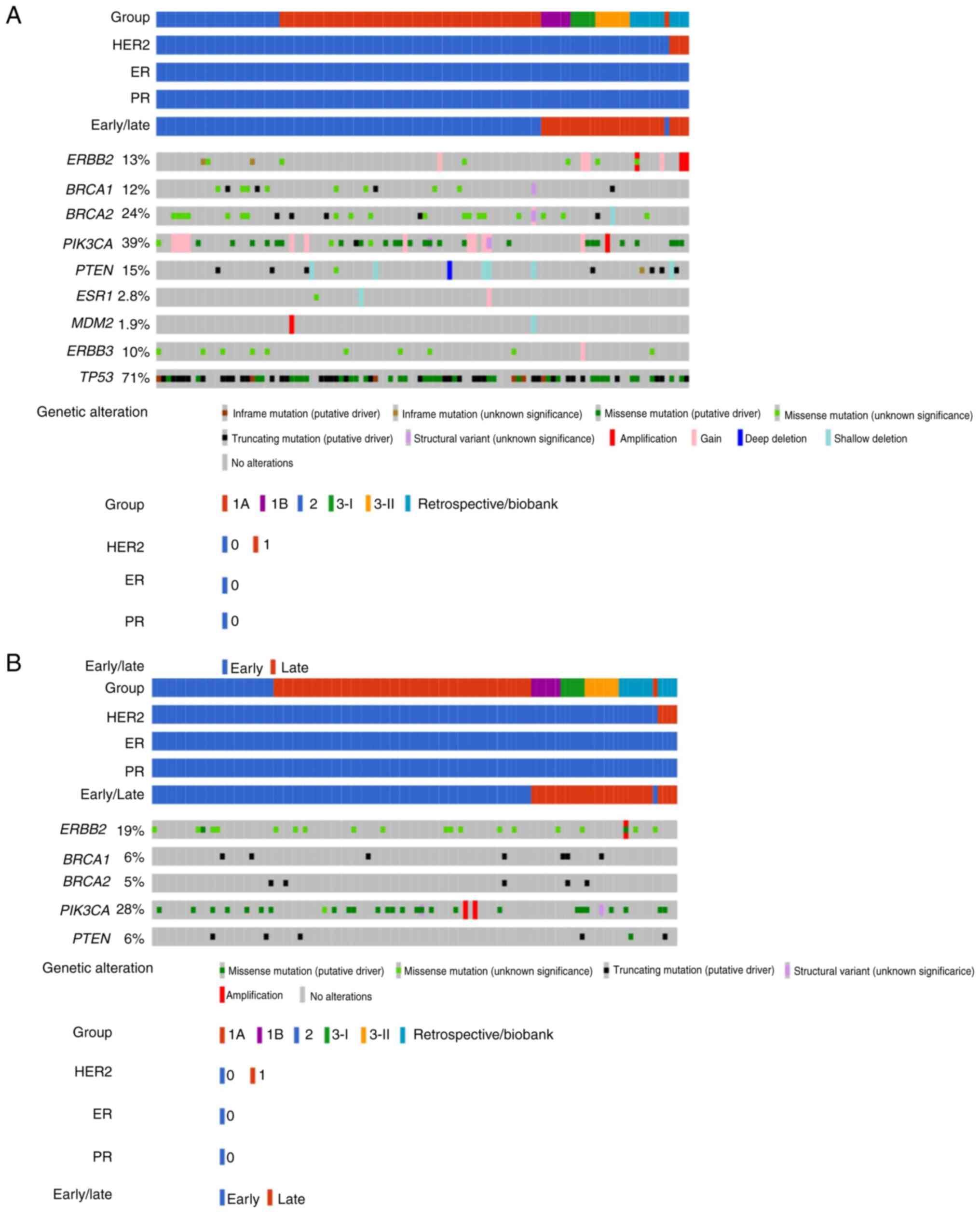
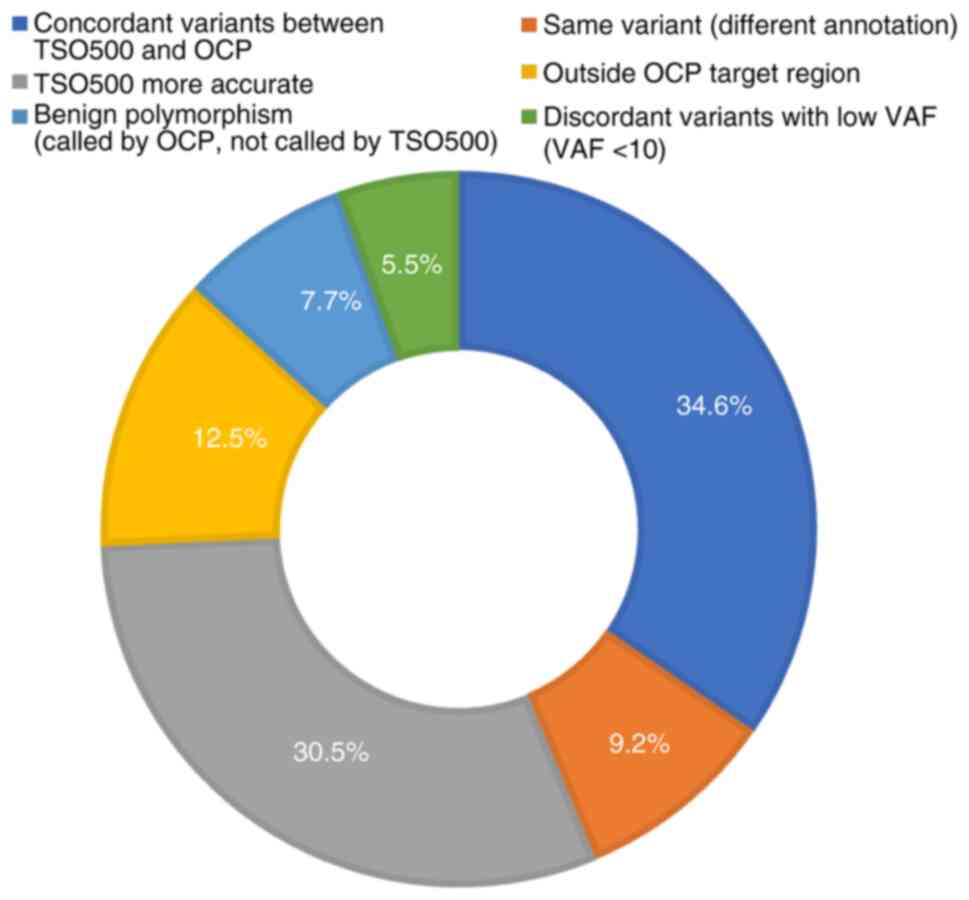
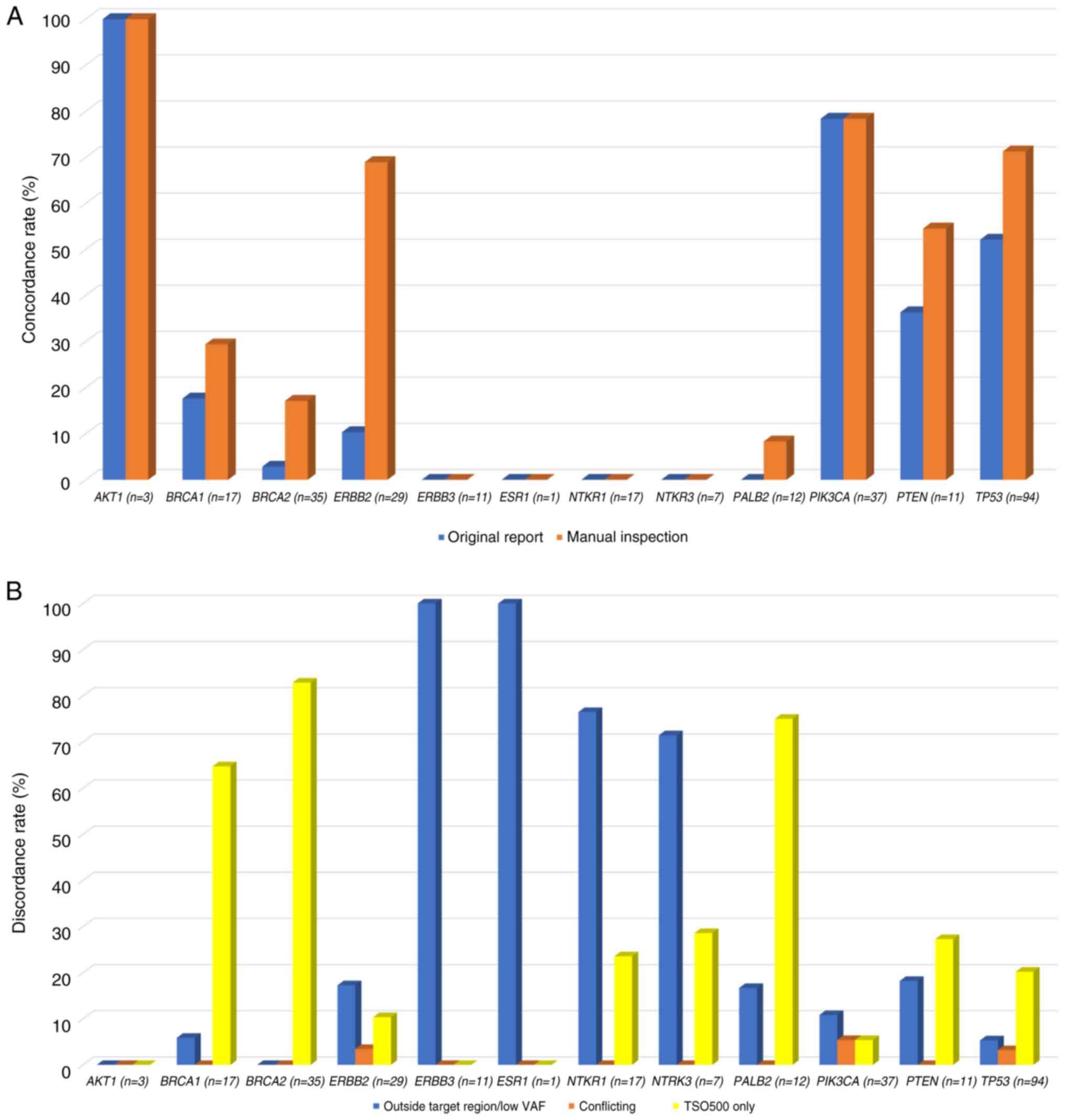
|
1
|
Huang CC, Yeh YC, Ho HL, Liu CY, Tsai YF
and Tseng LM: Comprehensive genomic profiling of Taiwanese patients
with breast cancer using a novel targeted panel: Preliminary
analyses from a prospective triple-negative cohort. J Clin Oncol.
41 (Suppl 16)(e12553)2023.
|
|
2
|
Wang L, Zhai Q, Lu Q, Lee K, Zheng Q, Hong
R and Wang S: Clinical genomic profiling to identify actionable
alterations for very early relapsed triple-negative breast cancer
patients in the Chinese population. Ann Med. 53:1358–1369.
2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tierno D, Grassi G, Scomersi S, Bortul M,
Generali D, Zanconati F and Scaggiante B: Next-generation
sequencing and triple-negative breast cancer: Insights and
applications. Int J Mol Sci. 24(9688)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
O'Haire S, Degeling K, Franchini F, Tran
B, Luen SJ, Gaff C, Smith K, Fox S, Desai J and IJzerman M:
Comparing survival outcomes for advanced cancer patients who
received complex genomic profiling using a synthetic control arm.
Target Oncol. 17:539–548. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Cifuentes C, Lombana M, Vargas H, Laguado
P, Ruiz-Patiño A, Rojas L, Navarro U, Vargas C, Ricaurte L, Arrieta
O, et al: Application of comprehensive genomic profiling-based
next-generation sequencing assay to improve cancer care in a
developing country. Cancer Control.
