Introduction
In China, congenital heart disease (CHD) is the most
prevalent type of birth defect, accounting for ~30% of all
congenital birth defects in 2023(1). It affects 17-32‰ perinatal births
(1). The incidence of prenatal CHD
is also increasing. CHD incidence in 2020 was 4.3 times that in
2010(1). CHD is divided into
non-syndromic and syndromic phenotypes. Approximately 1/3 cases
have definite genetic or chromosomal abnormalities, of which ~23%
of CHD cases have chromosomal aneuploidy or copy number variation,
and ~10% have new mutations or genetic mutations inherited from
their parents (2,3). The pathogenesis is unclear in most
cases, especially in non-syndromic phenotypes. It is well accepted
that CHD is attributed to the co-effects of genetics, epigenetics
and environment (2-5).
Nutritional imbalances, drinking, smoking, drugs, hypoxia and other
environmental factors can increase the incidence of CHD (4,5).
Prenatal diagnosis of CHD mainly relies on ultrasonography. Hence,
it cannot be discovered until the defect develops to a stage that
can be measured using ultrasound (6). Therefore, additional molecular markers
are required for early screening.
Metabolites are the end products of all processes
occurring in cells. Metabolomics provides methods for identifying
changes in metabolite profiles to support early biomarker
discovery, disease diagnosis and treatment. Metabolomics, which is
performed on a high-throughput platform, refers to the screening,
identification, quantification and characterization of biochemicals
that are <1,800 Da and are involved in various biological
pathways (7). Metabolomics
represents a bridge between the genome and phenotype and serves a
connecting role in the transmission of biological information. The
disease or physiological state can be better reflected in the
metabolomic profile than in the transcriptome or proteome profile
(8). Effective small changes in
gene and protein expression can be amplified by metabolites, making
detection easier. Metabolomics has a simple metabolite information
base, which is less complex than others (genomics, transcriptomics
and proteomics). Several metabolomics studies have focused on fetal
CHD, using maternal serum and urine and amniotic fluid samples
(7-9).
To the best of our knowledge, amniotic fluid cells have not been
used for metabonomic screening of CHD. The present study used
cultured amniotic fluid cells from patients carrying fetuses with
CHD for untargeted metabolomics screening to discover the potential
pathogenesis and screen new molecular markers.
Materials and methods
Sample collection and preparation
The cultured amniotic fluid cells [healthy controls
(no chromosomal or other disease), n=24; CHD cases (fetus with
ventricular septal defects), n=24] were obtained from singleton
pregnant patients who underwent amniocentesis during gestational
weeks 20.0-26.3. The patients were aged from 22 to 43 years old.
Inclusion criteria: Patients were willing to have amniocentesis. In
CHD group, the fetus was diagnosed as CHD by ultrasound. The
indication for amniocentesis in the control group was either older
than 35 years, a high risk in serum Down's screening or a high risk
of non-invasive prenatal screening. Exclusion criteria: The fetus
was affected with abnormalities chromosomes or abnormal copy number
variation. The samples were collected in Shengjing Hospital of
China Medical University (Shenyang, China) from April 2022 to
December 2022. All participants provided written informed consent,
and the study was approved by the Ethics Committee of Shengjing
Hospital of China Medical University (approval no. 2022PS307K). The
sample collection and experimental procedures conformed to the
Declaration of Helsinki. All patients were non-smokers and
non-alcoholics. The samples were matched for maternal and
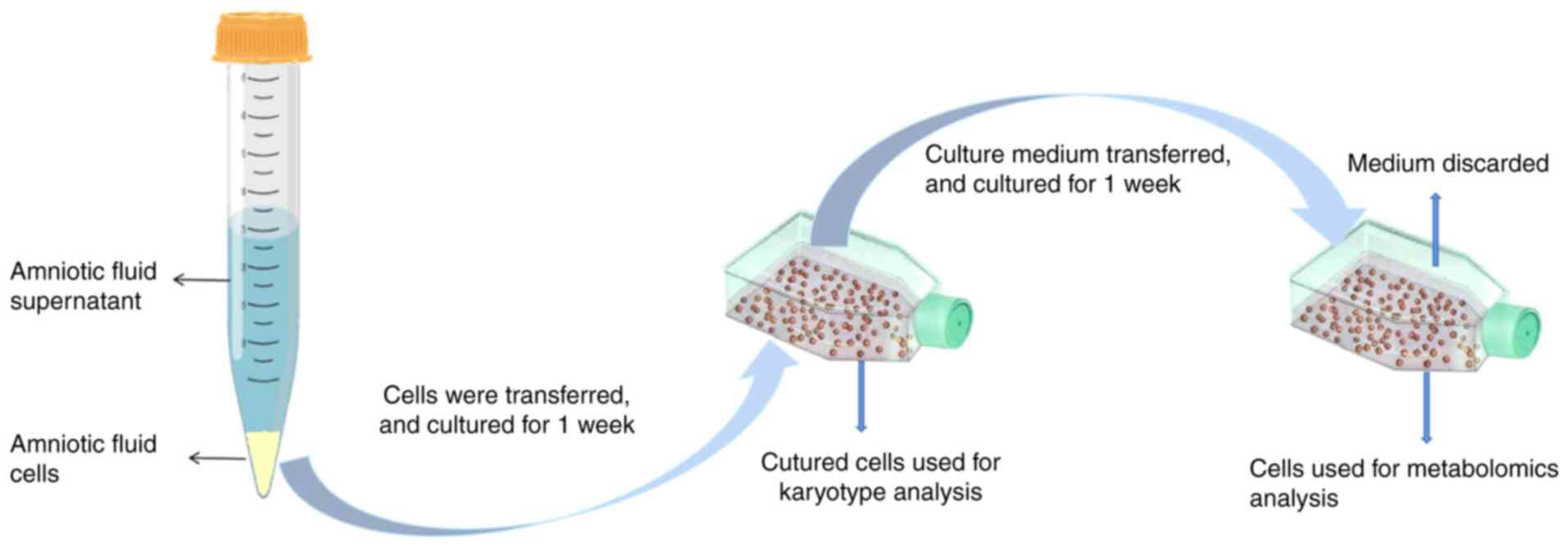
gestational age and sex of the fetus (Table I). The amniotic fluid cells were
cultured with amniotic fluid medium (FUJIFILM Biosciences; cat. no.
99473210805) for 2 weeks in 5% CO2 at 37˚C (Fig. 1).
 | Table IClinical information for amniotic
fluid cells. |
Table I
Clinical information for amniotic
fluid cells.
| Group | n | Mean maternal age,
years | Mean gestational
age, weeks | Fetus sex
(male/female) |
|---|
| Control | 24 | 31.6±5.45 | 22.4±1.88 | 11/13 |
| CHD | 24 | 30.9±4.65 | 25.0±1.35 | 12/12 |
The metabolites were extracted from the cell residue
with 1 ml precooled methanol/acetonitrile/water (v/v, 2:2:1) under
sonication every 10 min for 1 h in ice baths. The mixture was
incubated at -20˚C for 1 h, followed by centrifugation at 14,000 x
g at 4˚C for 20 min, and the supernatant was transferred to a
sampling vial. To ensure data quality for metabolic profiling,
quality control (QC) samples were prepared by pooling aliquots
representative of all samples for data normalization. The
supernatant was evaporated using a high-speed vacuum-concentration
centrifuge, 14,000 x g and 4˚C about for 1 h. Dried extracts were
redissolved in 50% acetonitrile. Each sample was centrifuged at
14,000 x g and 4˚C for 20 min, and the supernatant was used for
analysis.
