Introduction
The pathogeneses of osteoporosis and sarcopenia, the
two most common diseases encountered in older adults, are closely
related to age (1). Osteoporosis
refers to the deterioration of bone density and microarchitecture
(2), whereas sarcopenia refers to
defects in muscle quantity, quality and function (3). Numerous studies have demonstrated that
muscles and bones are spatially and metabolically connected
(4). Furthermore, individuals with
sarcopenia are five times more likely to develop osteoporosis than
healthy individuals and vice versa (5). Therefore, some studies have termed the
coexistence of osteoporosis and sarcopenia as osteosarcopenia
(6,7). The presence of OS leads to more
fractures, frailty, hospitalization, disability and mortality than
osteoporosis or sarcopenia alone (6).
Muscle and bone share a common origin of mesenchymal
precursors (8). Furthermore,
several molecular mechanisms regulate both muscles and bones
(9). Thus, finding the common
molecule that affects the progress of osteogenesis and myogenesis
could help determine the pathogenesis of osteosarcopenia. Bone
marrow mesenchymal stem cells (BMSC) are pluripotent stem cells
with trilineage differentiation abilities, which include
osteogenesis, lipogenesis and chondrogenesis (10,11).
L6 is a myoblast strain isolated from the skeletal muscle of rats
that be used to study myogenesis. Secreted frizzled-related protein
1 (SFRP1) is an antagonist of the Wnt signaling pathway that binds
to Wnt proteins and prevents it from interacting with their cell
surface receptors. This helps regulate the balance between cell
proliferation and differentiation, which is essential for normal
development and tissue homeostasis. Dysregulation of SFRP1 has been
implicated in various human diseases, including cancer, fibrosis
and degenerative disorders.
SFRP1, which modulates the Wnt signaling pathway,
plays a significant role in bone and muscle development,
homeostasis and regeneration. SFRP1 plays a role in the regulation
of osteoblast differentiation by inhibiting the Wnt signaling
pathway. Thus, a decrease in SFRP1 expression can increase bone
formation and reduce bone resorption, whereas an increase in SFRP1
expression can decrease bone mass and increase susceptibility to
fractures. SFRP1 also plays a role in myogenesis, the process of
muscle tissue formation and post-injury muscle regeneration. SFRP1
promotes the activation and differentiation of muscle satellite
cells, which are responsible for muscle repair and growth.
Additionally, SFRP1 inhibition may have a role in preventing muscle
atrophy, which is the loss of muscle mass due to various factors
such as aging or diseases. Thus, SFRP1 may be a potential
therapeutic target to prevent muscle atrophy.
Although these findings suggested that SFRP1 may be
an essential molecule in osteosarcopenia, its upstream regulator
remains unknown. microRNAs (miRNAs/miRs) are small non-coding RNA
molecules that play a crucial role in regulating gene expression at
the post-transcriptional level. They can modulate the stability and
translation of messenger RNA (mRNA) molecules to control protein
synthesis and cellular functions.
By binding to the 3'UTR of target mRNA, they enable
degradation or inhibition of translation. Additionally, they
participate in complex life processes such as cell proliferation,
differentiation, apoptosis and carcinogenesis, which are closely
related to the occurrence, development, prognosis, diagnosis and
treatment of several diseases (12,13).
miRNAs are involved in the proliferation and differentiation of
bones or muscles (14-16).
For example, miR-34a-5p promotes osteogenic differentiation by
targeting HDAC1(17), miR-29a-3p
promotes proliferation and inhibits osteogenic differentiation by
targeting FOXO3(18), miR-130b
inhibits proliferation and promotes myoblast differentiation by
targeting Sp1(19) and miR-452
promotes proliferation and inhibits myoblast differentiation by
targeting ANGPT1(20). However,
only a few studies have evaluated miRNAs that regulate both bone
and muscle actions through a single pathway.
Muscle and bone share a common origin and are
regulated by several molecular mechanisms (9). Osteoblasts and myoblasts originate
from the same mesenchymal stem cells (8). Among these, bone marrow mesenchymal
stem cells (BMSC)s are currently being studied (10). In osteoporosis, the osteogenic
capacity of BMSC is diminished, which decreases bone formation
(11).
The present study extracted bone marrow stem cells
from osteoporotic and control rats for transcriptome sequencing to
identify the differential gene SFRP1. The results indicated that
miR-206-3p might target SFRP1. miR-1, miR-133 and miR-206 are known
muscle-specific miRNAs that play an important role in muscle growth
and development (21). Therefore,
the regulatory roles of miR-206-3p was assessed in BMSC and
myogenic cells. The results demonstrated that miR-206-3p targets
SFRP1 through the Wnt/β-catenin signaling pathway, which in turn
affects muscle and bone differentiation.
Materials and methods
Animals
The experimental animal study protocol was approved
by the Animal Care and Use Committee of the Kunming Medical
University, Kunming, China (approval no. kmmu20230916). All animal
experiments and handling were performed in accordance with the
Guide for the Care and Use of Laboratory Animals published by the
National Institutes of Health [NIH Publication No. 8523, revised
1985(22)]. A total of
six-week-old, healthy, female Sprague-Dawley rats, each weighing
250-300 g, free of specific pathogens, were purchased from the
Department of Laboratory Animal Science, Kunming Medical
University. All animals were maintained under a standard 12-h
light/dark cycle with free access to food and water. Environmental
conditions were maintained with relative humidity ranging from
30-60% and a temperature of 23±2˚C. After adaptive feeding, the
rats were ovariectomized (OVX group) or subjected to a sham surgery
(sham group; n=3 per group). Subsequently, the rats were fed for 3
months and euthanized. Subsequently, the rats were fed for 3 months
and sacrificed. Sacrifice was by intraperitoneal injection of
excessive sodium pentobarbital (100 mg/kg).
Ovariectomization
Rats were anesthetized intraperitoneally with 3%
sodium pentobarbital (30 mg/kg) and ovariectomy (OVX) was performed
under anesthesia. After making incisions on both sides of the
abdominal midline, the proximal end of each fallopian tube was
ligated and the ovaries were isolated. After bilateral ovaries were
excised with scissors, the wound was sutured.
Microcomputed tomography scanning
After the rats were euthanized, the femurs were
isolated and scanned using micro-computed tomography (CT). The
distal femur was scanned to determine the trabecular bone
volume/tissue volume ratio (BV/TV), bone mineral density (BMD) and
trabecular thickness (TbTh).
Hematoxylin and eosin staining
The rat femurs were fixed with 4% paraformaldehyde
for 24 h. Thereafter, the specimens were soaked in 70% ethanol for
3 days at 4˚C and the liquid was changed every day. Subsequently,
the specimens were embedded in paraffin and cut into 5-µm sections.
Finally, the sections were deparaffinized, rehydrated and stained
with hematoxylin and eosin. The sections were stained 10 min at
room temperature and observed using a light microscope
(magnification, x20).
Von Kossa staining
The rat femurs were fixed with 4% paraformaldehyde
for 24 h then immersed in 70% ethanol with daily solution changes
for 3 days and stored at 4˚C. All subsequent procedures were
performed at 4˚C whenever possible. Thereafter, embedding and
sectioning were performed to obtain 5 µm thick hard tissue
sections. The sections were stained using Von Kossa solution
(Beijing Solarbio Science & Technology Co., Ltd.) following the
manufacturer's instructions: Von Kossa silver solution was added
dripwise to the sections which were then irradiated with strong
light for 15-60 min at room temperature before being washed with
distilled water for 1 min at room temperature. After sealing, the
sections were observed under a light microscope (magnification,
x20).
Toluidine blue O solution
After being fixed with 4% paraformaldehyde for 24 h,
the rat femurs were soaked in 70% ethanol for 3 days at 4˚C. The
ethanol was changed every day. Thereafter, the femur was sectioned
at 5 µm and stained with toluidine blue solution (Beijing Solarbio
Science & Technology Co., Ltd.). The sections were stained with
1% toluidine blue O solution for 10 min at room temperature and
observed using a light microscope (magnification x20).