30(10732748231175256)2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Almansour NM: Triple-negative breast
cancer: A brief review about epidemiology, risk factors, signaling
pathways, treatment and role of artificial intelligence. Front Mol
Biosci. 9(836417)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948.
2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Stewart RL, Updike KL, Factor RE, Henry
NL, Boucher KM, Bernard PS and Varley KE: A multigene assay
determines risk of recurrence in patients with triple-negative
breast cancer. Cancer Res. 79:3466–3478. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jie H, Ma W and Huang C: Diagnosis,
prognosis, and treatment of triple-negative breast cancer: A
review. Breast Cancer (Dove Med Press). 17:265–274. 2025.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mateo J, Chakravarty D, Dienstmann R,
Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, Ng CKY, Bedard PL,
Tortora G, Douillard JY, et al: A framework to rank genomic
alterations as targets for cancer precision medicine: The ESMO
scale for clinical actionability of molecular targets (ESCAT). Ann
Oncol. 29:1895–1902. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Condorelli R, Mosele F, Verret B, Bachelot
T, Bedard PL, Cortes J, Hyman DM, Juric D, Krop I, Bieche I, et al:
Genomic alterations in breast cancer: Level of evidence for
actionability according to ESMO scale for clinical actionability of
molecular targets (ESCAT). Ann Oncol. 30:365–373. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bianchini G, Balko JM, Mayer IA, Sanders
ME and Gianni L: Triple-negative breast cancer: Challenges and
opportunities of a heterogeneous disease. Nat Rev Clin Oncol.
13:674–690. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Masci D, Naro C, Puxeddu M, Urbani A,
Sette C, La Regina G and Silvestri R: Recent advances in drug
discovery for triple-negative breast cancer treatment. Molecules.
28(7513)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Baden J, Zhao C, Pratt J, Kirov S, Pant A,
Seminara A, Green G, Bilke S, Deras I, Fabrizio DA and Pawlowski T:
90PD-Comparison of platforms for determining tumour mutational
burden (TMB) in patients with non-small cell lung cancer (NSCLC).
Ann Oncol. 30 (Suppl 5)(v25)2019.
|
|
16
|
Wei B, Kang J, Kibukawa M, Arreaza G,
Maguire M, Chen L, Qiu P, Lang L, Aurora-Garg D, Cristescu R and
Levitan D: Evaluation of the TruSight oncology 500 assay for
routine clinical testing of tumor mutational burden and clinical
utility for predicting response to pembrolizumab. J Mol Diagn.
24:600–608. 2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu CY, Huang CC, Tsai YF, Chao TC, Lien
PJ, Lin YS, Feng CJ, Chen JL, Chen YJ, Chiu JH, et al: VGH-TAYLOR:
Comprehensive precision medicine study protocol on the
heterogeneity of Taiwanese breast cancer patients. Future Oncol:
Oct 19, 2021 (Epub ahead of print).
|
|
18
|
Huang CC, Tsai YF, Liu CY, Chao TC, Lien
PJ, Lin YS, Feng CJ, Chiu JH, Hsu CY and Tseng LM: Comprehensive
molecular profiling of Taiwanese breast cancers revealed potential
therapeutic targets: prevalence of actionable mutations among 380
targeted sequencing analyses. BMC Cancer. 21(199)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang CC, Tsai YF, Liu CY, Lien PJ, Lin
YS, Chao TC, Feng CJ, Chen YJ, Lai JI, Phan NN, et al: Prevalence
of tumor genomic alterations in homologous recombination repair
genes among taiwanese breast cancers. Ann Surg Oncol. 29:3578–3590.
2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Huang CC, Tsai YF, Liu CY, Lien PJ, Lin
YS, Chao TC, Feng CJ, Chen YJ, Lai JI, Cheng HF, et al: Concordance
of targeted sequencing from circulating tumor DNA and paired tumor
tissue for early breast cancer. Cancers (Basel).