Ultra-high performance liquid
chromatography (UHPLC)-tandem mass spectrometry (MS/MS)
analysis
Metabolomic profiling was performed on UPLC-MS/MS
system (1290 Infinity LC, Agilent Technologies, Inc.) and TripleTOF
5600 (SCIEX). Then, 5 µl Samples were separated with a 2.1x100.0 mm
ACQUIY UPLC BEH 1.7 µM column (Waters China Ltd.). The flow rate
was 0.5 ml/min and the mobile phase contained A (25 mM each
ammonium acetate and ammonium hydroxide in water) and B
(acetonitrile). The gradient was 95% B for 0.5 min and was linearly
reduced to 65% in 6.5 min, 40% in 2.0 min (maintained for 1.0 min)
and increased to 95% for 1.1 min, employing a 5-min
re-equilibration period. Both electrospray ionization (ESI)
positive and negative modes were applied for MS data acquisition.
The ESI source conditions were as follows: Ion source gas 1 and 2,
60; curtain gas, 30; source temperature, 600˚C; ion spray voltage
floating, ±5,500 V. In MS acquisition, the instrument was set to
acquire data over the m/z range of 60-1,200 Da, and the
accumulation time for MS scanning was 0.15 sec/spectra. In the auto
MS/MS acquisition, the instrument was set to acquire data over the
m/z range of 25-1,200 Da, and the accumulation time for the
production scan was 0.03 sec/spectra. Blank (50% acetonitrile in
water) and QC samples were injected every 12 samples during
acquisition. All the samples were run in one batch for the positive
and negative ion mode. Each sample was detected and analyzed using
UPLC-MS/MS, and two original files (positive and negative ion mode)
were obtained. MS total ion chromatogram (TIC) flow diagram was
generated.
Data preprocessing and filtering
Data processing and analysis were as previously
described with minor amendments (10). Raw MS data were converted to MzXML
files using ProteoWizard MS Convert and processed using XCMS for
feature detection, retention time correction and alignment. The
metabolites were identified by accurate MS (<25 ppm) and MS/MS
data, which were matched with a standard database (BaseDeepBP,
Shanghai Bioprofile Technology, Human Metabolome Database
(hmdb.ca/, MassBank, https://massbank.eu/MassBank/, MetaboBASE, http://metabase.com/, Global Natural Product Social
Molecular Networking, https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash-old.jsp).
In the extracted-ion features, only the variables with >50% of
the non-zero measurement values in ≥1 group were retained.
Multivariate statistical analyses
All multivariate data analyses and modeling were
conducted using the SIMCAP software (Version 14.0, Umetrics;
Sartorius AG). After mean-centering the data using Pareto scaling,
models were developed using principal component analysis (PCA),
orthogonal partial least squares discriminant analysis (OPLS-DA)
and PLS-DA. All models were evaluated for overfitting using
permutation tests. Descriptive performance was assessed using
cumulative R2X [ideal model: R2X (cumulative)=1 and R2Y (ideal
model: R2Y (cumulative)=1] values, while prediction performance was
measured using cumulative Q2 [ideal model: Q2 (cumulative)=1)] and
a permutation test. In the permuted model, R2 and Q2 values at the
Y-axis intercept should be lower than those of the non-permuted
model. OPLS-DA was used to identify discriminating metabolites
using variable importance on projection (VIP) scores, which
indicate a variable contribution to class discrimination. VIP
scores were calculated as the weighted sum of squares of the PLS
weights, with values >1 considered statistically significant.
High VIP scores indicate strong discriminatory ability and help in
selecting biomarkers. Discriminating metabolites were obtained
using a statistically significant threshold of VIP values from the
OPLS-DA model and two-tailed Student's t-test (P-value) on
normalized raw data. Metabolites with VIP values >1 and
P<0.05 were considered statistically significant. The
fold-change was calculated as the ratio of the average mass area of
the CHD group to that of the control group. Identified differential
metabolites were used for cluster analyses using the R package (R
package version 1.0.12, CRAN.R-project.org/package=pheatmap). Hierarchical
clustering for each group was performed for heatmap.
Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis
To identify perturbed biological pathways,
differential metabolite data were subjected to KEGG pathway
analysis using the KEGG database (kegg.jp/). KEGG
enrichment analyses were performed using the Fisher's exact test
and false discovery rate correction for multiple testing was also
performed.
Animals and arginine/lysine
administration
A total of 18 Female C57BL/6J mice (age, 8-10 weeks,
Shenyang Lan Pudas technology co., ltd) were used (mean weight,
22.2±0.81 g; range, 20.8-23.6 g). The mice were kept in standard
cages under a 12/12-h light/dark cycle, ad libitum food and
water, temperature was 20-26˚C and humidity was 40-70%. To obtain
pregnant mice, female mice mated with male mice (n=6, 8-10 weeks
age, 23.2-25.6 g weight, Shenyang Lan Pudas Technology co., ltd)
and underwent testing every morning. Females with a vaginal plug
were immediately separated from the males and indicated as
embryonic day 0.5 (E0.5). Pregnant mice were randomly assigned to
treatment (either arginine or lysine, Sigma) and control group
(both n=6). The treatment group received a single intraperitoneal
injection of 1.5 mg/g body weight/day arginine/lysine from E7.5 to
E14.5. The control group received an equivalent volume of saline.
Doses of arginine or lysine administration were determined as
previously described (11-13).
In addition to the routine feeding, mouse health and behavior were
checked every 12 h. The humane endpoints were cessation of eating
and movement and listlessness or lack of response when people
approached. No animals reached these endpoints. Pregnant mice at
E15.5 were anaesthetized with 1.5% isoflurane for 2-3 min. The
anesthetized mice had even heartbeat and breathing, relaxed muscles
and no limb activity or pedal reflex. A total of 100 µl peripheral
blood was collected from the inner canthus by capillary tube. Then,
mice were euthanized by cervical dislocation. Death was confirmed
when heartbeat and respiration stopped and the pupils dilated for
>5 min. Maternal plasma arginine concentration was determined by
LC-MS/MS. All experiments were approved by the Ethics Committee of
Shengjing Hospital of China Medical University (approval no.
2024PS202K).
Hematoxylin-eosin staining of
embryonic heart
The embryonic trunk was fixed in 4% paraformaldehyde
solution at 4˚C for ≥48 h. The samples were dehydrated in 75, 85,
95 and 100% ethanol for 1 h. Following xylene treatment at room
temperature, 100% paraffin was used for tissue embedding. Then,
continuous slices of the embryonic heart (4 µm) were cut from the
top of the aortic arch to the apex of the heart and stained with
hematoxylin and eosin at room temperature for 1 min using automatic
stainer (Leica GmbH) and images were captured under a light
microscope.
