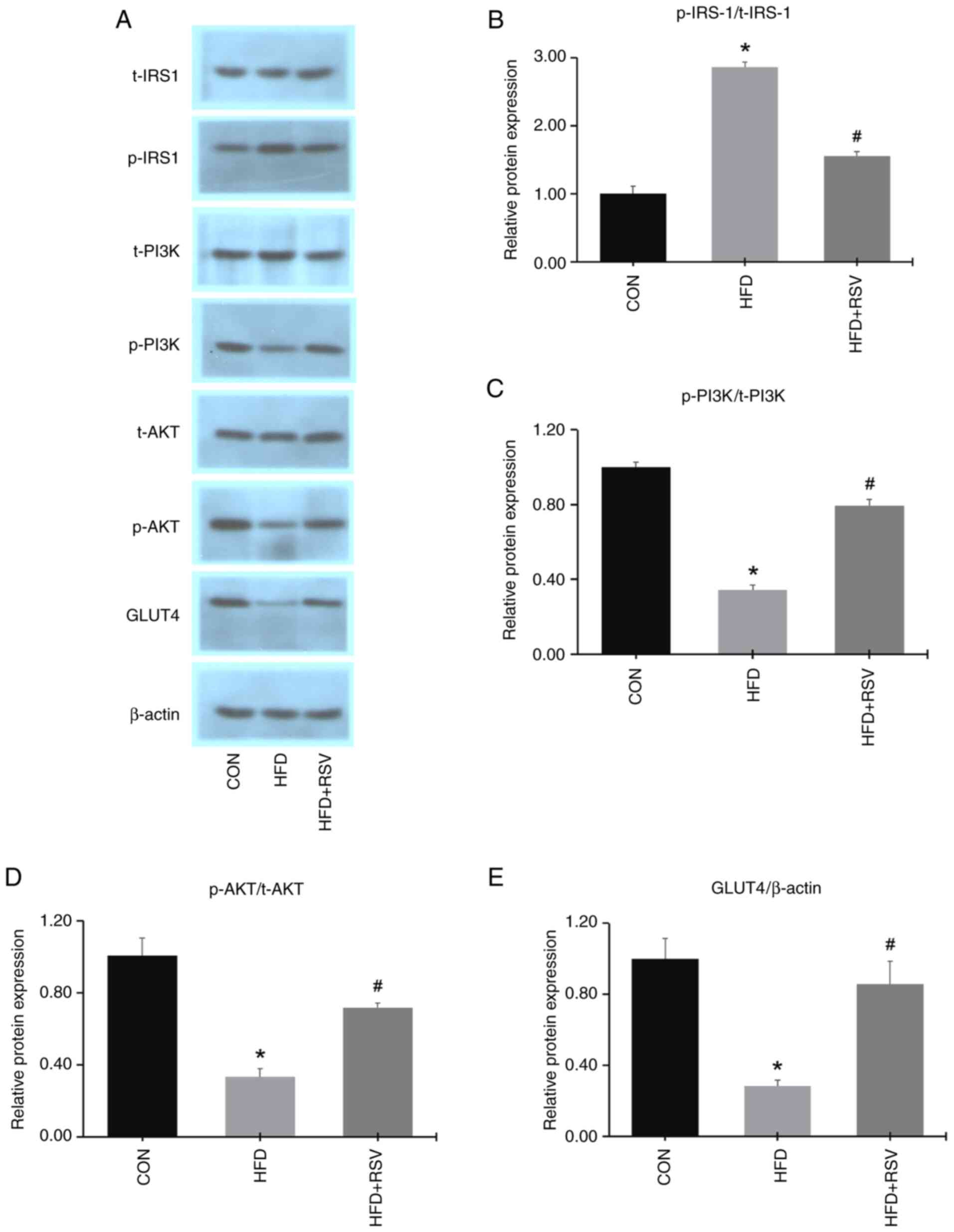 | Figure 5Effects of RSV on the protein
expression levels of insulin signaling pathway factors in skeletal
muscle. (A) Bands of various insulin signaling pathway proteins.
(B) p-IRS-1/t-IRS-1, (C) p-PI3K/t-PI3K, (D) p-AKT/t-AKT and (E)
GLUT4/β-actin protein expression levels. Data are presented as the
mean ± SD (n=11). *P<0.05 vs. the CON group;
#P<0.05 vs. the HFD group. RSV, resveratrol; p-,
phosphorylated; t-, total; IRS-1, insulin receptor substrate-1;
PI3K, phosphatidylinositol 3-kinase; AKT, protein kinase B; GLUT4,
glucose transporter 4; CON, control; HFD, high-fat diet. |
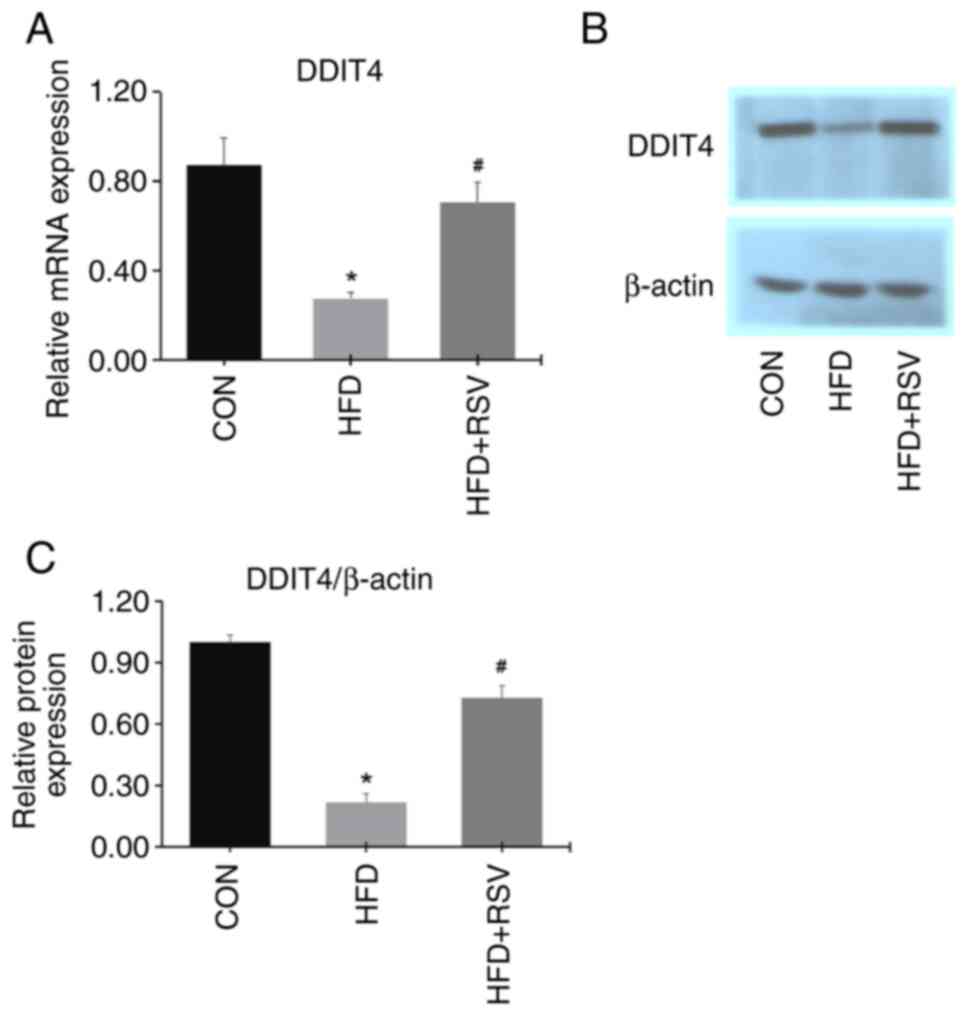
Effects of RSV on mRNA and protein
expression levels of DDIT4, mTOR and p70S6K in skeletal tissue of
mice with IR