15(4475)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chao TC, Tsai YF, Liu CY, Lien PJ, Lin YS,
Feng CJ, Chen YJ, Lai JI, Hsu CY, Lynn JJ, et al: Prevalence of
PIK3CA mutations in Taiwanese patients with breast cancer: A
retrospective next-generation sequencing database analysis. Front
Oncol. 13(1192946)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gradishar WJ, Moran MS, Abraham J,
Abramson V, Aft R, Agnese D, Allison KH, Anderson B, Burstein HJ,
Chew H, et al: NCCN guidelines® insights: Breast cancer,
version 4.2023. J Natl Compr Canc Netw. 21:594–608. 2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gennari A, André F, Barrios CH, Cortés J,
de Azambuja E, DeMichele A, Dent R, Fenlon D, Gligorov J, Hurvitz
SA, et al: ESMO clinical practice guideline for the diagnosis,
staging and treatment of patients with metastatic breast cancer.
Ann Oncol. 32:1475–1495. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
No authors listed. Pathologists' guideline
recommendations for immunohistochemical testing of estrogen and
progesterone receptors in breast cancer. Breast Care (Basel).
5:185–187. 2010.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wolff AC, Somerfield MR, Dowsett M,
Hammond MEH, Hayes DF, McShane LM, Saphner TJ, Spears PA and
Allison KH: Human epidermal growth factor receptor 2 testing in
breast cancer: ASCO-college of american pathologists guideline
update. J Clin Oncol. 41:3867–3872. 2023.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Schirmer M, Ijaz UZ, D'Amore R, Hall N,
Sloan WT and Quince C: Insight into biases and sequencing errors
for amplicon sequencing with the Illumina MiSeq platform. Nucleic
Acids Res. 43(e37)2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Li MM, Datto M, Duncavage EJ, Kulkarni S,
Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ,
Younes A and Nikiforova MN: Standards and guidelines for the
interpretation and reporting of sequence variants in cancer: A
joint consensus recommendation of the association for molecular
pathology, American society of clinical oncology, and college of
American pathologists. J Mol Diagn. 19:4–23. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6(pl1)2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chakravarty D, Gao J, Phillips SM, Kundra
R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et
al: OncoKB: A precision oncology knowledge base. JCO Precis Oncol.
2017(PO.17.00011)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Phan L, Zhang H, Wang Q, Villamarin R,
Hefferon T, Ramanathan A and Kattman B: The evolution of dbSNP: 25
Years of impact in genomic research. Nucleic Acids Res.
53:D925–D931. 2025.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chen S, Francioli LC, Goodrich JK, Collins
RL, Kanai M, Wang Q, Alföldi J, Watts NA, Vittal C, Gauthier LD, et
al: A genomic mutational constraint map using variation in 76,156
human genomes. Nature. 625:92–100. 2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Karczewski KJ, Weisburd B, Thomas B,
Solomonson M, Ruderfer DM, Kavanagh D, Hamamsy T, Lek M, Samocha
KE, Cummings BB, et al: The ExAC browser: Displaying reference data
information from over 60 000 exomes. Nucleic Acids Res.
45:D840–D845. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Robinson JT, Thorvaldsdottir H, Turner D
and Mesirov JP: igv.js: An embeddable JavaScript implementation of
the Integrative Genomics Viewer (IGV). Bioinformatics.
39(btac830)2023.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wolff L and Kiesewetter B: Applicability
of ESMO-MCBS and ESCAT for molecular tumor boards. Memo.
15:190–195. 2022.
|
|
36
|
Huang CC, Yeh YC, Cheng HF, Chen BF, Liu
CY, Tsai YF, Ho HL and Tseng LM: Comprehensive genomic profiling of
Taiwanese triple-negative breast cancer with a large targeted
sequencing panel. J Chin Med Assoc: Jun 20, 2025 (Epub ahead of
print).
|
|
37
|
Loderer D, Hornáková A, Tobiášová K,
Lešková K, Halašová E, Danková Z, Biringer K, Kúdela E, Rokos T,
Dzian A, et al: Comparison of next-generation sequencing quality
metrics and concordance in the detection of cancer-specific
molecular alterations between formalin-fixed paraffin-embedded and
fresh-frozen samples in comprehensive genomic profiling with the
Illumina® TruSight oncology 500 assay. Exp Ther Med.