Detection of amino acids in maternal
peripheral blood
Maternal peripheral blood (96 controls and 69 CHD
cases; gestational age, weeks 18.0-30.0, aged from 20 to 44 years
old, collected in Shengjing Hospital of China Medical University
from April 2022 to December 2022, the same Inclusion/exclusion
criteria as amniotic fluid cells collection) were collected to
detect the change of amino acids using a non-derivatized amino
acids detection kit (Neobase 2, Revvity, Inc.) on LC-MS/MS (QSight
210MD, PerkinElmer). The sample was detected with multiple reaction
monitoring mode scanning in positive ion mode. The ionization
conditions were as follows: Drying gas, 120 l/min; electrospray
voltage, 4,900 V; source temperature, 150˚C; nebulizer gas, 220
l/min; hot surface-induced desolvation temperature, 250˚C. The
injection volume was 10 µl. Chromatographic mobile phase A
(included in the detection kit) and the chromatographic gradient
elution procedure was as follows: 0.00-0.15 min, flow rate, 0.16
ml/min; 0.15-0.90 min, flow rate, 0.03 ml/min; 0.90-1.00 min, flow
rate, 0.7 ml/min and 1.0-1.15 min, flow rate, 0.16 ml/min. The
parameters of amino acid detection MS are listed in Table SI.
Statistical analysis
All data are presented as the mean ± standard
deviation. At least three independent experimental repeats. The
analysis was performed using SPSS 17.0 software (SPSS, Inc.).
Student's unpaired two-tailed t test, one-way ANOVA followed by
least significant difference post hoc test and χ2 test
were used for statistical analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
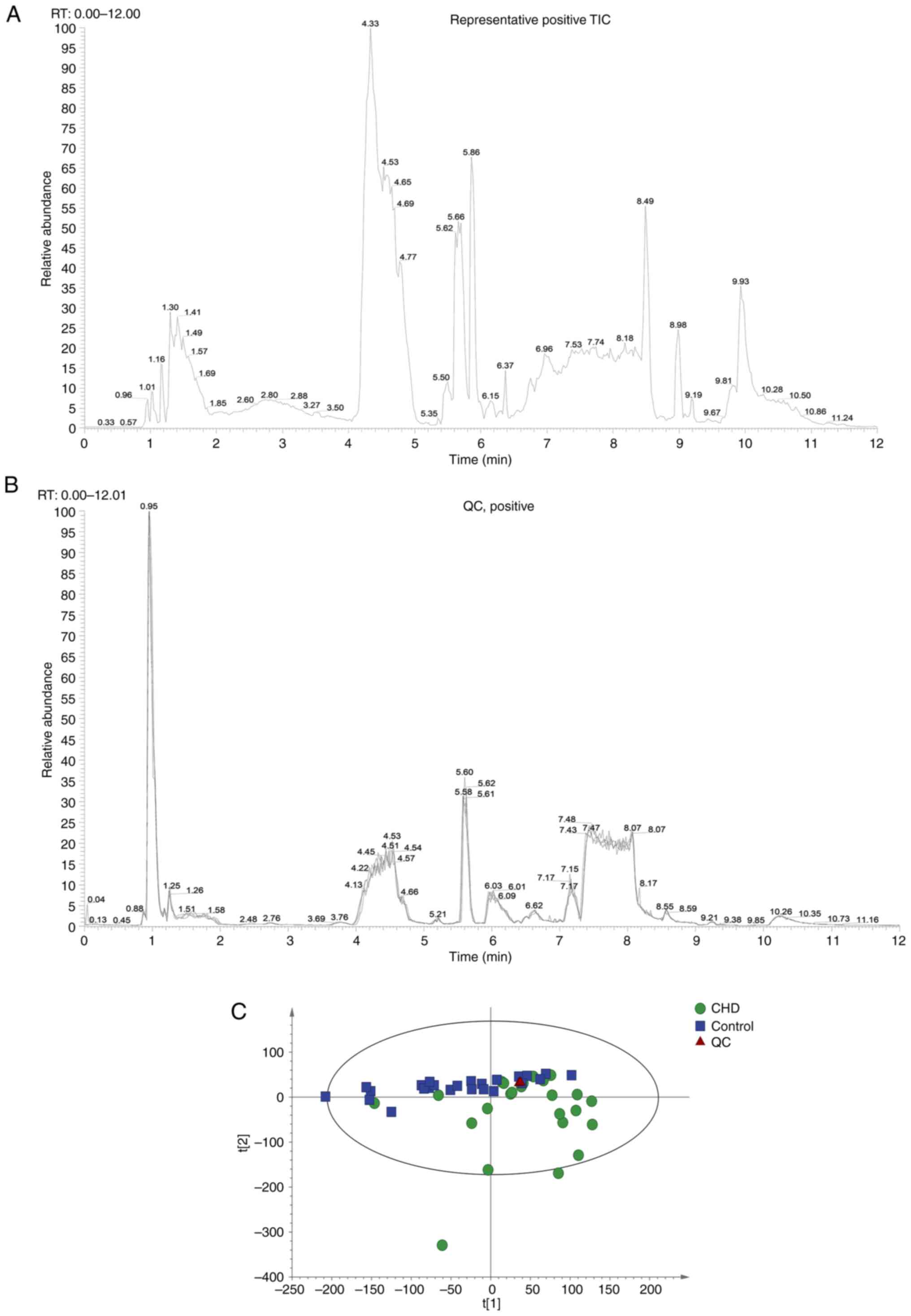
Experimental quality evaluation
Metabonomics analysis was performed on 48 samples
(24 controls and 24 CHD cases). Each sample was detected and
analyzed using UPLC-MS/MS, and two original files (positive and
negative ion mode) were obtained. Representative images are shown
in Fig. 2A. A total of 883
metabolites were identified (data not shown). The system stability
was evaluated using two strategies: MS total ion chromatogram (TIC)
flow diagram comparison of QC samples and PCA statistical analysis
of overall samples. A total of four QC TIC diagrams were overlaid
(Fig. 2B). The results suggested
that the response intensity and retention time of each color
spectrum peak overlapped, indicating that the variation caused by
instrument error was small and the data quality was reliable. The
ion peaks of metabolites extracted from all experimental and QC
samples were subjected to PCA. Samples were closely gathered
together, indicating that the experiment had good repeatability
(Fig. 2C). Thus, the instrument
analysis system had good stability, and the experimental data were
reliable. The metabolic spectrum difference reflected the
biological differences between the samples.