Tartrate-resistant acidic phosphatase
staining
After being fixed with 4% paraformaldehyde for 24 h,
the rats femurs were soaked in 70% ethanol for 3 days at 4˚C. The
ethanol was changed every day. Thereafter, the femur was sectioned
at 5 µm and stained using tartrate-resistant acidic phosphatase
(TRAP; Wuhan Servicebio Technology Co., Ltd.). The sections were
reacted with tartrate-resistant acidic phosphatase working solution
for 20 min at 37˚C in the dark and observed using a light
microscope (magnification x20).
BMSC isolation and culture
BMSCs were isolated from the bone marrow aspirate of
the rat femurs. In the OVX group, the BMSCs were isolated 3 months
after OVX. After being euthanized, the rats femurs were dissected
out aseptically and the external soft tissue was discarded. A
20-gauge needle was used to prepare the cell suspension by
repeatedly flushing the bone marrow lumen with minimum essential
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
The cells were inoculated into cell culture flasks and subsequently
inoculated in an α-minimal essential medium supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
1% penicillin/streptomycin at 37˚C in a humidified atmosphere of 5%
CO2.
Flow cytometry identification of
BMSC
Third-generation BMSCs were digested with TrypLE
(Gibco; Thermo Fisher Scientific, Inc.) and the cells were
resuspended in a complete medium to produce single-cell suspensions
containing 5x105-1x106 cells/ml. Thereafter,
the cells were washed twice with PBS containing 1% bovine serum
albumin (1% BSA/PBS; Beijing Solarbio Science & Technology Co.,
Ltd.). Thereafter, the cells were resuspended in 1% BSA/PBS before
being transplanted into Eppendorf tubes. CD29 (Invitrogen; Thermo
Fisher Scientific, Inc.; cat. no. 2282645), CD44 (Invitrogen;
Thermo Fisher Scientific, Inc.; cat. no. 2252597), CD45
(Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 2265339) and
CD90 (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 2227596)
cells and the antibody compensation beads were added to the
Eppendorf tubes according to the manufacturer's instructions. The
tubes were incubated at 4˚C for 60 min. Thereafter, the cells were
washed with 1%BSA/PBS twice at 4˚C before being resuspended.
Finally, the cells were analyzed by flow cytometry (BD
FACSCelesta™), and analyzed using FlowJov10.6.2 (FlowJo LLC).
Trilineage differentiation and
staining of BMSCs
BMSCs were placed in osteogenic, lipogenic and
chondrogenic induction media to induce differentiation. The
osteogenic, lipogenic and chondrogenic differentiation of BMSCs was
determined by staining with alizarin red, oil red O and Alcian
blue, respectively. For osteogenic differentiation, alizarin red
staining working solution stained cells for 6 min at room
temperature. For adipogenic differentiation, oil red O staining
working solution was used to stain cells for 30 min at room
temperature. For chondrogenic differentiation, Alcian blue staining
working solution was stained at 37˚C for 1 h.
Transcriptome sequencing of BMSCs
The BMSCs obtained from the OVX and Sham groups were
cultured up to P3 generation and four samples containing 5 million
cells each were obtained from each group. To extract the total RNA
from the cells, 1 ml of TRIzol® (Thermo Fisher
Scientific, Inc.) was added to each sample and the concentration
and purity of the extracted RNA were detected using NanoDrop 2000
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.). After the
RNA quality control had reached the standard, the ribosomal RNA was
removed and fragmented into small segments of approximately 200 bp.
Reverse transcription is then performed to generate cDNA. The
double-stranded cDNA structure has sticky ends, which are converted
to blunt ends by adding an End Repair Mix. Subsequently, a single A
base is added to the 3' end to facilitate the ligation of a
Y-shaped adapter, forming the adapter-ligated product. The product
is then purified and subjected to size selection. The selected
fragments are amplified by PCR, and the final library is purified.
The library is sequenced using the Illumina NovaSeq 6000 platform
(Illumina, Inc.).
L6 cell culture and myogenic
differentiation
L6 cells were obtained from the Fan Wenxing Research
Group of the First Affiliated Hospital of Kunming Medical
University. The L6 cells were inoculated in a high-glucose
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS and 1%
penicillin-streptomycin. These were then incubated in a
constant-temperature cell incubator at 37˚C and 5% CO2.
When the L6 cells had reached 60% confluence, the growth medium was
aspirated and the cells were washed twice with PBS. Thereafter, a
medium that induced myogenic differentiation [DMEM supplemented
with 2% horse serum (Gibco; Thermo Fisher Scientific, Inc.)] was
added. The differentiation medium was replaced daily.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from cells (1x106
cells/ml) using TRIzol® (Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. RNA (1 µg per sample) was
then reverse-transcribed into cDNA using a reverse transcription
kit (RR036A, Takara Biotechnology Co., Ltd.). qPCR was performed
with the TB Green Premix Ex Taq II kit (RR820A, Takara
Biotechnology Co., Ltd.) on a QuantStudio3 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). All
procedures were performed according to the manufacturer's
instructions. The PCR amplification conditions were 95˚C for 30 sec
(initial denaturation), next 40 cycles of 95˚C for 5 sec, and 60˚C
for 34 sec, then 95˚C for 5 sec and 60˚C for 60 sec (dissociation),
then cooled to 37˚C. Relative gene expression was determined using
the 2−ΔΔCq (23) method
and normalized to GAPDH mRNA expression. All mRNA primers (Table I) were synthesized by Sangon Biotech
Co., Ltd. To detect miRNA expression, total miRNA was extracted
from the cells using RNAiso for small RNA (9753A, Takara
Biotechnology Co., Ltd.). The miRNAs were reverse transcribed using
the Mir-X miRNA First-Strand Synthesis kit (638313, Takara
Biotechnology Co., Ltd.). RT-qPCR was performed using the miRNA
tailing reaction. The relative transcription levels of miRNA were
normalized to that of U6 RNA. All miRNA primers (Table I) were synthesized by Sangon Biotech
Co., Ltd.
 | Table IPrimers. |
Table I
Primers.
| Gene symbol | Forward | Reverse |
|---|
| SFRP1 |
TACTGGCCCGAGATGCTCAA |
TACTGGCCCGAGATGCTCAA |
| GAPDH |
CAGGTTGTCTCCTGTGACTT |
TTATGGGGTCTGGGATGGAA |
| U6 |
TGGAACGCTTCACGAATTTGCG |
GGAACGATACAGAGAAGATTAGC |
| miR-206-3p |
CGTGGAATGTAAGGAAGTGTGTGG | |
Western blotting
RIPA lysate (Beijing Solarbio Science &
Technology Co., Ltd.) was used to lyse the cells and extract the
proteins. A total of 20 µg (as measured by the BCA protein assay
kit) of the protein sample was electrophoresed on a 10% sodium
dodecyl sulfate-polyacrylamide gel. The isolated proteins were
transferred to a polyvinylidene difluoride membrane by wet
transfer. The PVDF membranes were blocked at room temperature in 5%
skimmed milk for 2 h and incubated overnight with the primary
antibody at 4˚C. Subsequently, the PVDF membranes were incubated
with horseradish peroxide-coupled secondary antibodies for 2 h at
25˚C. The results were visualized using an enhanced
chemiluminescence solution (MilliporeSigma) after washing the
membranes. The protein bands were quantified using ImageJ v1.8.0
(National Institutes of Health). The antibody and dilution ratios
were as follows: GAPDH, 1:50,000, Proteintech Group, Inc., cat. no.
10494-1-AP; osteopontin (OPN), 1:4,000, Proteintech Group, Inc.,
cat. no. 22952-1-AP; osteocalcin (OCN), 1:1,000, Proteintech Group,
Inc., cat. no. 23418-1-AP; SFRP1, 1:1,000, Bioss, bs-1303R; lycogen
synthase kinase 3 beta (GSK3β), 1:5,000, Proteintech Group, Inc.,
cat. no. 82061-1-RR; α-tubulin, 1:5,000, Proteintech Group, Inc.,
cat. no. 11224-1-AP; HRP-conjugated Goat Anti-Rabbit lgG(H+L),
1:10,000, Proteintech Group, Inc., cat. no. SA 00001-2; and
HRP-conjugated Goat Anti-Mouse IgG(H+L), 1:10,000, Proteintech
Group, Inc., cat. no. SA 00001-1. GAPDH was used as an internal
reference protein in Fig. 5, while
α-tubulin was used in Fig. 2,
Fig. 3 and Fig. 4.