Compared with CON mice, the mRNA and protein
expression levels of DDIT4 in skeletal muscle were significantly
decreased in the HFD mice (Fig.
6A-C), indicating that the expression of DDIT4 was decreased
under the IR conditions induced by the HFD. However, the mRNA and
protein expression levels of DDIT4 in skeletal muscle were
increased in the HFD + RSV group compared with the HFD group,
indicating that RSV promotes the expression of DDIT4 in mice with
IR.
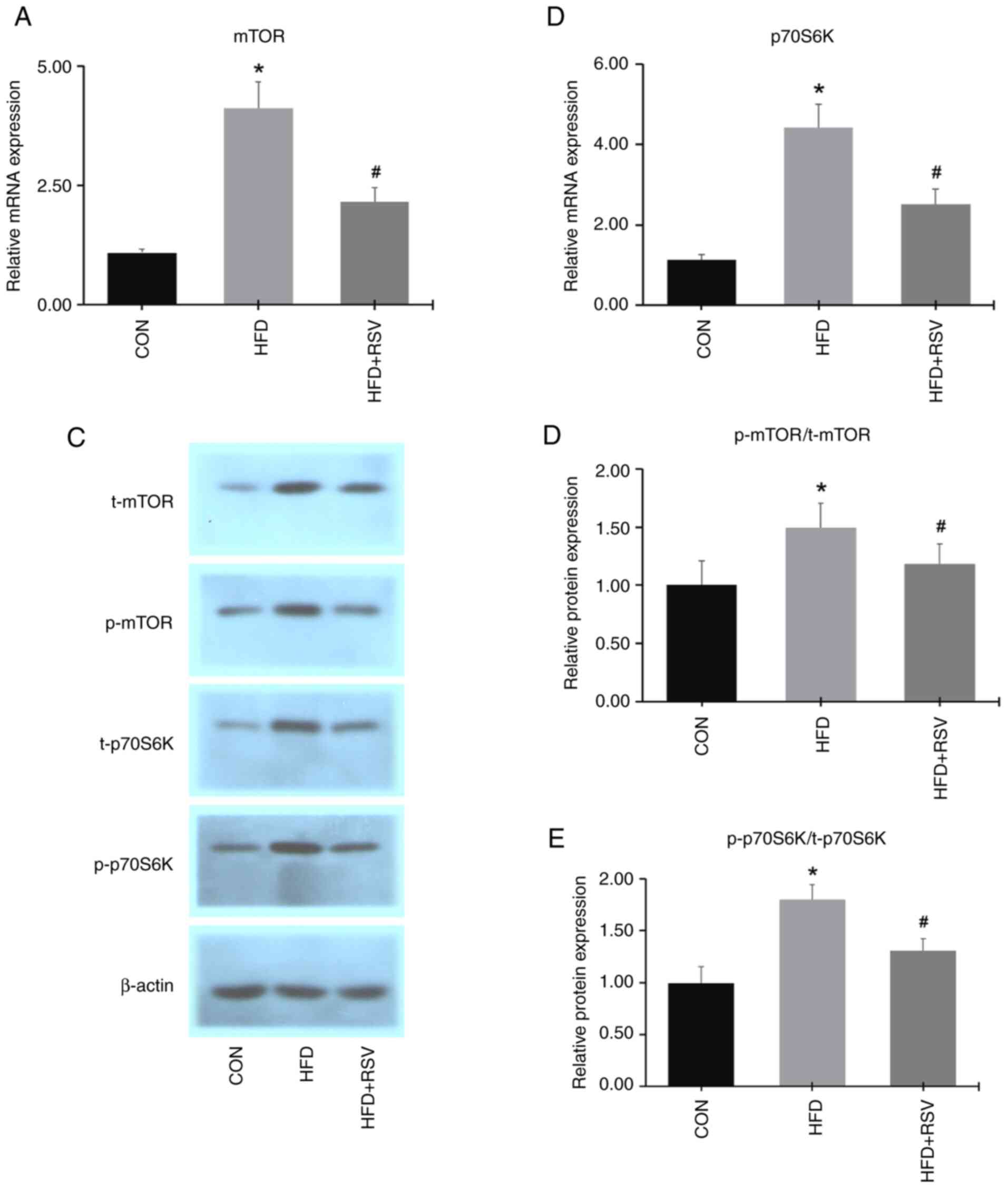
Compared with the CON group, the mRNA expression
levels of mTOR and p70S6K in skeletal muscle tissue were
significantly increased in the HFD group. However, the mRNA
expression levels of mTOR and p70S6K in skeletal muscle were
significantly decreased in the HFD + RSV group compared with the
HFD group (Fig. 7A and B).