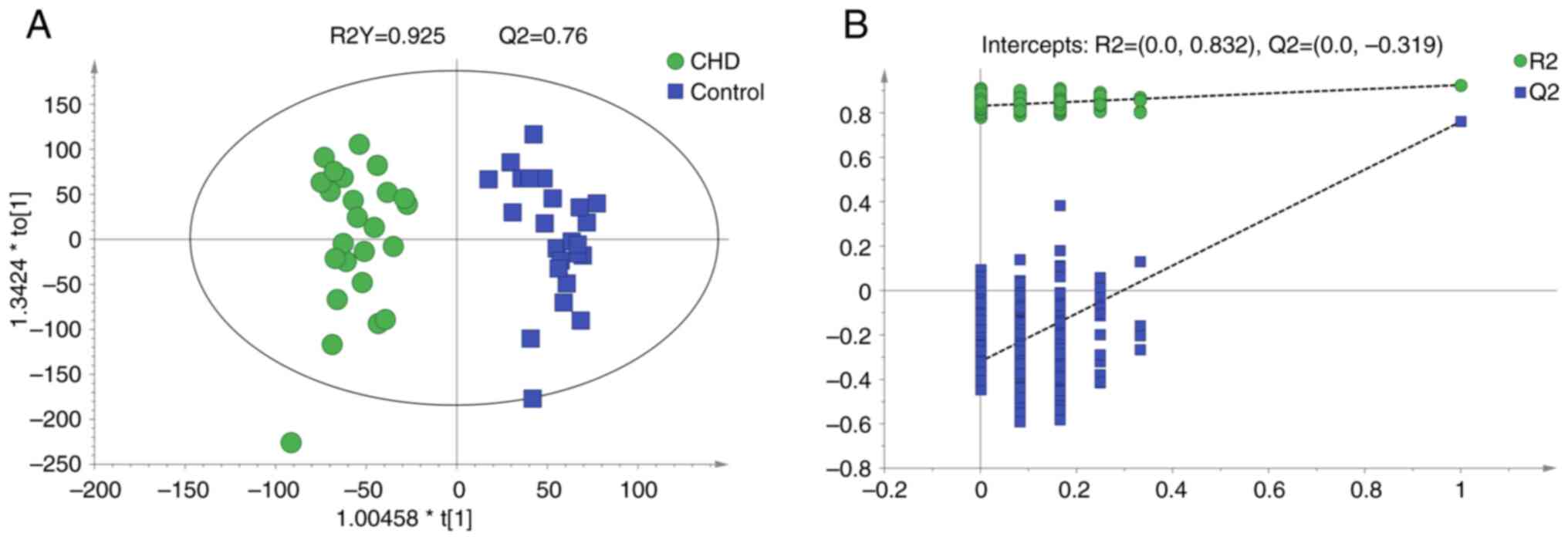
OPLS-DA
An OPLS-DA model was established to compare
metabolite profiles between the two groups. The model evaluation
parameters and score diagram are shown in Fig. 3. The OPLS-DA model distinguished the
two sample groups. The parameters (R2Y=0.925 and Q2=0.76) in the
OPLS-DA model suggested that the experimental data were stable and
reliable. The intercept of Q2 was -0.319, indicating there was no
overfitting in the OPLS-DA model established using the experimental
data. The OPLS-DA model showed separation in the metabolic profiles
between the CHD and control groups. A total of 292 different
metabolites (VIP>1 and P<0.05) were identified, of which 172
metabolites were up- and 120 metabolites were downregulated in the
CHD compared with the control group (supplementary data). The two
most changed metabolites were lysine
(L-lysine/lysine/N-methyllysine) and arginine
(homoarginine/L-arginine; Table
II).
 | Table IIDifferential metabolites in
congenital heart disease compared with controls. |
Table II
Differential metabolites in
congenital heart disease compared with controls.
| Metabolite | ESI mode | RT, min | m/z | VIP | Fold-change | P-value |
|---|
| L-lysine | Negative | 9.954 | 145.097 | 1.67 | 1,848.38 | 0.0005 |
| Lysine | Positive | 10.013 | 147.112 | 1.41 | 629.88 | 0.0042 |
| N-methyllysine | Negative | 9.886 | 159.112 | 1.76 | 50.56 | 0.0002 |
| Homoarginine | Negative | 9.853 | 187.119 | 1.74 | 766.51 | 0.0003 |
| L-arginine | Positive | 10.086 | 175.119 | 1.58 | 38.19 | 0.0015 |
| D-aspartate | Negative | 8.672 | 132.029 | 1.27 | 25.97 | 0.0034 |
| L-asparagine | Negative | 8.445 | 131.045 | 1.24 | 7.26 | 0.0049 |
| Proline | Positive | 10.139 | 116.071 | 1.51 | 22.71 | 0.0023 |
| L-ornithine | Negative | 10.061 | 131.081 | 1.49 | 11.66 | 0.0016 |
| Glutamine | Negative | 8.200 | 145.061 | 1.33 | 11.28 | 0.0038 |
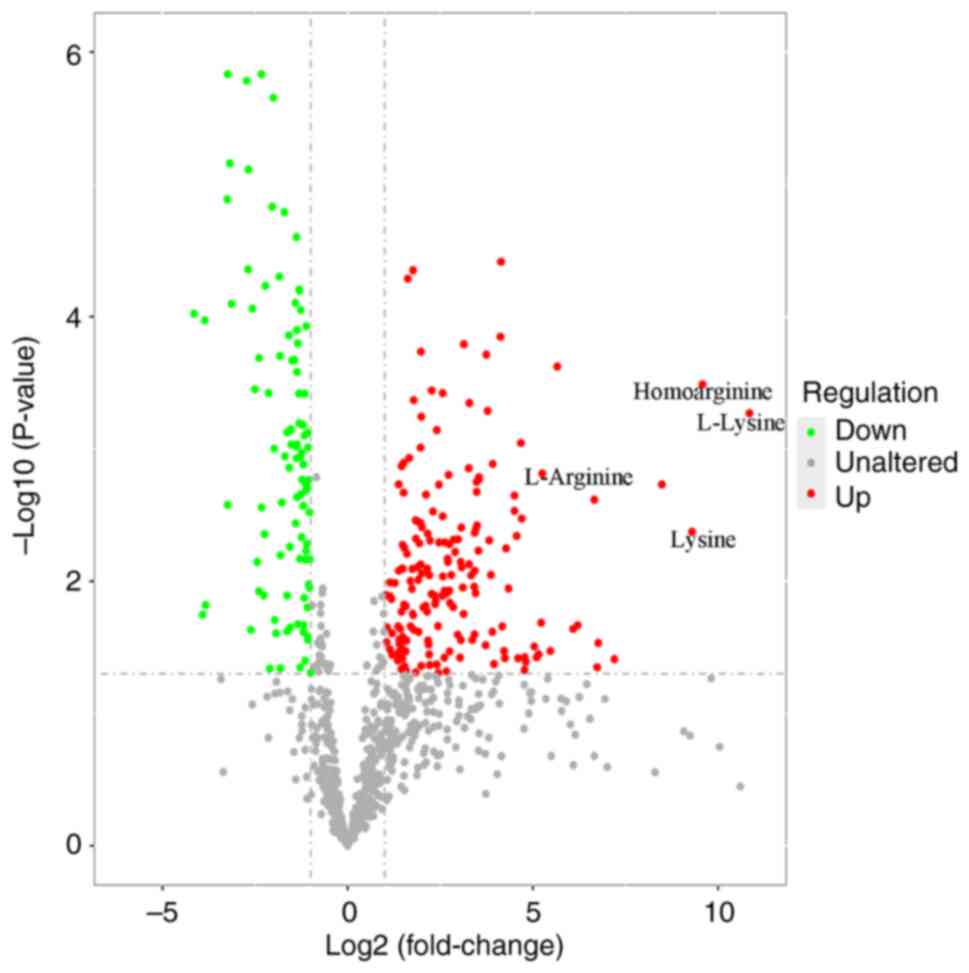
Univariate statistical analyses
To identify potential marker metabolites, univariate
analysis was performed (FC<0.5 or >2.0). Lysine and arginine
were the two most changed metabolites (Fig. 4). The fold-changes of L-lysine,
lysine, homoarginine and L-arginine were 1848.4, 629.9, 766.5 and
38.2, respectively.