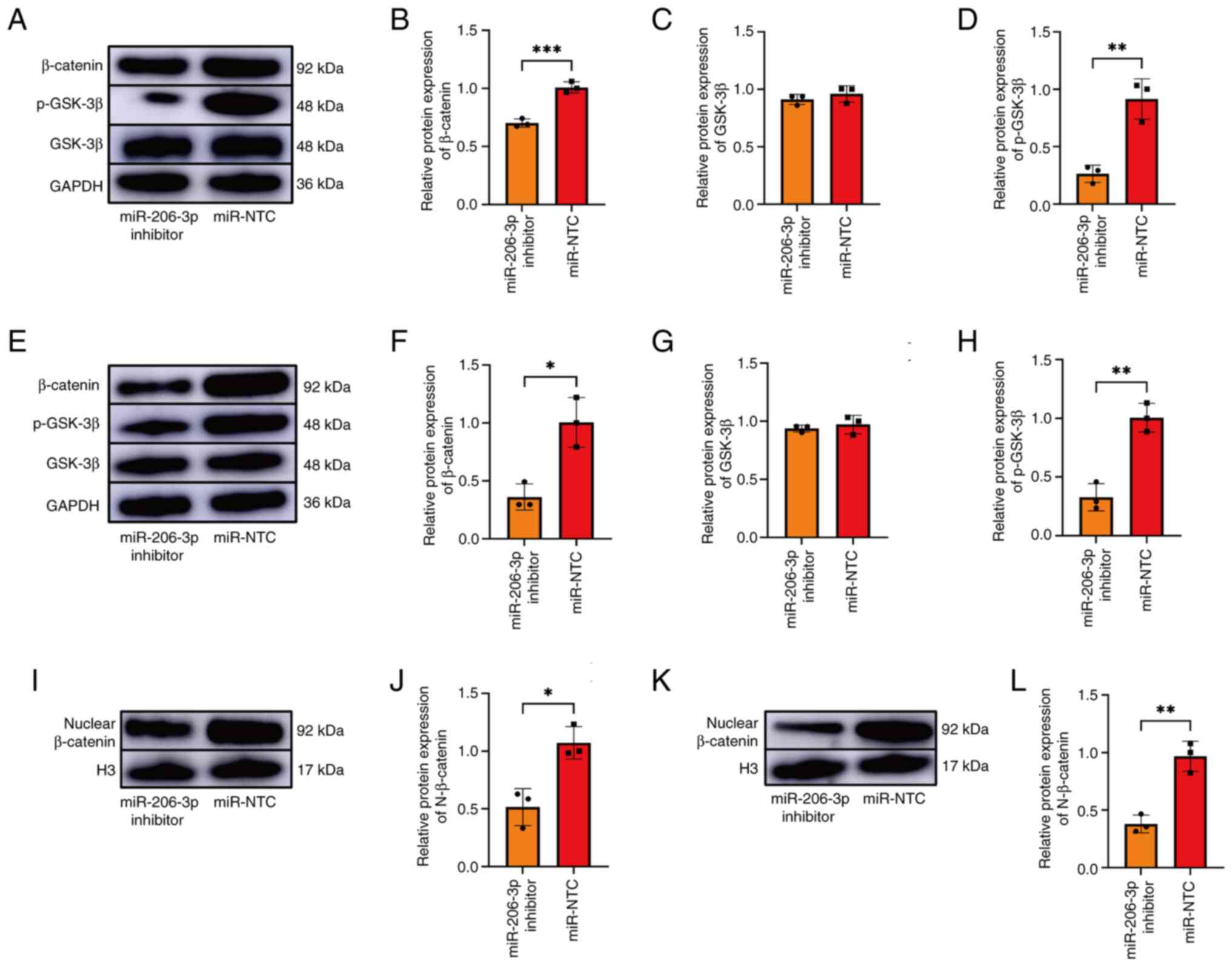 | Figure 5Expression of GSK-3β, p-GSK-3β,
β-catenin and nuclear β-catenin in the transfected BMSCs and L6
cells. (A-D) The expression of GSK-3β, p-GSK-3β, β-catenin, SFRP1
were detected by western blotting in the transfected group of BMSC.
(E-H) The expression of GSK-3β, p-GSK-3β, β-catenin and SFRP1 in
the transfected L6 cells was detected by western blotting. (I and
J) The expression of nuclear β-catenin in the transfected BMSCs. (K
and L) The expression of nuclear β-catenin in the transfected L6
cells. The presented gel blots have been cropped from different
parts of the gel and are separated with black lines. Data were
analyzed using Student's t-test (n=3). *P<0.05,
**P<0.01, ***P<0.001. GSK, glycogen
synthase kinase; p-, phosphorylated; BMSCs, bone marrow mesenchymal
stem cells; SFRP1, secreted frizzled-related protein 1; NTC,
negative control. |
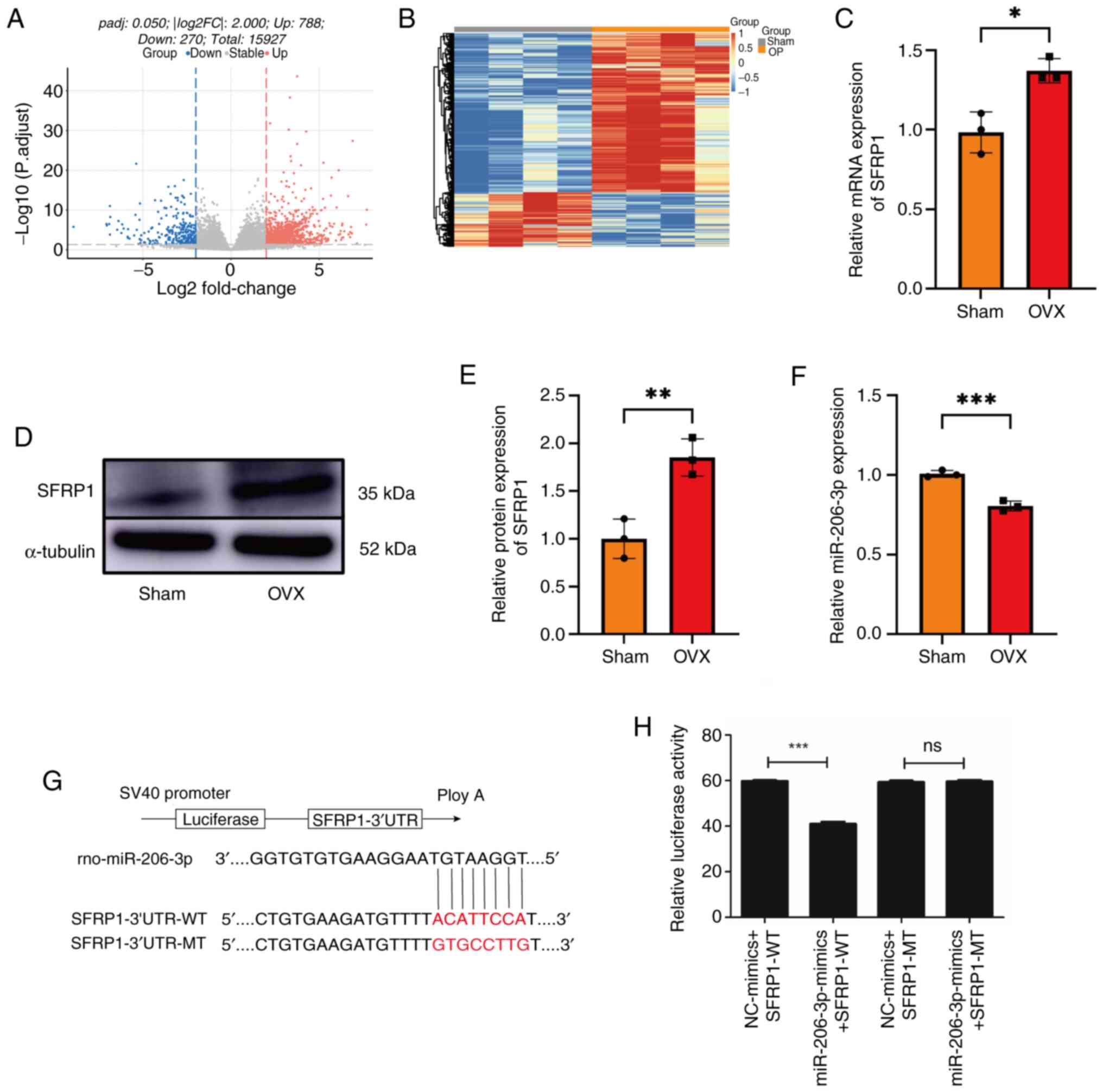 | Figure 2SFRP1 and miR-206-3p are expressed in
the BMSCs of the OVX and Sham groups. mRNA differential analysis in
the form a (A) volcano plot and (B) clustering heatmap. (C)
quantitative PCR revealed in BMSCs that compared with the Sham
group, the OVX group demonstrated an increase in the expression of
SFRP1. GAPDH was used as an internal reference in the PCR
experiments. (D) Western blotting results and (E) quantification in
BMSCs. Compared with the Sham group, the OVX group demonstrated an
increase in the expression of SFRP1. Gel blots cropped from
different parts of the gel and separated by black lines are
presented. α-tubulin was used as an internal reference in this
experiment. (F) PCR revealed that compared with the Sham group, the
OVX group demonstrated an increase in the expression of miR-206-3p.