Western blotting results showed that the expression
levels of total and phosphorylated mTOR and p70S6K proteins in
skeletal muscle tissue were increased in the HFD group compared
with the CON group (Fig. 7C). The
expression levels of total and phosphorylated mTOR and p70S6K
proteins in skeletal muscle from the HFD + RSV group were
significantly decreased compared with the HFD group (Fig. 7C and E). Furthermore, RSV reduced the increasing
trend of the phosphorylated protein-to-total protein ratios for
mTOR and p70S6K induced by the HFD (Fig. 7D and E). These findings indicated that RSV
inhibited the expression levels of mTOR and p70S6K in mice with
IR.
Expression of DDIT4, mTOR, PI3K, AKT,
and GLUT4 in MHY1485-treated C2C12 cells
Compared with those in the PA group, the protein
expression levels of DDIT4, p-PI3K/t-PI3K, p-AKT/t-AKT and GLUT4
were increased, whereas those of p-mTOR/t-mTOR were decreased in
the PA + RSV group (Fig. 8A and
B). There were no differences in
the protein expression levels of DDIT4 between the PA + RSV group
and PA + RSV + MHY1485 group (Fig.
8A and B). Compared with the PA
+ RSV group, the protein expression levels of p-PI3K/t-PI3K,
p-AKT/t-AKT and GLUT4 were decreased, whereas those of
p-mTOR/t-mTOR were increased in the PA + RSV + MHY1485 group
(Fig. 8A and B). Compared with the PA group, the glucose
concentration was significantly decreased in the culture medium
from the PA + RSV group (Fig. 8C).
Compared with the PA + RSV group, the glucose concentration was
significantly increased in the culture medium from the PA + RSV +
MHY1485 group (Fig. 8C). The TG
content was decreased in the PA + RSV group cells compared with
those in the PA group (Fig. 8D).
The TG content was increased in the PA + RSV + MHY1485 group cells
compared with those in the PA + RSV group (Fig. 8D).
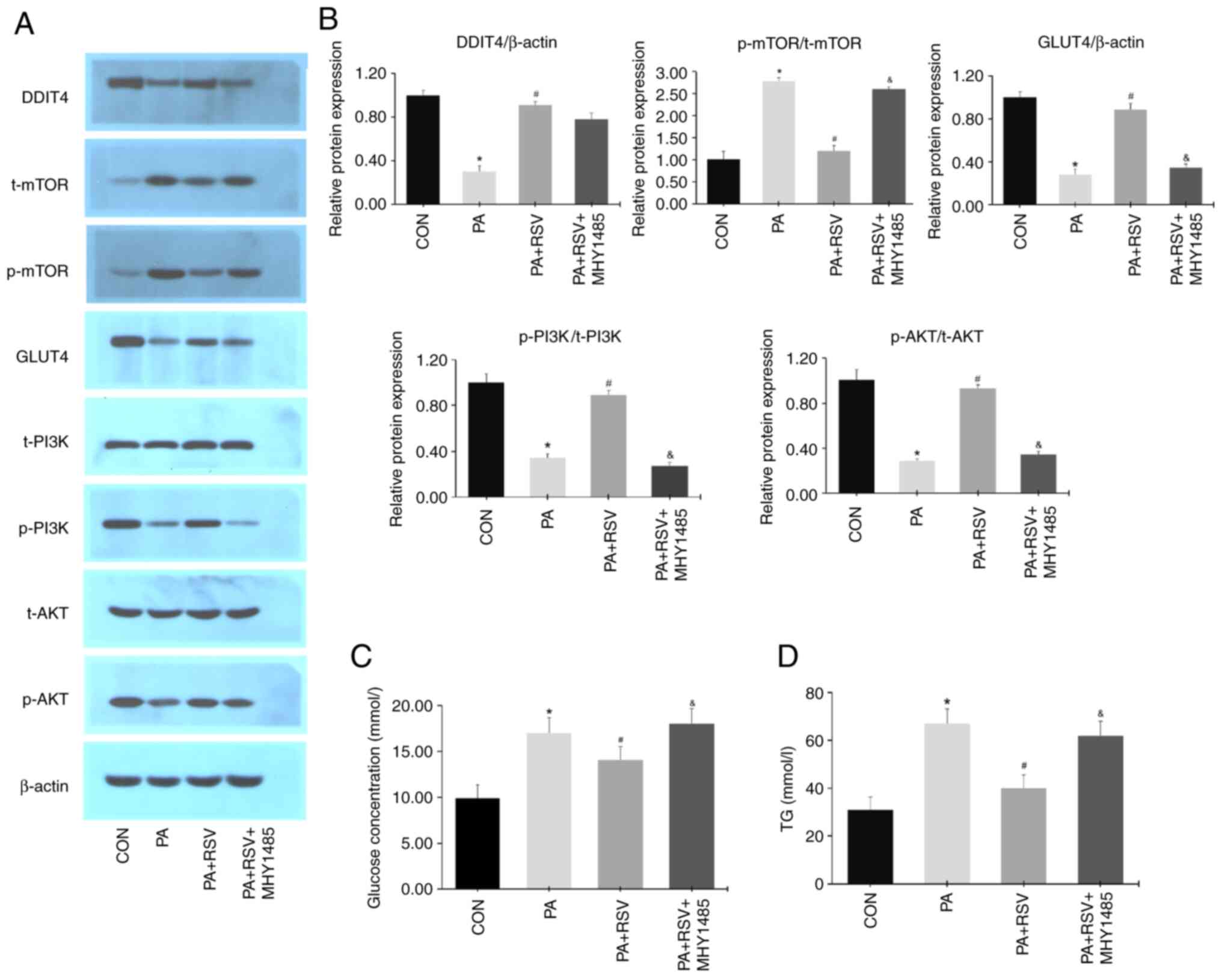 | Figure 8Expression of DDIT4, mTOR, GLUT4,
PI3K, and AKT in MHY1485-treated IR C2C12 cells. (A) Protein
expression levels of DDIT4, mTOR, GLUT4, PI3K, AKT after the
activation of mTOR. (B) Semi-quantification of p-protein/t-protein
ratios for mTOR, PI3K, AKT and protein expression levels of DDIT4,
GLUT4 after MHY1485 treatment. (C) Glucose levels in culture medium
after MHY1485 treatment. (D) Cell TG levels after MHY1485
treatment. Data are presented as the mean ± SD (n=3).