Hierarchical cluster analysis
To evaluate the potential differential metabolites,
hierarchical clustering for each group was performed (Fig. S1). Generally, when the candidate
metabolites screened are reasonable and accurate, the same group of
samples appear in the same cluster. Metabolites in the same cluster
have similar expression patterns. Although some samples strayed
from their groups, most were concentrated in their respective
groups.
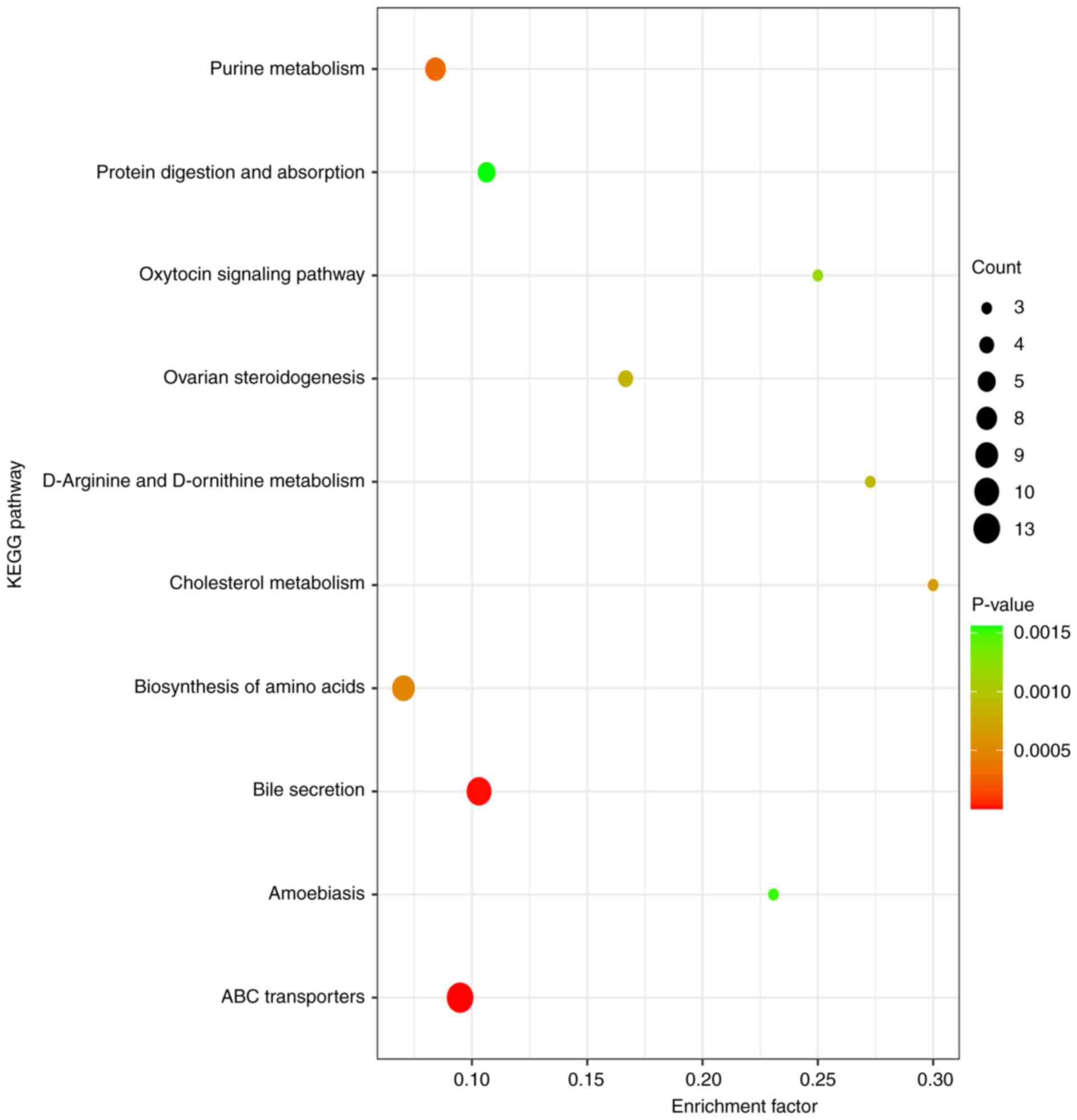
KEGG pathway enrichment analysis
The differential metabolites were analyzed using
KEGG (Fig. 5; supplementary data).
The results demonstrated that amino acid metabolism (involved in
‘ABC transporters’, ‘biosynthesis of amino acids’ and ‘D-arginine
and D-ornithine metabolism’) may play an important role in CHD
development.