U6 was used as an internal reference in the PCR experiments. (G,H)
Dual-luciferase reporter assay demonstrated that miR-206-3p could
bind to SFRP1. Data were analyzed using Student's t-test (n=3).
*P<0.05, **P<0.01,
***P<0.001. SFRP1, secreted frizzled-related protein
1; miR, microRNA; BMSCs, bone marrow mesenchymal stem cells; OVX,
ovariectomized. |
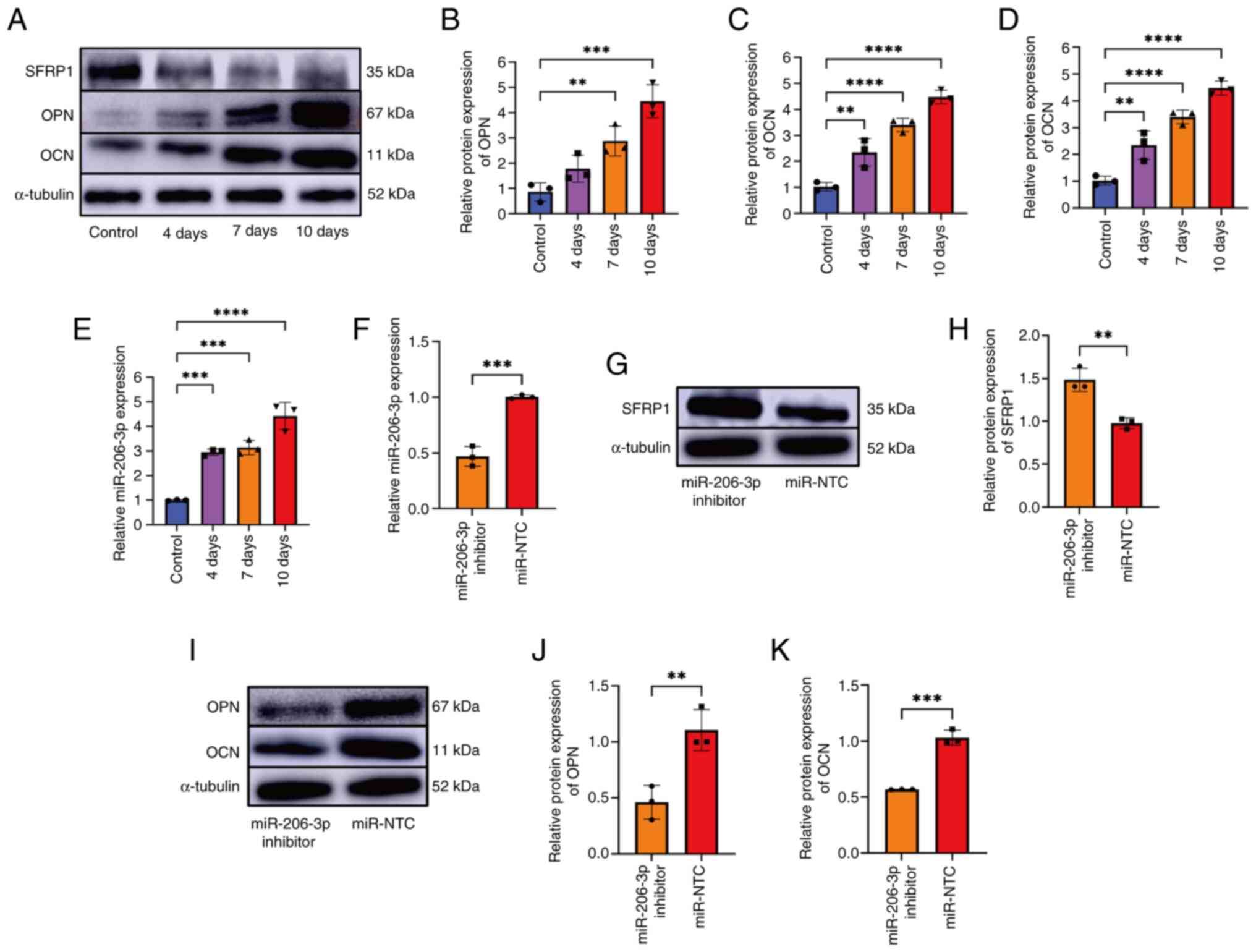 | Figure 3Detection of the osteogenic ability
of miR-206-3p inhibitor-transfected BMSCs. (A-D) Expression levels
of SFRP1, OPN and OCN at 0, 4, 7 and 10 days during osteogenic
differentiation. (E) PCR revealed that the expression of miR-206-3p
was upregulated at 0, 4, 7 and 10 days, respectively. (F) PCR
revealed that compared with the miR-NTC group, the miR-206-3p
inhibitor group demonstrated a reduction in the expression of
miR-206-3p. U6 was used as an internal reference in PCR. (G and H)
The expression level of SFRP1 increased following the silencing of
miR-206-3p. (I-K) The expression of OPN and OCN were detected using
western blotting. The presented gel blots have been cropped from
different parts of the gel and are separated with black lines. Data
were analyzed using the Welch and Brown-Forsy versions of one-way
ANOVA (B-E) or Student's t-test (F-K) and compared using the
χ2 test. **P<0.01,
***P<0.001, ****P<0.0001. miR,
microRNA; BMSCs, bone marrow mesenchymal stem cells; SFRP1,
secreted frizzled-related protein 1; OPN, osteopontin; OCN,
osteocalcin; NTC, negative control. |
Plasmid transfection
The miR-206-3p inhibitor and its negative control
(miR-NTC) were purchased from Shanghai GeneChem Co., Ltd. The cells
were transfected with Lipofectamine® 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. The transfected nucleic acid concentration was 50 nm.
Transfection was performed for 20 min at room temperature to form
complexes and then cells were incubated at 37˚C in a 5%
CO2 incubator. The transfected cells were subjected to
follow-up experiments 24-48 h after transfection. Negative control
sequence: TTCTCCGAACGTGTCACGT.
Dual-luciferase reporter assay
A total of 293 cells were cultured to a density of
2x104 cells/well in 96-well culture plates. The cells
were transfected with dual-luciferase reporter (DLR) construct p1
(0.2 µg) or co-transfected with DLR construct p2 (0.2 µg) and the
internal control vector pRL-TK, pRL-SV40, or pRL-CMV (Promega
Corporation) at a ratio of 20:1 using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 5 h following
transfection, the transfection medium was removed and replenished
with a medium containing 6 µM of curcumin (MilliporeSigma)
solubilized in 100% dimethylsulfoxide (MilliporeSigma). At 48 h
following transfection, luciferase activity was measured using a
DLR assay (Promega Corporation). Renilla luciferase activity was
normalized to firefly luciferase activity in cells transfected with
the DLR construct p1 and firefly luciferase activity was normalized
to Renilla luciferase activity in cells co-transfected with DLR
construct p2 and the control vector.