*P<0.05 vs. the CON group; #P<0.05 vs.
the PA group; &P<0.05 vs. the PA + RSV group.
DDIT4, DNA-damage-inducible transcript 4; mTOR, mammalian target of
rapamycin; GLUT4, glucose transporter 4; PI3K, phosphatidylinositol
3-kinase; AKT, protein kinase B; IR, insulin resistance; p-,
phosphorylated; t-, total; TG, triglyceride; CON, control; PA,
palmitic acid; RSV, resveratrol. |
Expression of DDIT4, mTOR, PI3K, AKT,
and GLUT4 in C2C12 cells with silencing of DDIT4
Plasmid validation experiments revealed that
DDIT4-siRNA-1 had the most significant silencing effect (Fig. 9A). The cells were then divided into
four groups: CON, PA, PA + RSV, and PA + RSV + DDIT4 siRNA
groups.
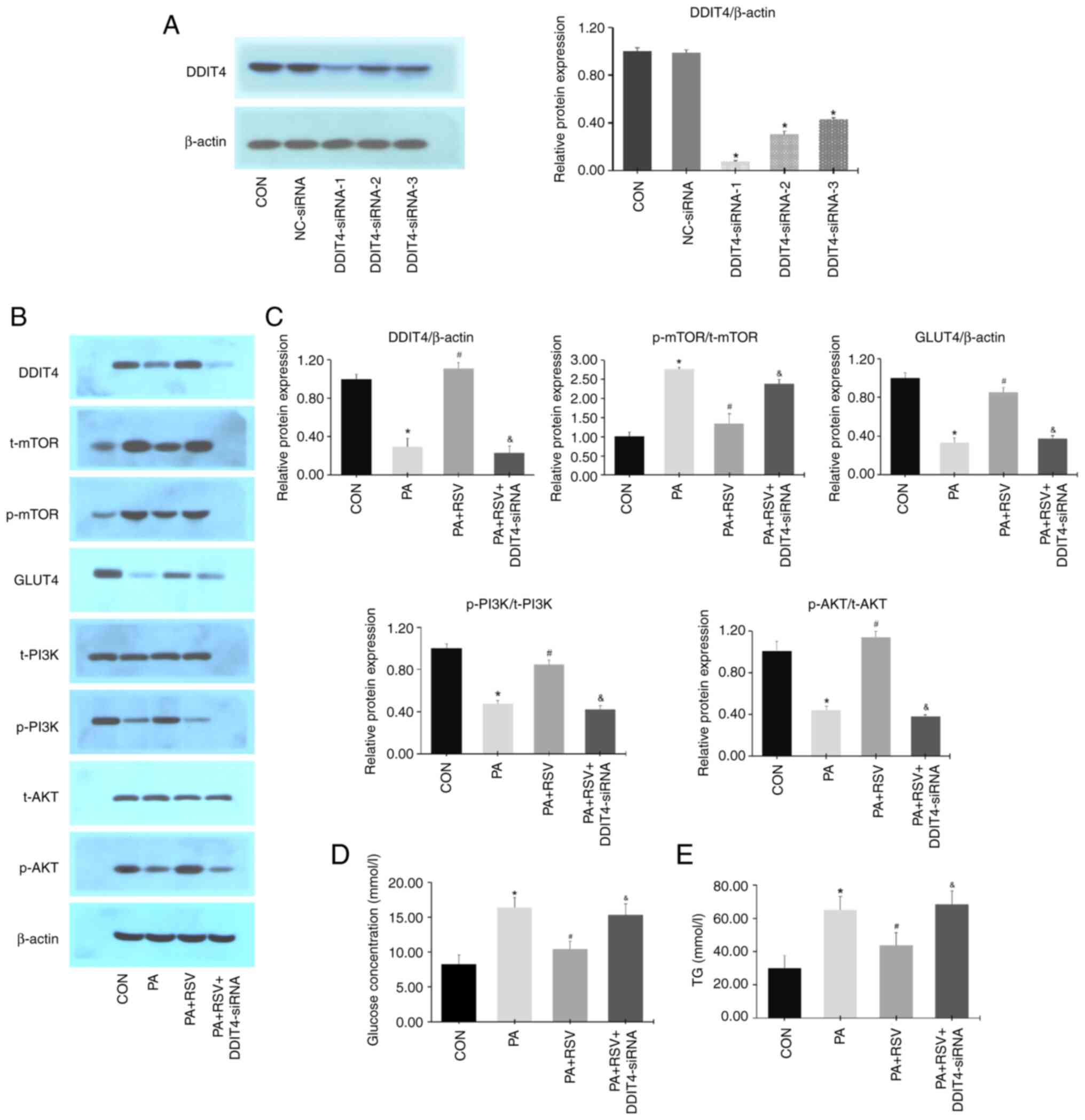 | Figure 9Expression levels of DDIT4, mTOR,
GLUT4, PI3K, and AKT in DDIT4-siRNA-treated IR C2C12 cells. (A)
DDIT4-siRNA-1 had the most significant silencing effect. (B)
Protein expression levels of DDIT4, mTOR, GLUT4, PI3K, and AKT
after DDIT4-siRNA treatment. (C) Semi-quantification of
p-protein/t-protein ratios for mTOR, PI3K, and AKT and protein
expression levels of DDIT4 and GLUT4 after DDIT4-siRNA treatment.
(D) Glucose levels in culture medium after DDIT4-siRNA treatment.