29(64)2025.
|
|
38
|
Carpten JD, Faber AL, Horn C, Donoho GP,
Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage
S, et al: A transforming mutation in the pleckstrin homology domain
of AKT1 in cancer. Nature. 448:439–444. 2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kalinsky K, Hong F, McCourt CK, Sachdev
JC, Mitchell EP, Zwiebel JA, Doyle LA, McShane LM, Li S, Gray RJ,
et al: Effect of capivasertib in patients with an AKT1 E17K-mutated
tumor: NCI-MATCH subprotocol EAY131-Y nonrandomized trial. JAMA
Oncol. 7:271–278. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Turner NC, Oliveira M, Howell SJ, Dalenc
F, Cortes J, Gomez Moreno HL, Hu X, Jhaveri K, Krivorotko P, Loibl
S, et al: Capivasertib in hormone receptor-positive advanced breast
cancer. N Engl J Med. 388:2058–2070. 2023.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Daly GR, AlRawashdeh MM, McGrath J,
Dowling GP, Cox L, Naidoo S, Vareslija D, Hill ADK and Young L:
PARP inhibitors in breast cancer: A short communication. Curr Oncol
Rep. 26:103–113. 2024.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Szabo C, Masiello A, Ryan JF and Brody LC:
The breast cancer information core: Database design, structure, and
scope. Hum Mutat. 16:123–131. 2000.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chenevix-Trench G, Milne RL, Antoniou AC,
Couch FJ, Easton DF and Goldgar DE: CIMBA. An international
initiative to identify genetic modifiers of cancer risk in BRCA1
and BRCA2 mutation carriers: The consortium of investigators of
modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res.
9(104)2007.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Spain BH, Larson CJ, Shihabuddin LS, Gage
FH and Verma IM: Truncated BRCA2 is cytoplasmic: Implications for
cancer-linked mutations. Proc Natl Acad Sci USA. 96:13920–13925.
1999.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Tung NM, Robson ME, Ventz S, Santa-Maria
CA, Nanda R, Marcom PK, Shah PD, Ballinger TJ, Yang ES, Vinayak S,
et al: TBCRC 048: Phase II study of olaparib for metastatic breast
cancer and mutations in homologous recombination-related genes. J
Clin Oncol. 38:4274–4282. 2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Cheng HF, Tsai YF, Liu CY, Hsu CY, Lien
PJ, Lin YS, Chao TC, Lai JI, Feng CJ, Chen YJ, et al: Prevalence of
BRCA1, BRCA2, and PALB2 genomic alterations among 924 Taiwanese
breast cancer assays with tumor-only targeted sequencing: Extended
data analysis from the VGH-TAYLOR study. Breast Cancer Res.
25(152)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hyman DM, Piha-Paul SA, Won H, Rodon J,
Saura C, Shapiro GI, Juric D, Quinn DI, Moreno V, Doger B, et al:
HER kinase inhibition in patients with HER2- and HER3-mutant
cancers. Nature. 554:189–194. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
André F, Ciruelos EM, Juric D, Loibl S,
Campone M, Mayer IA, Rubovszky G, Yamashita T, Kaufman B, Lu YS, et
al: Alpelisib plus fulvestrant for PIK3CA-mutated, hormone
receptor-positive, human epidermal growth factor
receptor-2-negative advanced breast cancer: Final overall survival
results from SOLAR-1. Ann Oncol. 32:208–217. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Choudhury AD, Higano CS, de Bono JS, Cook
N, Rathkopf DE, Wisinski KB, Martin-Liberal J, Linch M, Heath EI,
Baird RD, et al: A phase I study investigating AZD8186, a potent
and selective inhibitor of PI3Kβ/δ, in patients with advanced solid
tumors. Clin Cancer Res. 28:2257–2269. 2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chen HJ, Romigh T, Sesock K and Eng C:
Characterization of cryptic splicing in germline PTEN intronic
variants in Cowden syndrome. Hum Mutat. 38:1372–1377.