TargetScan
The candidate target miRNA of SFRP1 was predicted
using TargetScan (http://www.targetscan.org/).
Statistical analysis
Experimental data were expressed as means ± standard
deviation. Statistical analysis was performed using SPSS (version
19.0; IBM Corp.). Data were analyzed using Welch and Brown-Forsythe
versions of one-way ANOVA or Student's t-test and compared using
the χ2 test. All experiments were repeated at least
three times. P<0.05 was considered to indicate a statistically
significant difference.
Results
Establishment and identification of
osteoporosis model in the OVX group
An osteoporosis rat model was established by
ovariectomy and the degree of osteoporosis was evaluated using
histological staining. Quantitative analysis of micro-CT data
demonstrated that BMD, BV/TV and TbTh were lower in the OVX group
than in the Sham group (Figs. 1A-C
and S1). Furthermore,
immunohistochemical staining of the rat femurs demonstrated an
imbalance in bone metabolism in the OVX group. The resultant
reduction in bone mineral density and damage to bone
microarchitecture confirmed the presence of osteoporosis in the OVX
group (Fig. 1D-G).
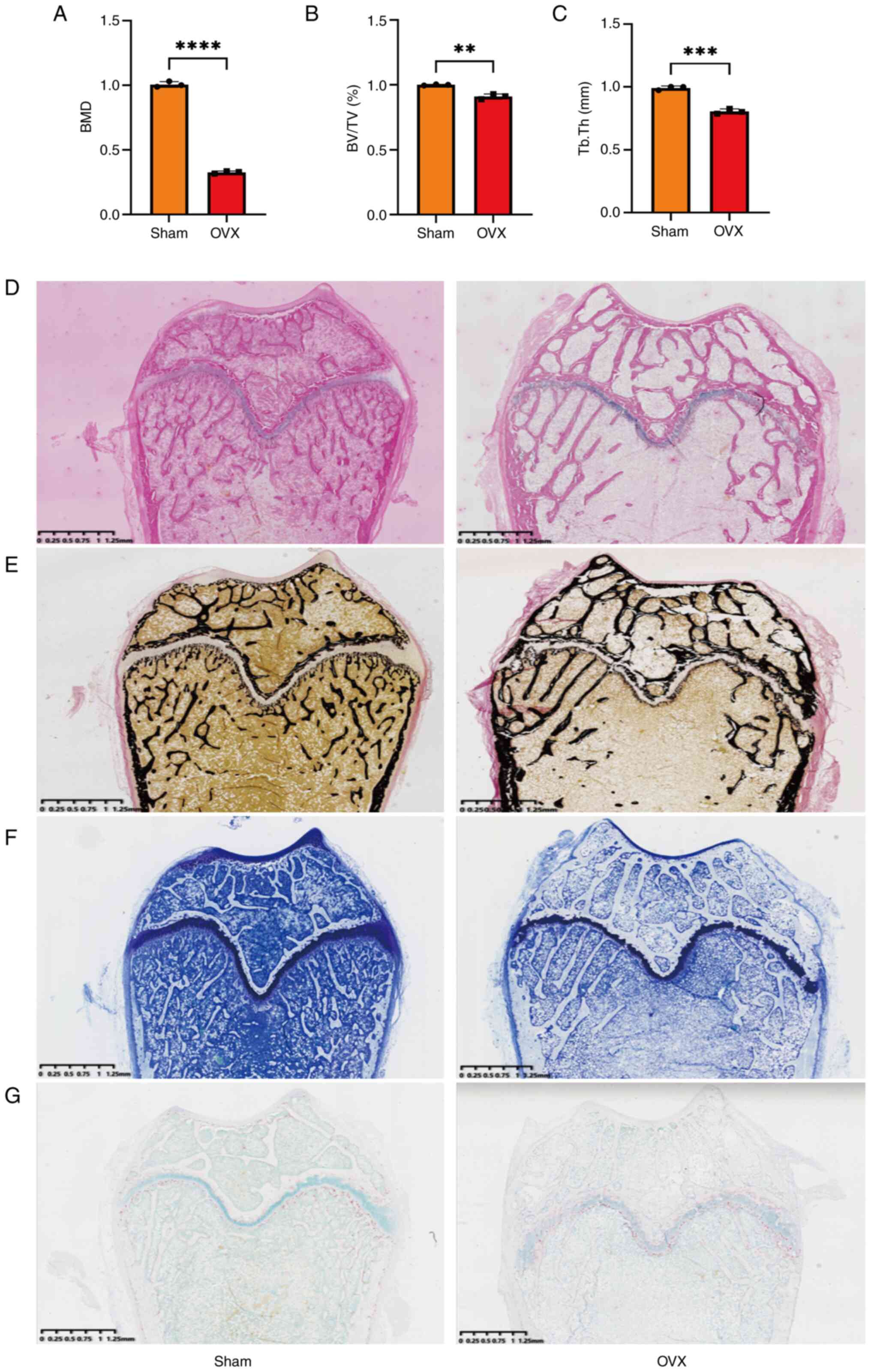 | Figure 1Quantitative analysis of micro-CT
data and histological staining results. Quantitative analysis of
(A) BMD, (B) BV/TV and (C) TbTh. Results of (D) H&E, (E) Von
Kossa, (F) toluidine blue and (G) TRAP staining. Data were analyzed
using Student's t-test (n=3). **P<0.01,
***P<0.001, ****P<0.0001. CT, computed
tomography; BMD, bone mineral density; BV/TV, bone volume/tissue
volume ratio; TbTh, trabecular thickness; H&E, hematoxylin and
eosin; TRAP, tartrate-resistant acidic phosphatase. |
Isolation, culture, identification and
three-line differentiation of the BMSCs from the rat bone
marrow
The differentiation of the extracted BMSCs into
osteogenic, lipogenic and chondrogenic cells was confirmed using
alizarin red, oil red O and Alcian blue staining, respectively
(Fig. S2). Flow cytometry
demonstrated that BMSCs displaying CD29+,
CD44+, CD45- and CD90+ had a
purity of >98.6% (Fig. S3).
This indicates that the cells extracted using the method of the
present study were rat BMSCs. Western blotting demonstrated that
the expression of OPN and OCN were significantly lower in the BMSC
of the OVX group than in the BMSC of the Sham group (Fig. S4). This indicated that the
osteogenic differentiation ability of BMSCs in the OVX group was
lower than that in the Sham group.
SFRP1 increases in the OVX group's
BMSCs and decreases during osteogenesis
RNA sequencing revealed that, compared with the Sham
group, 788 mRNAs were upregulated and 270 mRNAs were downregulated
in the OVX group (Table SI).
Fig. 2A and B shows the mRNA differential analysis
volcano plot and clustering heatmap. Among the markedly upregulated
mRNA, SFRP1 negatively regulates osteoblasts and osteoblast
functions and it plays an important role in maintaining bone
stability. Therefore, SFRP1 was selected as the research object.
Results of the RT-qPCR and western blotting for the detection of
SFRP1 expression were consistent with those of RNA sequencing
(Fig. 2C-E). TargetScan (http://www.targetscan.org/) predicted that miR-206-3p
directly targeted the 3'UTR region of SFRP1 (Fig. S5). RT-qPCR revealed that miR-206-3p
expression was downregulated in the BMSCs of the OVX group and the
expression of miR-206-3p was negatively associated with that of
SFRP1 (Fig. 2F). DLR assay was
performed to detect the binding sites between miR-206-3p and SFRP1
(Fig. 2G and H). These findings indicated that
miR-206-3p may directly target SFRP1 mRNA.
miR-206-3p regulates the osteogenic
differentiation of BMSCs by targeting SFRP1
The expression levels of miR-206-3p during
osteogenic differentiation after transfection were measured at 4, 7
and 10 days (Fig. 3A-E). The
expression level of miR-206-3p increased markedly during osteogenic
differentiation. To explore the function of miR-206-3p during
osteogenic differentiation of the BMSCs, the BMSCs were transfected
with an miR-206-3p inhibitor or a negative control (miR-NTC).