(E) TG levels of C2C12 cells after DDIT4-siRNA treatment. Data are
presented as the mean ± SD (n=3). *P<0.05 vs. the CON
group; #P<0.05 vs. the PA group;
&P<0.05 vs. the PA + RSV group. DDIT4,
DNA-damage-inducible transcript 4; mTOR, mammalian target of
rapamycin; GLUT4, glucose transporter 4; PI3K, phosphatidylinositol
3-kinase; AKT, protein kinase B; siRNA, small interfering RNA; IR,
insulin resistance; p-, phosphorylated; t-, total; TG,
triglyceride; CON, control; PA, palmitic acid; RSV,
resveratrol. |
Compared with the PA + RSV group, the protein
expression levels of DDIT4, p-PI3K/t-PI3K, p-AKT/t-AKT and GLUT4
were decreased, whereas those of p-mTOR/t-mTOR were increased in
the PA + RSV + DDIT4-siRNA group (Fig.
9B and C). Compared with the PA
+ RSV group, the glucose concentration in the culture medium and TG
content of the cells were increased in the PA + RSV + DDIT4-siRNA
group (Fig. 9D and E).
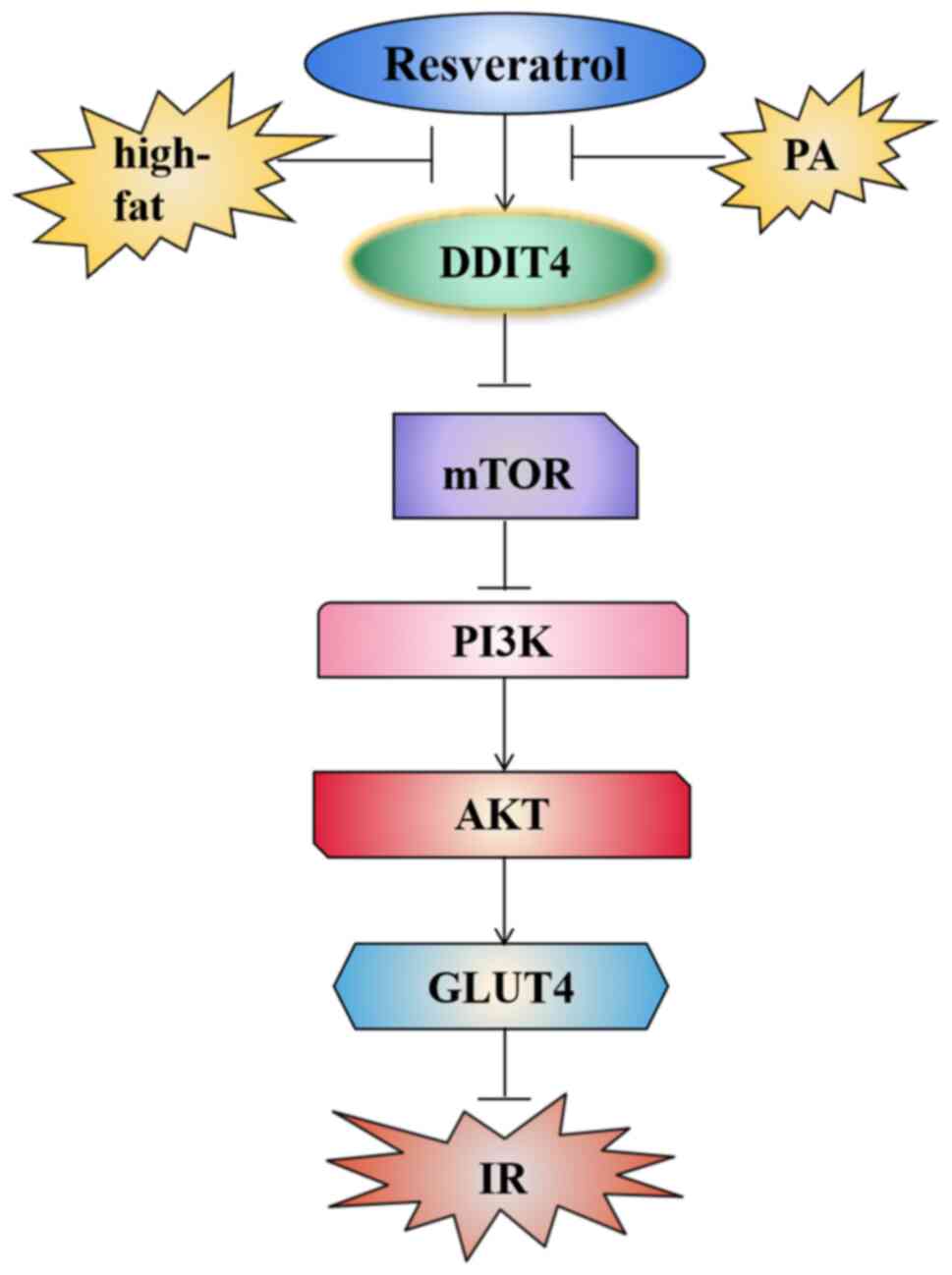
The results showed that RSV could activate DDIT4
expression, thereby inhibiting the mTOR pathway and then reducing
IR and lipid deposition in high-fat induced C2C12 cells (Fig. 10).
Discussion
In the present study, a mouse model with IR was
established by an HFD. This model closely simulated manifestations
of IR, including weight gain, hyperglycemia, hyperlipidemia,
impaired glucose tolerance and a low QUICKI index. In the present
study, treatment with RSV markedly reduced the concentration of
blood lipid and glucose, and decreased the AUC of IPGTT and body
weight of mice with HFD-induced IR. RSV also increased the QUICKI
index, improved the morphology and structure of skeletal muscle
tissue, and significantly reduced lipid droplet vacuoles in
skeletal muscle cells. It was determined that there was no
difference in weight loss among the groups after fasting for 12 h
with a maximum weight loss of 10.6%, which was similar to the
results found in another study (27). These results showed that RSV reduced
IR and lipid deposition in the skeletal muscle of mice fed an HFD.
Cell experiments also confirmed that PA intervention could reduce
glucose uptake and increase lipid levels in C2C12 cells. RSV could
reverse this condition, while the mTOR agonist MHY1485 or siRNA
against DDIT4 intervention further confirmed that RSV inhibited
mTOR by activating DDIT4, thereby affecting the activation of the
insulin pathway. Previous research also revealed that RSV plays a
beneficial role in glucose and lipid metabolism in animal models
and individuals with obesity and type 2 diabetes mellitus (28-30),
and reduced IR in various models (30,32).