2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Mester JL, Ghosh R, Pesaran T, Huether R,
Karam R, Hruska KS, Costa HA, Lachlan K, Ngeow J, Barnholtz-Sloan
J, et al: Gene-specific criteria for PTEN variant curation:
Recommendations from the ClinGen PTEN expert panel. Hum Mutat.
39:1581–1592. 2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Nykamp K, Anderson M, Powers M, Garcia J,
Herrera B, Ho YY, Kobayashi Y, Patil N, Thusberg J, Westbrook M, et
al: Sherloc: A comprehensive refinement of the ACMG-AMP variant
classification criteria. Genet Med. 19:1105–1117. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Garrido-Navas MC, García-Díaz A,
Molina-Vallejo MP, González-Martínez C, Alcaide Lucena M,
Cañas-García I, Bayarri C, Delgado JR, González E, Lorente JA and
Serrano MJ: The polemic diagnostic role of TP53 mutations in liquid
biopsies from breast, colon and lung cancers. Cancers (Basel).
12(3343)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhou W, Chen T, Chong Z, Rohrdanz MA,
Melott JM, Wakefield C, Zeng J, Weinstein JN, Meric-Bernstam F,
Mills GB and Chen K: TransVar: A multilevel variant annotator for
precision genomics. Nat Methods. 12:1002–1003. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Mosteiro M, Azuara D, Villatoro S, Alay A,
Gausachs M, Varela M, Baixeras N, Pijuan L, Ajenjo-Bauza M,
Lopez-Doriga A, et al: Molecular profiling and feasibility using a
comprehensive hybrid capture panel on a consecutive series of
non-small-cell lung cancer patients from a single centre. ESMO
Open. 8(102197)2023.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Fox EJ, Reid-Bayliss KS, Emond MJ and Loeb
LA: Accuracy of Next generation sequencing platforms. Next Gener
Seq Appl. 1(1000106)2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Song P, Chen SX, Yan YH, Pinto A, Cheng
LY, Dai P, Patel AA and Zhang DY: Selective multiplexed enrichment
for the detection and quantitation of low-fraction DNA variants via
low-depth sequencing. Nat Biomed Eng. 5:690–701. 2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Apostoli AJ and Ailles L: Clonal evolution
and tumor-initiating cells: New dimensions in cancer patient
treatment. Crit Rev Clin Lab Sci. 53:40–51. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Edsjö A, Gisselsson D, Staaf J, Holmquist
L, Fioretos T, Cavelier L and Rosenquist R: Current and emerging
sequencing-based tools for precision cancer medicine. Mol Aspects
Med. 96(101250)2024.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cocco E, Scaltriti M and Drilon A: NTRK
fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin
Oncol. 15:731–747. 2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Mosele MF, Westphalen CB, Stenzinger A,
Barlesi F, Bayle A, Bièche I, Bonastre J, Castro E, Dienstmann R,
Krämer A, et al: Recommendations for the use of next-generation
sequencing (NGS) for patients with advanced cancer in 2024: A
report from the ESMO precision medicine working group. Ann Oncol.
35:588–606. 2024.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Gradishar WJ, Moran MS, Abraham J,
Abramson V, Aft R, Agnese D, Allison KH, Anderson B, Bailey J,
Burstein HJ, et al: Breast cancer, version 3.2024, NCCN clinical
practice guidelines in oncology. J Natl Compr Canc Netw.
22:331–357. 2024.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kang J, Na K, Kang H, Cho U, Kwon SY,
Hwang S and Lee A: Prediction of homologous recombination
deficiency from oncomine comprehensive assay plus correlating with
SOPHiA DDM HRD solution. PLoS One. 19(e0298128)2024.PubMed/NCBI View Article : Google Scholar
|