Subsequently, RT-qPCR demonstrated that the expression of
miR-206-3p was significantly inhibited following silencing
(Fig. 3F). Furthermore, the SFRP1
expression increased after the miR-206-3p was silenced (Fig. 3G and H). After the transfected BMSCs were
induced to undergo osteogenic differentiation for 7 days,
osteoblast-specific markers (OPN and OCN) were detected by western
blotting. The results demonstrated that the osteogenic ability of
BMSC in the miR-206-3p inhibition group was weaker compared with
the control group (Fig. 3I-K).
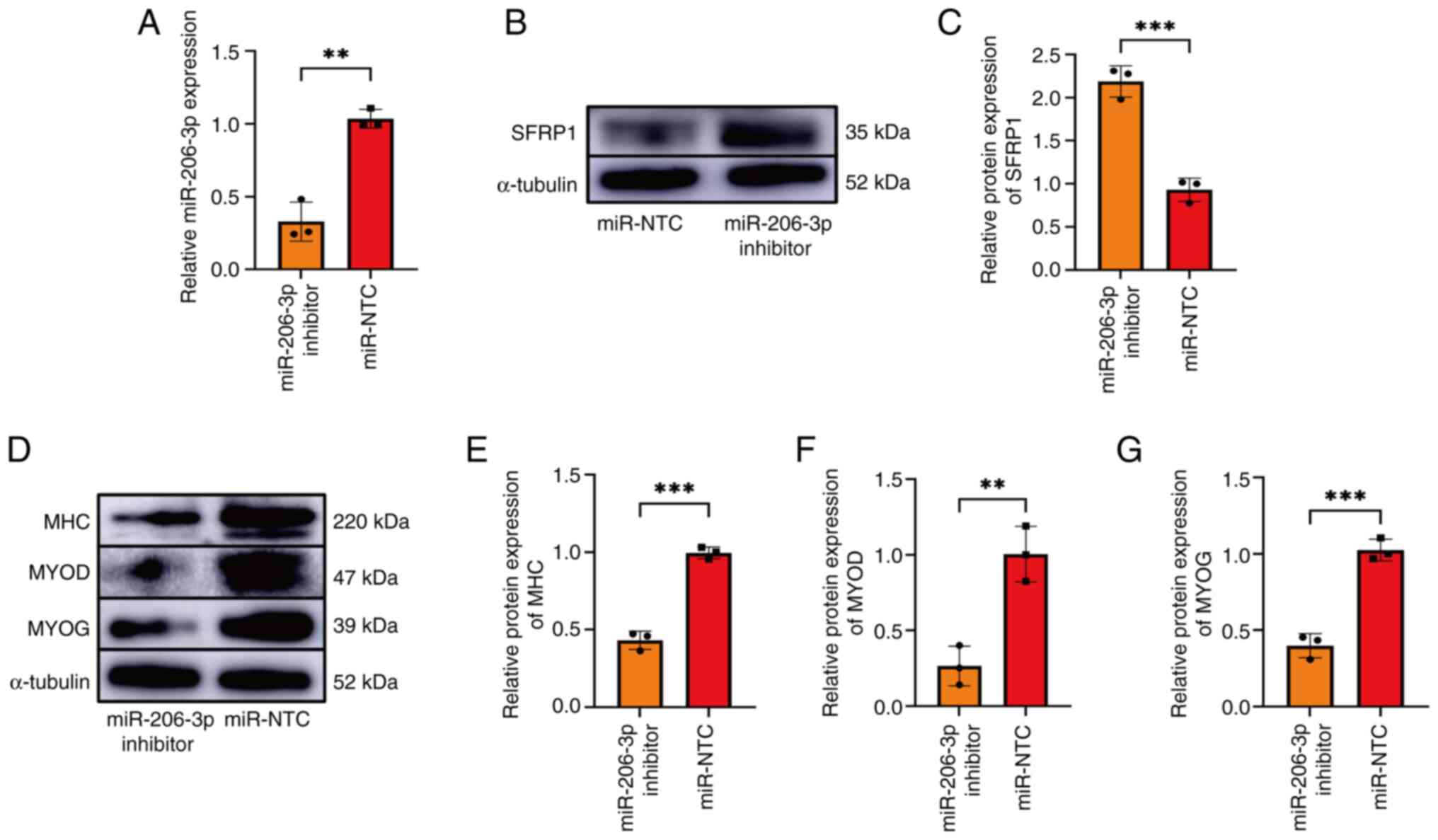
miR-206-3p regulates the myogenic
differentiation of L6 cells by targeting SFRP1
To investigate the role of miR-206-3p in myoblast
differentiation, L6 cells were transfected with an miR-206-3p
inhibitor or miR-NTC at 70% density. Thereafter, RT-qPCR revealed
that compared with the control group, miR-206-3p expression was
significantly inhibited in the miR-206-3p inhibitor group (Fig. 4A). Furthermore, an increase in SFRP1
expression in the miR-206-3p inhibition group was detected
(Fig. 4B and C). Following the induction of myogenic
differentiation in the transfected L6 cells, the expression of
myogenic differentiation 1 decreased in the miR-206-3p inhibition
group. Furthermore, the myogenin and myosin heavy chain expression
levels decreased in the miR-206-3p inhibition group (Fig. 4D-G).
miR-206-3p regulates osteogenic and
myogenic differentiation via the classical β-catenin signaling
pathway
As SFRP1 is the main antagonist of β-catenin
signaling, the expression of GSK-3β, p-GSK-3β, β-catenin and
nuclear β-catenin was detected after transfecting the BMSC and L6
cells. Compared with the control group, the protein levels of
p-GSK-3β and β-catenin were lower (Fig.
5A-D) and nuclear β-catenin was significantly downregulated
(Fig. 5I and J) during the differentiation process in
the miR-206-3p inhibition group. During the myogenesis of L6 cells,
the expression levels of p-GSK-3β, β-catenin (Fig. 5E-H) and nuclear β-catenin decreased
in the miR-206-3p inhibition group (Fig. 5K-L). There was no difference in the
expression of GSK-3β between the inhibition and control groups.
Discussion
miR-206-3p is expressed in a number of cells, such
as skeletal muscle cells, heart cells mesenchymal stem cells and
brown adipocytes, but not in white adipocytes (24,25). A
study in tumor cells proved the specific expression of miR-206-3p
and inhibit cell migration and invasion in various cancers
(26). Bone mineral density is
regulated by osteoclasts and osteoblasts in the bone
microenvironment within the basic multicellular unit, as well as
osteocytes, bone lining cells, osteoma cells and vascular
endothelial cells (27). For
example, miR-206-3p promotes osteogenic differentiation by
negatively regulating BMP3 expression in osteoblasts and can be
transferred to osteoclasts to inhibit osteoclast bone resorption
(28,29). Previous studies have found that a
number of miRNAs have multiple roles in bone diseases; miRNAs play
important regulatory roles in bone formation, bone healing and
osteoblastogenesis and can regulate osteoclastogenesis or enter
them to regulate cell functions (30,31).
Mechanical and chemical crosstalk exist between
muscles and bones (32).
Furthermore, some biochemical pathways, such as Dickkopf-1 and
IL-6, affect both muscle and bone (32,33).
The present study demonstrated that miR-206-3p affected muscle and
bone differentiation by targeting SFRP1 and acting via the
Wnt/β-catenin signaling pathway.
The present study performed transcriptome sequencing
of the BMSCs in both groups, which revealed that SFRP1 was markedly
elevated in the osteoporosis group. The SFRP gene family has five
members (SFRP1-SFRP5) in the human and mouse genomes and it plays
an important role in the development of cancer and inflammatory
diseases (34). SFRP1 is involved
in bone metabolism and homeostasis. For example, overexpression of
SFRP1 in osteoblasts inhibits the parathyroid hormone-induced bone
formation as well as the differentiation and maturation of
osteoblasts (35). Simultaneously,
SFRP1 plays a role in muscle proliferation and differentiation. For
example, the addition of recombinant Sfrp1 to C2C12 or satellite
cells inhibits muscle duct formation (36). Therefore, identifying the regulatory
factors of the upstream and downstream pathways of SFRP1 may help
determine the therapeutic targets of osteosarcopenia.