To determine the molecular mechanism of the effects
of RSV, the present study examined gene and protein expression
levels of PI3K, AKT and GLUT4 in murine skeletal muscle and C2C12
cells. Biologically, insulin can combine with the insulin receptor
of peripheral target organs, such as skeletal muscle, and then
interact with IRS to activate the PI3K/AKT signaling pathway,
resulting in the downstream translocation of GLUT4 to the cell
membrane, thereby increasing glucose uptake and metabolism in
peripheral target organs (33). RSV
can increase the expression of GLUT4 in peripheral tissues by
activating the PI3K/AKT signaling pathway, thereby increasing
tissue uptake of glucose and lowering blood glucose levels
(34,35). Similarly, the present study found
that an HFD inhibited the IRS-1/PI3K/AKT/GLUT4 insulin signaling
pathway. However, RSV partially corrected the aberrant protein
levels of PI3K, AKT and GLUT4 in mice with IR, suggesting that RSV
exerts a regulatory effect by alleviating IR in skeletal muscle
through modulation of the PI3K/AKT/GLUT4 signaling pathway.
The present study found that the mTOR pathway was
activated in mice with HFD-induced IR and in C2C12 cells with
PA-induced IR, and that the expression levels of mTOR were
increased in skeletal muscle and in C2C12 cells. The continuous
activation of the mTOR pathway has been revealed to induce lipid
deposition, leading to tissue and cell IR (36,37).
The activation of mTOR can trigger a negative feedback pathway
involving the downstream signal p70S6K, which when activated leads
to the phosphorylation of IRS-1 and promotion of the degradation of
IRS-1, thus blocking the PI3K/AKT signal transduction pathway,
leading to IR (7). The inhibition
of mTOR and p70S6K can inhibit serine phosphorylation of
IRS-1(38), restore the
insulin-mediated glucose transport of GLUT4(39), and improve insulin sensitivity and
reduce obesity in mice (40). The
present study found that RSV inhibited activation of mTOR, then
promoted the PI3K/AKT/GLUT4 insulin signaling pathway, increased
insulin sensitivity and reduced the abnormal glucose and lipid
metabolism in PA-treated C2C12 cells and mice fed an HFD.
The present study determined the expression levels
of DDIT4, which is a negative regulator of the mTOR signaling
pathway (41-43).
High expression levels of DDIT4 in rat gastrocnemius muscle cells
were demosntrated to inhibit the activity of the mTOR signaling
pathway (44). Regazzetti et
al (45) showed that silencing
DDIT4 in 3T3-L1 adipocytes induced an increase in mTOR activity,
and inhibited the insulin signaling pathway and adipogenesis in
adipocytes. Consistent with the aforementioned studies, the current
results indicated that the expression of DDIT4 was inhibited, the
expression levels of mTOR and p70S6K were increased, and the
PI3K/AKT/GLUT4 insulin pathway was inhibited in mice or C2C12 cells
with IR. Wang et al (46)
found that the expression of DDIT4 was reduced in a rat model of
diabetic nephropathy, and the administration of
1,25-dihydroxyvitamin D3 increased DDIT4 expression and caused a
significant decrease in the phosphorylation levels of mTOR and
p70S6K, resulting in the inhibition of mesangial cell
proliferation, hypertrophy, and disordered blood glucose and lipid
metabolism (46). Notably, the
present study showed that administration of RSV to mice or cells
with IR increased the expression of DDIT4, inhibited the mTOR
pathway, activated the PI3K/AKT/GLUT4 pathway and reduced the
HFD-induced IR, and glucose and lipid metabolism disorders in
skeletal muscle and cells. Therefore, RSV may reduce IR by
regulating the DDIT4/mTOR signaling pathway.
Limited data from a human-based study revealed that
RSV improved insulin sensitivity and fasting glucose levels in
patients with type 2 diabetes mellitus and may improve inflammatory
status in human obesity (28). A
meta-analysis revealed that RSV significantly improved glucose
control and insulin sensitivity in individuals with diabetes, but
did not affect glycemic levels in nondiabetic individuals (47). A randomized controlled study
revealed that RSV treatment improved inflammation, renal function,
blood glucose parameters, inflammation, IR, and nutrient sensing
systems in elderly patients with type 2 diabetes mellitus,
indicating that RSV may be a potential therapeutic drug for the
treatment of elderly patients with type 2 diabetes mellitus
(48). Another randomized
controlled study showed that natural polyphenol, RSV, represents a
potential new treatment for management of nonalcoholic fatty liver
disease due to its anti-inflammatory and antioxidant properties
(49). There is limited research on
the mechanism of RSV on IR, as well as glucose and lipid metabolism
disorders in human studies. Therefore, in the present study, the
mechanism of RSV in improving IR, as well as glucose and lipid
metabolism through animal and cell experiments was explored, which
generated data for drug preclinical research and laid a foundation
for future clinical studies.
Treatment with RSV decreased body weight, blood
glucose and lipid levels, and reduced IR in mice. It is proposed
that RSV activated the PI3K/AKT/GLUT4 signaling pathway by
regulating the DDIT4/mTOR pathway, resulting in significant
anti-hyperglycemic and anti-hyperlipidemic activities in skeletal
muscle and C2C12 cells. Therefore, RSV can potentially be used as
an effective drug for the treatment of type 2 diabetes, as well as
a nutritional supplement for the prevention of type 2 diabetes and
cardiovascular diseases.
Acknowledgements
Not applicable.