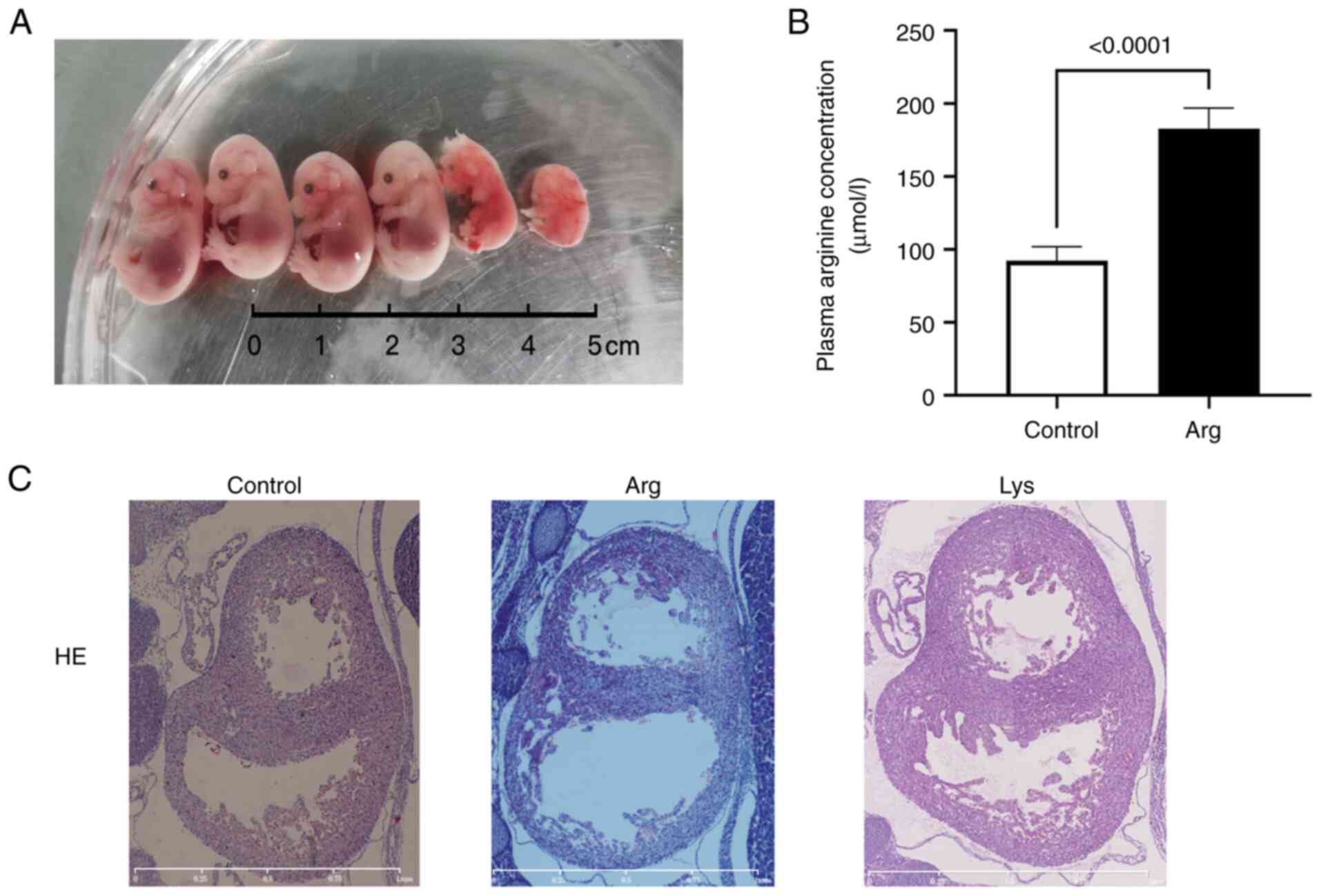
Arginine results in heart
malformation
To test whether arginine/lysine affect heart
development, arginine/lysine was intraperitoneal injected in
pregnant mice. Litter size decreased, and the naked deformity rate
increased after arginine treatment (Fig. 6A; Table III). The concentration of maternal
plasma arginine increased following arginine treatment (182.6±14.4
vs. 92.3±9.6 µmol/l, Fig. 6B). In
the arginine-treated group, the hearts of the embryos with no
notable gross abnormality demonstrated enlarged heart cavities and
thinner heart walls (Fig. 6C),
compared with the control. In the lysine-treated group, no obvious
deformity was observed.
 | Table IIIEffects of arginine/lysine treatment
on mice. |
Table III
Effects of arginine/lysine treatment
on mice.
| Group | Pregnant mice,
n | Embryos, n | Mean litter
size | Deformity
ratea, % (n) |
|---|
| Control | 6 | 39 | 6.5±1.05 | 0 (0) |
| Arginine | 6 | 26 |
4.3±1.03b |
30.8(8)c |
| Lysine | 6 | 36 | 6.0±1.41 | 0 (0) |
Amino acids in maternal peripheral
blood are not significantly altered between CHD and control
group
As there were several amino acids changes in
amniotic fluid cells metabonomics, the present study investigated
whether the amino acids change in human maternal peripheral blood
between CHD and control group. Compared with control group, glycine
and glutamine were downregulated by 17.3% (503.8±147.1 vs.
608.7±266.8 µmol/l, Table IV) and
upregulated by 6.7% (381.0±70.1 vs. 357.8±77.30 µmol/l),
respectively. Therefore, these amino acids cannot be used as
non-invasive markers for CHD screening.
 | Table IVMean concentration of amino acids in
maternal peripheral blood. |
Table IV
Mean concentration of amino acids in
maternal peripheral blood.
| Amino acid | Control (n=96),
µM | CHD (n=69), µM | T-value | P-value |
|---|
| Ala | 504.3±142.0 | 472.9±127.2 | 1.489 | 0.138 |
| Val | 177.7±34.7 | 176.6±33.8 | 0.217 | 0.828 |
| Gly | 608.7±266.8 | 503.8±147.1 | 3.230 | 0.002 |
| Gln | 357.8±77.3 | 381.0±70.1 | -2.011 | 0.046 |
| Glu | 159.1±48.9 | 144.7±54.6 | 1.740 | 0.084 |
| Orn | 106.5±18.9 | 105.6±17.1 | 0.314 | 0.754 |
| Leu | 181.0±40.7 | 174.1±43.9 | 1.013 | 0.313 |
| Arg | 11.8±3.9 | 12.4±4.8 | -0.75 | 0.455 |
| Met | 13.4±5.2 | 13.3±5.5 | 0.041 | 0.968 |
| Phe | 73.1±26.4 | 72.2±19.5 | 0.248 | 0.804 |
| Tyr | 57.5±12.6 | 54.7±15.7 | 1.220 | 0.225 |
| Cit | 16.8±3.2 | 17.4±4.5 | -0.913 | 0.363 |
| Pro | 145.2±37.6 | 135.9±33.0 | 1.692 | 0.093 |
| HArg | 11.4±3.1 | 11.2±2.8 | 0.518 | 0.605 |
| Lys | 49.5±9.5 | 49.9±7.8 | -0.289 | 0.773 |
Discussion
Samples from fetuses with CHD had different
metabolic profiles compared with controls. It was determined that
amino acids serve an important role in heart development. When
arginine was used to treat the pregnant mice, the embryos
demonstrated increased malformation rate. The embryonal heart
cavity became larger and the heart wall became thinner. The present
study may provide insight into the pathophysiology of CHD.
The high incidence of CHD have prompted
investigation into its pathogenesis to identify early screening
indicators (1). With the rapid
development of high-throughput sequencing technology, an increasing
number of copy number variations and gene mutations have been
identified in CHD (2,3). However, only one-third of the cases
can be explained by genetics (2,3). The
wide application of metabolomics facilitates investigation of
potential CHD pathogenesis or early screening biomarker.
There are three primary metabolomics detection
platforms, namely LC-MS, gas chromatography-MS (GC-MS) and nuclear
magnetic resonance (NMR) (14,15).
Because NMR has defects, such as relatively low sensitivity, poor
selectivity and limited metabolite coverage, MS platforms with high
sensitivity and specificity are more commonly used (14-16).
Compared with GC-MS, LC-MS has advantages: The samples are
separated at room temperature without special treatment (GC-MS
often requires derivatization). In addition, the range of the
molecular weights of the tested substances is wider. Moreover, it
has higher sensitivity and can detect more trace substances. Thus,
LC-MS has a wide range of applications (14-16).
Several types of biological samples (amniotic fluid, plasma, urine)
have been used for metabolomics screening (9). The present study used cells; as the
basic functional unit of life, cells directly reflect the levels of
intracellular metabolism and are more conducive to discover the
potential pathogenic mechanisms.