The present study hypothesized that miR-206-3p may
bind to SFRP1 and found that SFRP1 and miR-206-3p were negatively
associated in the BMSCs of the OVX and Sham groups. Subsequently, a
DLR assay and transfection experiment indicated that miR-206-3p may
directly target SFRP1 and affect the osteogenic differentiation of
BMSC. This indicated that SFRP1 plays an important role in the
occurrence and development of osteoporosis. These findings are
consistent with a previous study in which miR-206-3p was
demonstrated as a positive regulator of muscle differentiation and
SFRP1 has inhibited myoblast differentiation (37). The present study further
demonstrated that miR-206-3p affected the Wnt pathway by negatively
regulating SFRP1, which promoted the myogenic differentiation of L6
cells.
The SFRPs family includes Wnt signaling antagonists
that play a role in various diseases by regulating Wnt signaling
(38). SFRP1 antagonizes Wnt
signaling by competitively binding to the cysteine-rich Wnt
receptor frizzled (39). The
Wnt/β-catenin signaling pathway is closely associated with bone and
muscle (40). The Wnt/β-catenin
signaling pathway can regulate bone formation and absorption,
muscle fiber formation and the activation of adult muscle stem
cells (41-43).
The typical Wnt/β-catenin signaling pathway inhibits the expression
of GSK3β in BMSCs via the low density lipoprotein receptor-related
protein co-receptor and it can promote the nuclear translocation of
β-catenin in the cytoplasm. An increase in the β-catenin-T-cell
factor (TCF) 4/lymphoid enhancer factor-1 (LEF1) complex in the
nucleus activates the transcription of downstream osteogenic genes
(44). In skeletal muscle,
activation of Wnt signaling leads to an increase in the nuclear
translocation of β-catenin. This in turn increases the interaction
of β-catenin with TCF/LEF1 transcription factors, which promotes
the expression of myogenesis-related target genes (45). The present study found that the
protein levels of p-GSK-3β and nuclear β-catenin were significantly
decreased in BMSCs treated with the miR-206-3p inhibitor. However,
in L6 cells treated with the miR-206-3p inhibitor, the protein
levels of p-GSK-3β and nuclear β-catenin were significantly
decreased. These findings indicate that miR-206-3p promotes
β-catenin signaling by negatively regulating SFRP1, which affects
bone and muscle differentiation. This result is consistent with a
previous study (40).
The present study found that mir-206-3p and SFRP1
has indeed been reported in the literature, but it was the first
time to study and report the regulatory relationship between
mir-206-3p, SFRP1 and β-catenin in osteoporosis and sarcopenia at
the same time. The present study has several limitations. First, it
only focused on SFRP1 and its upstream target, miR-206-3p, after
obtaining multiple differential genes via transcriptome sequencing.
However, miR-206-3p may simultaneously affect other target genes
and SFRP1 may be regulated by other miRNAs. Thus, a comprehensive
study on the biological functions of miR-206-3p and SFRP may
provide a deeper understanding of the development of diseases.
Second, the present study used rat BMSCs and the verified myoblasts
L6 cells, which do not fully represent the processes of bone and
muscle formation in humans. Third, rats in the osteoporosis model
group were ovariectomized to mimic postmenopausal osteoporosis in
humans. The present study results need to be verified in studies
with larger samples of human patients with osteoporosis. Mouse
models of accelerated aging have been shown to be important animal
models for studying musculoskeletal aging and related disorders
(46,47). In the future, studies will model
animals in osteosarcopenia and continue to explore the regulatory
pathways of miR-206-3p and SFRP1 genes in osteosarcopenia.
In conclusion, miR-206-3p affects the Wnt/β-catenin
signaling pathway by functionally targeting SFRP1. This in turn
regulates bone and muscle differentiation. Therefore, miR-206-3p
and SFRP1 may be novel targets for the treatment of
osteosarcopenia.
Supplementary Material
Micro-CT images in the (A) Sham group
and (B) OVX group. CT, computed tomography; OVX,
ovariectomized.
Differentiation of bone marrow
mesenchymal stem cells into three cell lines via osteogenesis,
lipogenesis and chondrogenesis. (A) Bone Marrow Stromal Cells. (B)
Alizarin Red staining. (C) Oil Red O staining. (D) Alixin Blue
staining. Scale bars represent (A, C and D) 200 μm and (B)
100 μm.
Flow cytometry identification of
BMSCs. BMSCs, bone marrow mesenchymal stem cells.
Detection of the ability of the cells
to differentiate into bone in the OVX and Sham groups. (A) The
quantitative western blotting results demonstrate that the
expression of (B) OPN and (C) OCN were significantly lower in the
OVX than in the Sham group. Data were analyzed using Student's
t-test (n=3). **P<0.01, ***P<0.001.
OVX, ovariectomized; OPN, osteopontin; OCN, osteocalcin.
TargetScan website (http://www.targetscan.org/) prediction that miR-206-3p
directly targets SFRP1 3'UTR. miR, microRNA; SFRP1, secreted
frizzled-related protein 1.
Differentially expressed genes in the
normal and OVX group.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the National Natural
Science Foundation of China (grant no. 82172442), the Yunnan
Provincial Department of Science and Technology Social Development
Special Project (grant no. 202403AC100003), Major Science and
Technology Special Project of Yunnan Provincial Science and
Technology Program (grant no. 202102AA310042) and Opening Subjects
of Clinical Medical Research Center of the First People's Hospital
of Yunnan Province (grant no. 2022YJZX-GK17).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
CY and ZL jointly conceptualized this study,
designed and conducted experiments and wrote and edited the
manuscript. DY conceived this study, designed and conducted
experiments and edited the manuscript. YL wrote the manuscript and
contributed to the analysis and interpretation of the data. JF and
YZ designed and implemented the experiment, and wrote the
manuscript. ZP and SL made critical revisions to the manuscript. CY
and SL confirm the authenticity of all raw data. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental animal study protocol was approved
by the Animal Care and Use Committee of the Kunming Medical
University, Kunming, China (approval no. kmmu20230916). All animal
experiments and handling were performed in accordance with the
Guide for the Care and Use of Laboratory Animals published by the
National Institutes of Health (NIH Publication No. 8523, revised
1985) (22).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kirk B, Zanker J and Duque G:
Osteosarcopenia: Epidemiology, diagnosis and treatment-facts and
numbers. J Cachexia Sarcopenia Muscle. 11:609–618. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Li GH, Cheung CL, Tan KC, Kung AW, Kwok
TC, Lau WC, Wong JS, Hsu WWQ, Fang C and Wong IC: Development and
validation of sex-specific hip fracture prediction models using
electronic health records: a retrospective, population-based cohort
study. EClinicalMedicine. 58(101876)2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yuan S and Larsson SC: Epidemiology of
sarcopenia: Prevalence, risk factors and consequences. Metabolism.
144(155533)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shimada H, Suzuki T, Doi T, Lee S,
Nakakubo S, Makino K and Arai H: Impact of osteosarcopenia on
disability and mortality among Japanese older adults. J Cachexia
Sarcopenia Muscle. 14:1107–1116. 2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Locquet M, Beaudart C, Reginster JY and
Bruyère O: Association between the decline in muscle health and the
decline in bone health in older individuals from the SarcoPhAge
cohort. Calcif Tissue Int. 104:273–284. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Clynes MA, Gregson CL, Bruyère O, Cooper C
and Dennison EM: Osteosarcopenia: where osteoporosis and sarcopenia
collide. Rheumatology (Oxford). 60:529–537. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Inoue T, Maeda K, Nagano A, Shimizu A,
Ueshima J, Murotani K, Sato K, Hotta K, Morishita S and Tsubaki A:
Related factors and clinical outcomes of osteosarcopenia: A
narrative review. Nutrients. 13(291)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Prockop DJ: Marrow stromal cells as stem
cells for nonhematopoietic tissues. Science. 276:71–74.
1997.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Loh KM, Chen A, Koh PW, Deng TZ, Sinha R,
Tsai JM, Barkal AA, Shen KY, Jain R, Morganti RM, et al: Mapping
the pairwise choices leading from pluripotency to human bone, heart
and other mesoderm cell types. Cell. 166:451–467. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yin L, Yang Z, Wu Y, Denslin V, Yu CC, Tee
CA, Lim CT, Han J and Lee EH: Label-free separation of mesenchymal
stem cell subpopulations with distinct differentiation potencies
and paracrine effects. Biomaterials. 240(119881)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Feng Z, Jin M, Liang J, Kang J, Yang H,
Guo S and Sun X: Insight into the effect of biomaterials on
osteogenic differentiation of mesenchymal stem cells: A review from
a mitochondrial perspective. Acta Biomater. 164:1–14.