Funding
Funding: This study was partially supported by the Scientific
Research Project of Hebei Provincial Administration of Traditional
Chinese Medicine (grant no. 2023010).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XP performed the data collection and analysis and
wrote the manuscript. GX, MZ, ZX, DL and ZD performed the sample
collection and detection. CW designed the study, performed the
primary research, and supported manuscript revision. XP and CW
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
2023-LW-055) by the Ethics Committee of Hebei General Hospital
(Shijiazhuang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhu X, Wu C, Qiu S, Yuan X and Li L:
Effects of resveratrol on glucose control and insulin sensitivity
in subjects with type 2 diabetes: Systematic review and
meta-analysis. Nutr Metab (Lond). 14(60)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Duan TT, Hu XB and Cao ZH: Effect of
resveratrol in treatment of diabetes mellitus. Prog Microbiol
Immunol. 48:99–103. 2020.(In Chinese).
|
|
3
|
Vlavcheski F and Tsiani E: Attenuation of
free fatty acid-induced muscle insulin resistance by rosemary
extract. Nutrients. 10(1623)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang L, Gao P, Zhang M, Huang Z, Zhang D,
Deng Q, Li Y, Zhao Z, Qin X, Jin D, et al: Prevalence and ethnic
pattern of diabetes and prediabetes in China in 2013. JAMA.
317:2515–2523. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Khamzina L, Veilleux A, Bergeron S and
Marette A: Increased activation of the mammalian target of
rapamycin pathway in liver and skeletal muscle of obese rats:
Possible involvement in obesity-linked insulin resistance.
Endocrinology. 146:1473–1481. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tremblay F and Marette A: Amino acid and
insulin signaling via the mTOR/p70 S6 kinase pathway. A negative
feedback mechanism leading to insulin resistance in skeletal muscle
cells. J Biol Chem. 276:38052–38060. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Holz MK, Ballif BA, Gygi SP and Blenis J:
mTOR and S6K1 mediate assembly of the translation preinitiation
complex through dynamic protein interchange and ordered
phosphorylation events. Cell. 123:569–580. 2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tinline-Goodfellow CT, Lees MJ and Hodson
N: The skeletal muscle fiber periphery: A nexus of mTOR-related
anabolism. Sports Med Health Sci. 5:10–19. 2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ellisen LW, Ramsayer KD, Johannessen CM,
Yang A, Beppu H, Minda K, Oliner JD, McKeon F and Haber DA: REDD1,
a developmentally regulated transcriptional target of p63 and p53,
links p63 to regulation of reactive oxygen species. Mol Cell.
10:995–1005. 2002.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Brugarolas J, Lei K, Hurley RL, Manning
BD, Reiling JH, Hafen E, Witters LA, Ellisen LW and Kaelin WG Jr:
Regulation of mTOR function in response to hypoxia by REDD1 and the
TSC1/TSC2 tumor suppressor complex. Genes Dev. 18:2893–2904.
2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Reiling JH and Hafen E: The
hypoxia-induced paralogs Scylla and Charybdis inhibit growth by
down-regulating S6K activity upstream of TSC in Drosophila. Gene
Dev. 18:2879–2892. 2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang H, Kubica N, Ellisen LW, Jefferson LS
and Kimball SR: Dexamethasone represses signaling through the
mammalian target of rapamycin in muscle cells by enhancing
expression of REDD1. J Biol Chem. 281:39128–39134. 2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Jin HO, Seo SK, Woo SH, Kim ES, Lee HC,
Yoo DH, An S, Choe TB, Lee SJ, Hong SI, et al: Activating
transcription factor 4 and CCAAT/enhancer-binding protein-beta
negatively regulate the mammalian target of rapamycin via Redd1
expression in response to oxidative and endoplasmic reticulum
stress. Free Radical Bio Med. 46:1158–1167. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Whitney ML, Jefferson LS and Kimball SR:
ATF4 is necessary and sufficient for ER stress-induced upregulation
of REDD1 expression. Biochem Biophys Res Commun. 379:451–455.
2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
McGhee NK, Jefferson LS and Kimball SR:
Elevated corticosterone associated with food deprivation
upregulates expression in rat skeletal muscle of the mTORC1
repressor, REDD1. J Nutr. 139:828–834. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Dennis MD, Coleman CS, Berg A, Jefferson
LS and Kimball SR: REDD1 enhances protein phosphatase 2A-mediated
dephosphorylation of Akt to repress mTORC1 signaling. Sci Signal.
7(ra68)2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Williamson DL, Li Z, Tuder RM, Feinstein
E, Kimball SR and Dungan CM: Altered nutrient response of mTORC1 as
a result of changes in REDD1 expression: Effect of obesity vs REDD1
deficiency. J Appl Physiol (1985). 117:246–256. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gordon BS, Steiner JL, Williamson DL, Lang
CH and Kimball SR: Emerging role for regulated in development and
DNA damage 1 (REDD1) in the regulation of skeletal muscle
metabolism. Am J Physiol Endocrinol Metab. 311:E157–E174.
2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang ZM, Liu ZH, Nie Q, Zhang XM, Yang
LQ, Wang C, Yang LL and Song GY: Metformin improves high-fat
diet-induced insulin resistance in mice by downregulating the
expression of long noncoding RNA NONMMUT031874.2. Exp Ther Med.
23(332)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ayala JE, Samuel VT, Morton GJ, Obici S,
Croniger CM, Shulman GI, Wasserman DH and McGuinness OP: NIH Mouse
Metabolic Phenotyping Center Consortium. Standard operating
procedures for describing and performing metabolic tests of glucose
homeostasis in mice. Dis Model Mech. 3:525–534. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bowe JE, Franklin ZJ, Hauge-Evans AC, King
AJ, Persaud SJ and Jones PM: Metabolic phenotyping guidelines:
Assessing glucose homeostasis in rodent models. J Endocrinol.
222:G13–G25. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Benedé-Ubieto R, Estévez-Vázquez O,
Ramadori P, Cubero FJ and Nevzorova YA: Guidelines and
considerations for metabolic tolerance tests in mice. Diabetes
Metab Syndr Obes. 13:439–450. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jørgensen MS, Tornqvist KS and Hvid H:
Calculation of glucose dose for intraperitoneal glucose tolerance
tests in lean and obese mice. J Am Assoc Lab Anim Sci. 56:95–97.
2017.PubMed/NCBI
|
|
24
|
Shu L, Hou G, Zhao H, Huang W, Song G and
Ma H: Resveratrol improves high-fat diet-induced insulin resistance
in mice by downregulating the lncRNA NONMMUT008655.2. Am J Transl
Res. 12:1–18. 2020.PubMed/NCBI
|
|
25
|
Katz A, Nambi SS, Mather K, Baron AD,
Follmann DA, Sullivan G and Quon MJ: Quantitative insulin
sensitivity check index: A simple, accurate method for assessing
insulin sensitivity in humans. J Clin Endocrinol Metab.