To explore the possibility of arginine and other
amino acids as metabolic markers, the present study detected the
concentration of amino acids in maternal peripheral blood. There
was no significant difference between CHD and control group,
indicating plasma amino acids cannot be used as non-invasive
markers for CHD screening. Although the present study demonstrated
a number of differential metabolites in amniotic fluid cell
metabonomics, there were limitations. First, the present study
could not determine whether the altered metabolites were the result
or the cause of CHD. More studies are needed to clarify this.
Second, the sample size was not sufficiently large, and the type of
CHD was only concentrated in the ventricular septal defect. A
comprehensive, large-scale multi-center study is needed.
L-lysine, an essential amino acid, was the most
notable differential metabolite identified in CHD. Humans receive
L-lysine from daily food such as meat, cereal grains or legumes
(17). L-lysine is taken up into
cells by cationic transport system y+ (18). Lysine has important functions in the
promotion of human physiological development and fatty acid
oxidation (19). It promotes brain
development and fat metabolism and prevents cell degeneration.
Shimomura et al (20) found
that dietary L-lysine exerts protective effects against vascular
calcification in uremic rats. The addition of lysine to the diet
can manage osteoporosis (21).
L-lysine also has beneficial hemodynamic effects and decreases
nitric oxide levels in endotoxemic rats (22). Lysine acetylsalicylate can recover
sepsis-induced lung tissue damage in rats (23), and L-lysine suppresses acute
pancreatitis in mice (24).
L-lysine ameliorates sepsis-induced acute lung injury in a
lipopolysaccharide-induced mouse model (25). Abnormalities in lysine degradation
are involved in the early development of cardiomyocyte hypertrophy
in pressure-overloaded rats (19).
In the present study, lysine did not cause any difference in the
litter size and deformity rate from the controls and embryonal
hearts also did not show obvious malformation (the data not shown
in this study). Although lysine has a range of functions, more
studies are needed to clarify its role in heart development.
L-arginine, homoarginine, L-ornithine, L-asparagine
and L-glutamine belong to the arginine family and participate in
the urea cycle (26). Glutamine,
glutamate and arginine can regulate nutrient intake and neonatal
development through norepinephrine, glucagon-like peptide-1,
mammalian target of rapamycin, mitogen-activated protein kinase and
autophagy (26,27). Arginine is an important nutrient
with regulatory roles in neonatal growth (28,29).
Rat studies have indicated that supplementing the maternal diet
with arginine and glutamine improves embryo implantation and
survival, enhances litter size and increases the number and birth
weight of surviving embryos (30,31).
In addition, arginine and glutamine affect gene expression to
improve antioxidant responses and DNA transcription (32,33).
Plasma homoarginine levels have been reported to show a declining
trend in patients with complex CHD compared with healthy controls
(34). Therefore, homoarginine may
be a prognostic indicator of CHD. Cedars found that plasma
concentrations of multiple amino acids differ between adult
patients with CHD and healthy controls (35). Although the numerical difference is
not large, a metabolite cluster containing amino acids and
metabolites is associated with negative clinical outcomes (35). A total of 11 amino acids (including
arginine, lysine, asparagine and glutamine) are upregulated in the
cardiac tissues of children with cyanotic CHD (36) compared with those with acyanotic
CHD. Yu et al (37) found
that glutamine and glutamate have considerable diagnostic value for
pediatric patients with CHD patients. Li et al (9) found that the concentrations of uric
acid and proline in the amniotic fluid are significantly elevated
in patients with CHD. In the present study, when pregnant mice were
treated with arginine at the dose of 1.5 mg/g body weight, the
litter size decreased and the embryo deformity rate increased. The
embryonal hearts demonstrated enlarged heart cavities and thinner
heart walls. The present study only tested the maternal plasma
arginine concentration following arginine treatment and did not
test the concentration in the placenta and heart in the embryo.
Thus, changes in amino acid levels may serve an important role in
the occurrence and development of CHD. Future studies should focus
on metabolic reprogramming to clarify the potential mechanism of
arginine-induced cardiac malformation.
In summary, differential metabolites were identified
in CHD using metabolomics. The levels of numerous types of amino
acids were significantly elevated, indicating that they may serve
an important role in heart development. Arginine treatment of
pregnant mice resulted in embryonal heart defects.
Supplementary Material
Hierarchical clustering results of the
differential metabolites. Each line represents a metabolite. The
deeper the red, the higher its content in the measured sample; the
deeper the blue, the lower its content in the measured sample. CHD,
congenital heart disease.
Amino acid detection mass spectrometry
parameters.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the People's
Livelihood Science and Technology Project of Liaoning Province
(grant nos. 2022020806-JH2/1015 and 2021JH2/10300123).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ZL collected the samples and wrote the manuscript.
CC performed animal experiments. JL analyzed the data. GL
interpreted the data, performed experiments and revised the
manuscript. CQ designed the study and revised the manuscript. ZL
and GL confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shengjing Hospital of China Medical University
(approval nos. 2022PS307K and 2024PS202K). All participants
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang Y, Wang J, Zhao J, Huang G, Liu K,
Pan W, Sun L, Li J, Xu W, He C, et al: Current status and
challenges in prenatal and neonatal screening, diagnosis, and
management of congenital heart disease in China. Lancet Child
Adolesc Health. 7:479–489. 2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zaidi S and Brueckner M: Genetics and
genomics of congenital heart disease. Circ Res. 120:923–940.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jin SC, Homsy J, Zaidi S, Lu Q, Morton S,
DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al: Contribution
of rare inherited and de novo variants in 2,871 congenital heart
disease probands. Nat Genet. 49:1593–1601. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Lai T, Xiang L, Liu Z, Mu Y, Li X, Li N,
Li S, Chen X, Yang J, Tao J and Zhu J: Association of maternal
disease and medication use with the risk of congenital heart
defects in offspring: A case-control study using logistic
regression with a random-effects model. J Perinat Med. 47:455–463.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu X, Nie Z, Chen J, Guo X, Ou Y, Chen G,
Mai J, Gong W, Wu Y, Gao X, et al: Does maternal environmental
tobacco smoke interact with social-demographics and environmental
factors on congenital heart defects? Environ Pollut. 234:214–222.
2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rocha LA, Araujo Júnior E, Nardozza LM and
Moron AF: Screening of fetal congenital heart disease: The
challenge continues. Rev Bras Cir Cardiovasc. 28:V–VII.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Menon R, Jones J, Gunst PR, Kacerovsky M,
Fortunato SJ, Saade GR and Basraon S: Amniotic fluid metabolomic
analysis in spontaneous preterm birth. Reprod Sci. 21:791–803.