2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shivdasani RA: MicroRNAs: Regulators of
gene expression and cell differentiation. Blood. 108:3646–3653.
2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang E: MicroRNA, the putative molecular
control for mid-life decline. Ageing Res Rev. 6:1–11.
2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sun Y, Kuek V, Liu Y, Tickner J, Yuan Y,
Chen L, Zeng Z, Shao M, He W and Xu J: MiR-214 is an important
regulator of the musculoskeletal metabolism and disease. J Cell
Physiol. 234:231–245. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Fariyike B, Singleton Q, Hunter M, Hill
WD, Isales CM, Hamrick MW and Fulzele S: Role of MicroRNA-141 in
the aging musculoskeletal system: A current overview. Mech Ageing
Dev. 178:9–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Shang Q, Shen G, Chen G, Zhang Z, Yu X,
Zhao W, Zhang P, Chen H, Tang K, Yu F, et al: The emerging role of
miR-128 in musculoskeletal diseases. J Cell Physiol. 236:4231–4243.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sun D, Chen Y, Liu X, Huang G, Cheng G, Yu
C and Fang J: miR-34a-5p facilitates osteogenic differentiation of
bone marrow mesenchymal stem cells and modulates bone metabolism by
targeting HDAC1 and promoting ER-α transcription. Connect Tissue
Res. 64:126–138. 2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang C, Zhu M, Yang D, Hu X, Wen X and Liu
A: MiR-29a-3p inhibits proliferation and osteogenic differentiation
of human bone marrow mesenchymal stem cells via targeting FOXO3 and
repressing Wnt/β-catenin signaling in steroid-associated
osteonecrosis. Int J Stem Cells. 15:324–333. 2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang YC, Yao X, Ma M, Zhang H, Wang H,
Zhao L, Liu S, Sun C, Li P, Wu Y, et al: miR-130b inhibits
proliferation and promotes differentiation in myocytes via
targeting Sp1. J Mol Cell Biol. 13:422–432. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Yang L, Qi Q, Wang J, Song C, Wang Y, Chen
X, Chen H, Zhang C, Hu L and Fang X: MiR-452 regulates C2C12
myoblast proliferation and differentiation via targeting
ANGPT1. Front Genet. 12(640807)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
van Rooij E, Liu N and Olson EN: MicroRNAs
flex their muscles. Trends Genet. 24:159–166. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
U.S. Office of Science and Technology
Policy. Laboratory animal welfare; U.S. government principles for
the utilization and care of vertebrate animals used in testing,
research and training; notice. Fed Regist. 50:20864–20865.
1985.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Townley-Tilson WH, Callis TE and Wang D:
MicroRNAs 1, 133 and 206: critical factors of skeletal and cardiac
muscle development, function and disease. Int J Biochem Cell Biol.
42:1252–1255. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Walden TB, Timmons JA, Keller P,
Nedergaard J and Cannon B: Distinct expression of muscle-specific
microRNAs (myomirs) in brown adipocytes. J Cell Physiol.
218:444–449. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu K, Li J, Qi Y, Zhang C, Zhu D, Liu D
and Zhao S: SNHG14 confers gefitinib resistance in non-small cell
lung cancer by up-regulating ABCB1 via sponging miR-206-3p. Biomed
Pharmacother. 116(108995)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kular J, Tickner J, Chim SM and Xu J: An
overview of the regulation of bone remodelling at the cellular
level. Clin Biochem. 45:863–873. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guo S, Gu J, Ma J, Xu R, Wu Q, Meng L, Liu
H, Li L and Xu Y: GATA4-driven miR-206-3p signatures control
orofacial bone development by regulating osteogenic and
osteoclastic activity. Theranostics. 11:8379–8395. 2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang B, Yu P, Li T, Bian Y and Weng X:
MicroRNA expression in bone marrow mesenchymal stem cells from mice
with steroid-induced osteonecrosis of the femoral head. Mol Med
Rep. 12:7447–7454. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li H, Zhai Z, Qu X, Xu J, Qin A and Dai K:
MicroRNAs as potential targets for treatment of osteoclast-related
diseases. Curr Drug Targets. 19:422–431. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li D, Liu J, Guo B, Liang C, Dang L, Lu C,
He X, Cheung HY, Xu L, Lu C, et al: Osteoclast-derived exosomal
miR-214-3p inhibits osteoblastic bone formation. Nat Commun.
7(10872)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tagliaferri C, Wittrant Y, Davicco MJ,
Walrand S and Coxam V: Muscle and bone, two interconnected tissues.
Ageing Res Rev. 21:55–70. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Bakker AD and Jaspers RT: IL-6 and IGF-1
signaling within and between muscle and bone: How important is the
mTOR pathway for bone metabolism? Curr Osteoporos Rep. 13:131–139.
2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
van Loon K, Huijbers EJM and Griffioen AW:
Secreted frizzled-related protein 2: A key player in noncanonical
Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev.
40:191–203. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bodine PVN, Seestaller-Wehr L, Kharode YP,
Bex FJ and Komm BS: Bone anabolic effects of parathyroid hormone
are blunted by deletion of the Wnt antagonist secreted
frizzled-related protein-1. J Cell Physiol. 210:352–357.
2007.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Descamps S, Arzouk H, Bacou F, Bernardi H,
Fedon Y, Gay S, Reyne Y, Rossano B and Levin J: Inhibition of
myoblast differentiation by Sfrp1 and Sfrp2. Cell Tissue Res.
332:299–306. 2008.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yang Y, Sun W, Wang R, Lei C, Zhou R, Tang
Z and Li K: Wnt antagonist, secreted frizzled-related protein 1, is
involved in prenatal skeletal muscle development and is a target of
miRNA-1/206 in pigs. BMC Mol Biol. 16(4)2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jiang P, Wei K, Chang C, Zhao J, Zhang R,
Xu L, Jin Y, Xu L, Shi Y, Guo S, et al: SFRP1 negatively modulates
pyroptosis of fibroblast-like synoviocytes in rheumatoid arthritis:
A review. front immunol. 13(903475)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Bodine PVN and Komm BS: Wnt signaling and
osteoblastogenesis. Rev Endocr Metab Disord. 7:33–39.
2006.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yang YJ and Kim DJ: An overview of the
molecular mechanisms contributing to musculoskeletal disorders in
chronic liver disease: Osteoporosis, sarcopenia and osteoporotic
sarcopenia. Int J Mol Sci. 22(2604)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Cossu G and Borello U: Wnt signaling and
the activation of myogenesis in mammals. EMBO J. 18:6867–6872.
1999.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hoeppner LH, Secreto FJ and Westendorf JJ:
Wnt signaling as a therapeutic target for bone diseases. Expert
Opin Ther Targets. 13:485–496. 2009.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Boudin E, Fijalkowski I, Piters E and Van
Hul W: The role of extracellular modulators of canonical Wnt
signaling in bone metabolism and diseases. Semin Arthritis Rheum.
43:220–240. 2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Law SM and Zheng JJ: Premise and peril of
Wnt signaling activation through GSK-3β inhibition. iScience.
25(104159)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Shevtsov SP, Haq S and Force T: Activation
of beta-catenin signaling pathways by classical G-protein-coupled
receptors: Mechanisms and consequences in cycling and non-cycling
cells. Cell Cycle. 5:2295–2300. 2006.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yılmaz D, Mathavan N, Wehrle E, Kuhn GA
and Müller R: Mouse models of accelerated aging in musculoskeletal
research for assessing frailty, sarcopenia and osteoporosis-A
review. Ageing Res Rev. 93(102118)2024.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang N, Chow SKH, Leung KS, Lee HH and
Cheung WH: An animal model of co-existing sarcopenia and
osteoporotic fracture in senescence accelerated mouse prone 8
(SAMP8). Exp Gerontol. 97:1–8. 2017.PubMed/NCBI View Article : Google Scholar
|