85:2402–2410. 2000.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jensen TL, Kiersgaard MK, Sørensen DB and
Mikkelsen LF: Fasting of mice: A review. Lab Anim. 47:225–240.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Barber TM, Kabisch S, Randeva HS, Pfeiffer
AFH and Weickert MO: Implications of resveratrol in obesity and
insulin resistance: A state-of-the-art review. Nutrients.
14(2870)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Luo J, Chen S, Wang L, Zhao X and Piao C:
Pharmacological effects of polydatin in the treatment of metabolic
diseases: A review. Phytomedicine. 102(154161)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ariyanto EF, Danil AS, Rohmawaty E,
Sujatmiko B and Berbudi A: Effect of resveratrol in melinjo seed
(Gnetum gnemon L.) extract on type 2 diabetes mellitus patients and
its possible mechanism: A review. Curr Diabetes Rev.
19(e280222201512)2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shahwan M, Alhumaydhi F, Ashraf GM, Hasan
PMZ and Shamsi A: Role of polyphenols in combating type 2 diabetes
and insulin resistance. Int J Biol Macromol. 206:567–579.
2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhou Q, Wang Y, Han X, Fu S, Zhu C and
Chen Q: Efficacy of resveratrol supplementation on glucose and
lipid metabolism: A meta-analysis and systematic review. Front
Physiol. 13(795980)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chadt A and Al-Hasani H: Glucose
transporters in adipose tissue, liver, and skeletal muscle in
metabolic health and disease. Pflugers Arch. 472:1273–1298.
2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Vlavcheski F, Den Hartogh DJ, Giacca A and
Tsiani E: Amelioration of high-insulin-induced skeletal muscle cell
insulin resistance by resveratrol is linked to activation of AMPK
and restoration of GLUT4 translocation. Nutrients.
12(914)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kang BB and Chiang BH: A novel phenolic
formulation for treating hepatic and peripheral insulin resistance
by regulating GLUT4-mediated glucose uptake. J Tradit Complement
Med. 12:195–205. 2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Bar-Tana J: Type 2 diabetes-unmet need,
unresolved pathogenesis, mTORC1-centric paradigm. Rev Endocr Metab
Dis. 21:613–629. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bodur C, Kazyken D, Huang K, Tooley AS,
Cho KW, Barnes TM, Lumeng CN, Myers MG and Fingar DC: TBK1-mTOR
signaling attenuates obesity-linked hyperglycemia and insulin
resistance. Diabetes. 71:2297–2312. 2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Huang SL, Xie W, Ye YL, Liu J, Qu H, Shen
Y, Xu TF, Zhao ZH, Shi Y, Shen JH and Leng Y: Coronarin A modulated
hepatic glycogen synthesis and gluconeogenesis via inhibiting
mTORC1/S6K1 signaling and ameliorated glucose homeostasis of
diabetic mice. Acta Pharmacol Sin. 44:596–609. 2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Den Hartogh DJ, Vlavcheski F, Giacca A and
Tsiani E: Attenuation of free fatty acid (FFA)-induced skeletal
muscle cell insulin resistance by resveratrol is linked to
activation of AMPK and inhibition of mTOR and p70S6K. Int J Mol
Sci. 21(4900)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Meng W, Liang X, Chen H, Luo H, Bai J, Li
G, Zhang Q, Xiao T, He S, Zhang Y, et al: Rheb inhibits beiging of
white adipose tissue via PDE4D5-dependent down regulation of the
cAMP-PKA signaling pathway. Diabetes. 66:1198–1213. 2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Dungan CM and Williamson DL: Regulation of
skeletal muscle insulin-stimulated signaling through the
MEK-REDD1-mTOR axis. Biochem Biophys Res Commun. 482:1067–1072.
2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lee Y, Song MJ, Park JH, Shin MH, Kim MK,
Hwang D, Lee DH and Chung JH: Histone deacetylase 4 reverses
cellular senescence via DDIT4 in dermal fibroblasts. Aging (Albany
NY). 14:4653–4672. 2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhidkova EM, Lylova ES, Grigoreva DD,
Kirsanov KI, Osipova AV, Kulikov EP, Mertsalov SA, Belitsky GA,
Budunova I, Yakubovskaya MG and Lesovaya EA: Nutritional sensor
REDD1 in cancer and inflammation: Friend or foe? Int J Mol Sci.
23(9686)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Murakami T, Hasegawa K and Yoshinaga M:
Rapid induction of REDD1 expression by endurance exercise in rat
skeletal muscle. Biochem Biophys Res Commun. 405:615–619.
2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Regazzetti C, Dumas K, Le Marchand-Brustel
Y, Peraldi P, Tanti JF and Giorgetti-Peraldi S: Regulated in
development and DNA damage responses-1 (REDD1) protein contributes
to insulin signaling pathway in adipocytes. PLoS One.
7(e52154)2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Wang H, Wang J, Qu H, Wei H, Ji B, Yang Z,
Wu J, He Q, Luo Y, Liu D, et al: In vitro and in vivo inhibition of
mTOR by 1,25-dihydroxyvitamin D3 to improve early diabetic
nephropathy via the DDIT4/TSC2/mTOR pathway. Endocrine. 54:348–359.
2016.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Delpino FM and Figueiredo LM: Resveratrol
supplementation and type 2 diabetes: A systematic review and
meta-analysis. Crit Rev Food Sci Nutr. 62:4465–4480.
2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ma N and Zhang Y: Effects of resveratrol
therapy on glucose metabolism, insulin resistance, inflammation,
and renal function in the elderly patients with type 2 diabetes
mellitus: A randomized controlled clinical trial protocol. Medicine
(Baltimore). 101(e30049)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Karimi M, Abiri B, Guest PC and Vafa M:
Therapeutic effects of resveratrol on nonalcoholic fatty liver
disease through inflammatory, oxidative stress, metabolic, and
epigenetic modifications. Methods Mol Biol. 2343:19–35.
2022.PubMed/NCBI View Article : Google Scholar
|