2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Romero R, Espinoza J, Gotsch F, Kusanovic
JP, Friel LA, Erez O, Mazaki-Tovi S, Than NG, Hassan S and Tromp G:
The use of highdimensional biology (genomics, transcriptomics,
proteomics, and metabolomics) to understand the preterm parturition
syndrome. BJOG 113 Suppl. 3:118–135. 2006.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li Y, Sun Y, Yang L, Huang M, Zhang X,
Wang X, Guan X, Yang P, Wang Y, Meng L, et al: Analysis of
biomarkers for congenital heart disease based on maternal amniotic
fluid metabolomics. Front Cardiovasc Med. 8(671191)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Du Y, Chang W, Gao L, Deng L and Ji WK:
Tex2 is required for lysosomal functions at TMEM55-dependent ER
membrane contact sites. J Cell Biol. 222(e202205133)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ishitobi K, Kotani H, Iida Y, Taniura T,
Notsu Y, Tajima Y and Harada M: A modulatory effect of L-arginine
supplementation on anticancer effects of chemoimmunotherapy in
colon cancer-bearing aged mice. Int Immunopharmacol.
113(109423)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Siriviriyakul P, Sriko J, Somanawat K,
Chayanupatkul M, Klaikeaw N and Werawatganon D: Genistein
attenuated oxidative stress, inflammation, and apoptosis in
L-arginine induced acute pancreatitis in mice. BMC Complement Med
Ther. 22(208)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bianchini Narde M, Belli Cassa Domingues
EL, Ribeiro Gonçalves K, Lomar Viana M, Santos Zanini M, Geraldo de
Lima W, Bahia MT and Matos Dos Santos F: L-arginine supplementation
increases cardiac collagenogenesis in mice chronically infected
with Berenice-78 trypanosoma cruzi strain. Parasitol Int.
83(102345)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Emwas AH, Roy R, McKay RT, Tenori L,
Saccenti E, Gowda GAN, Raftery D, Alahmari F, Jaremko L, Jaremko M
and Wishart DS: NMR spectroscopy for metabolomics research.
Metabolites. 9(123)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Marchev AS, Vasileva LV, Amirova KM,
Savova MS, Balcheva-Sivenova ZP and Georgiev MI: Metabolomics and
health: From nutritional crops and plant-based pharmaceuticals to
profiling of human biofluids. Cell Mol Life Sci. 78:6487–6503.
2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Azad RK and Shulaev V: Metabolomics
technology and bioinformatics for precision medicine. Brief
Bioinform. 20:1957–1971. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Tomé D and Bos C: Lysine requirement
through the human life cycle. J Nutr. 137(Suppl 2):1642S–1645S.
2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Boldt A, Gergs U, Frenker J, Simm A,
Silber RE, Klöckner U and Neumann J: Inotropic effects of L-lysine
in the mammalian heart. Naunyn Schmiedebergs Arch Pharmacol.
380:293–301. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liu J, Hu J, Tan L, Zhou Q and Wu X:
Abnormalities in lysine degradation are involved in early
cardiomyocyte hypertrophy development in pressure-overloaded rats.
BMC Cardiovasc Disord. 21(403)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Shimomura A, Matsui I, Hamano T, Ishimoto
T, Katou Y, Takehana K, Inoue K, Kusunoki Y, Mori D, Nakano C, et
al: Dietary L-lysine prevents arterial calcification in
adenine-induced uremic rats. J Am Soc Nephrol. 25:1954–1965.
2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Fürst P: Dietary L-lysine supplementation:
A promising nutritional tool in the prophylaxis and treatment of
osteoporosis. Nutrition. 9:71–72. 1993.PubMed/NCBI
|
|
22
|
Liaudet L, Gnaegi A, Rosselet A, Markert
M, Boulat O, Perret C and Feihl F: Effect of L-lysine on nitric
oxide overproduction in endotoxic shock. Br J Pharmacol.
122:742–748. 1997.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang WD, Wang JZ, Lu YQ, DI YM, Jiang JK
and Zhang Q: Lysine acetylsalicylate ameliorates lung injury in
rats acutely exposed to paraquat. Chin Med J (Engl). 124:2496–2501.
2011.PubMed/NCBI
|
|
24
|
Al-Malki AL: Suppression of acute
pancreatitis by L-lysine in mice. BMC Complement Altern Med.
15(193)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang Y, Yu W, Han D, Meng J, Wang H and
Cao G: L-lysine ameliorates sepsis-induced acute lung injury in a
lipopolysaccharide-induced mouse model. Biomed Pharmacother.
118(109307)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu G: Functional amino acids in nutrition
and health. Amino Acids. 45:407–411. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kim J, Song G, Wu G, Gao H, Johnson GA and
Bazer FW: Arginine, leucine, and glutamine stimulate proliferation
of porcine trophectoderm cells through the MTOR-RPS6K-RPS6-EIF4EBP1
signal transduction pathway. Biol Reprod. 88(113)2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wu G, Bazer FW, Dai Z, Li D, Wang J and Wu
Z: Amino acid nutrition in animals: Protein synthesis and beyond.
Annu Rev Anim Biosci. 2:387–417. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lin G, Wang X, Wu G, Feng C, Zhou H, Li D
and Wang J: Improving amino acid nutrition to prevent intrauterine
growth restriction in mammals. Amino Acids. 46:1605–1623.
2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wu G, Bazer FW, Satterfield MC, Li X, Wang
X, Johnson GA, Burghardt RC, Dai Z, Wang J and Wu Z: Impacts of
arginine nutrition on embryonic and fetal development in mammals.
Amino Acids. 45:241–256. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zeng X, Huang Z, Mao X, Wang J, Wu G and
Qiao S: N-carbamylglutamate enhances pregnancy outcome in rats
through activation of the PI3K/PKB/mTOR signaling pathway. PLoS
One. 7(e41192)2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Jobgen W, Fu WJ, Gao H, Li P, Meininger
CJ, Smith SB, Spencer TE and Wu G: High fat feeding and dietary
L-arginine supplementation differentially regulate gene expression
in rat white adipose tissue. Amino Acids. 37:187–198.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wang J, Chen L, Li P, Li X, Zhou H, Wang
F, Li D, Yin Y and Wu G: Gene expression is altered in piglet small
intestine by weaning and dietary glutamine supplementation. J Nutr.
138:1025–1032. 2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Raedle-Hurst T, Mueller M, Meinitzer A,
Maerz W and Dschietzig T: Homoarginine-A prognostic indicator in
adolescents and adults with complex congenital heart disease? PLoS
One. 12(e0184333)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cedars A, Manlhiot C, Ko JM, Bottiglieri
T, Arning E, Weingarten A, Opotowsky A and Kutty S: Metabolomic
profiling of adults with congenital heart disease. Metabolites.
11(525)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Dong S, Wu L, Duan Y, Cui H, Chen K, Chen
X, Sun Y, Du C, Ren J, Shu S, et al: Metabolic profile of heart
tissue in cyanotic congenital heart disease. Am J Transl Res.
13:4224–4232. 2021.PubMed/NCBI
|
|
37
|
Yu M, Sun S, Yu J, Du F, Zhang S, Yang W,
Xiao J and Xie B: Discovery and validation of potential serum
biomarkers for pediatric patients with congenital heart diseases by
metabolomics. J Proteome Res. 17:3517–3525. 2018.PubMed/NCBI View Article : Google Scholar
|