Introduction
Rheumatoid arthritis-associated interstitial lung
disease (RA-ILD) is a serious extra-articular manifestation of RA
that significantly increases morbidity and mortality.
Epidemiological studies indicate that 10-40% of patients with RA
develop ILD, with mortality rates up to 3-fold higher than those
with RA without pulmonary involvement (1,2).
RA-ILD is marked by progressive pulmonary fibrosis and
inflammation, ultimately leading to respiratory failure and reduced
survival. It accounts for 10-20% of RA-related deaths (3,4). The
high prevalence and poor prognosis highlight the urgent need for
improved understanding of its pathogenesis and more effective
management strategies.
The pathogenesis of RA-ILD remains poorly
understood. Diagnostic delays are common due to nonspecific early
symptoms and the absence of validated screening protocols, which
often result in treatment initiation only after irreversible lung
damage has occurred (5-7).
Current evidence points to a multifactorial etiology involving
genetic susceptibility (for example, HLA-DRB1 alleles),
environmental exposures (for example, smoking), and autoimmune
dysregulation (8,9). Molecular mechanisms driving RA-ILD
include abnormal immune activation (for example, TNF-α and IL-6),
fibroblast proliferation and dysregulated extracellular matrix
deposition-pathways that parallel those in idiopathic pulmonary
fibrosis (IPF) (10-12).
Emerging evidence implicates neutrophil extracellular traps (NETs),
YKL-40 and KL-6 in lung injury, while signaling pathways such as
AKT/TMEM175 and JAK-STAT may contribute to fibrosis progression
(13-16).
However, the exact molecular drivers linking RA to ILD remain
unclear, and currently available biomarkers (for example, KL-6 and
YKL-40) lack sufficient specificity for broad clinical application
(13). Management primarily relies
on immunosuppressive agents (for example, methotrexate and
rituximab) and antifibrotic therapies (for example, nintedanib and
pirfenidone), but their effectiveness is limited by heterogeneous
patient responses and the lack of RA-ILD-specific targeted
treatments (12,17). For instance, antifibrotic agents
have demonstrated benefit in IPF, their effectiveness in RA-ILD has
been inconsistent, likely due to distinct underlying molecular
pathways (18). These challenges
highlight the urgent need for precision medicine strategies
tailored to the unique pathogenesis of RA-ILD (19,20).
Elucidating the molecular mechanisms underlying
RA-ILD is crucial for the discovery of novel therapeutic targets.
Emerging evidence implicates that dysregulated immune pathways
(such as IL-36 and NETs), aberrant fibroblast activation and
interactions between the gut and lungs play a role in disease
progression (11,21). For example, targeting NETs may help
mitigate lung injury, while modulation of the gut microbiota could
attenuate systemic inflammation (9,16).
Despite these advances, our understanding of RA-ILD pathogenesis
remains incomplete, limiting the development of effective targeted
therapies. The present study aimed to identify novel molecular
mechanisms that could inform the design of disease-modifying
strategies for RA-ILD.
Materials and methods
Study design and population
The present study was conducted as part of a
multi-center cohort that includes participants from Beijing Haidian
Hospital and Beijing Shunyi Hospital. The study population
consisted of three groups: (i) Patients diagnosed with RA-ILD, (ii)
patients with RA but without ILD and (iii) healthy individuals. All
patients with RA met the 2010 American College of
Rheumatology/European League Against Rheumatism classification
criteria for RA (22), while
patients with RA-ILD also met the diagnostic criteria for ILD
established by the American Thoracic Society/European Respiratory
Society (23). Health controls were
selected from individuals without inflammatory or rheumatic
diseases. All participants underwent pulmonary function tests and
chest high-resolution computed tomography (HRCT). Patients with RA
but without ILD and healthy controls showing pulmonary symptoms or
abnormal HRCT findings were excluded to ensure specificity. The
study protocol was approved by the Research Ethics Committees of
Beijing Haidian Hospital (approval no. 2024-006; Beijing, China)
and Beijing Shunyi Hospital (approval no. 2023k-021; Beijing,
China). Written informed consent was obtained from all participants
prior to enrollment. Blood samples were collected for further
analysis.
Antibody array assay
Serum proteins were analyzed in three cohorts:
Healthy controls (n=7), RA (n=40, with 10 subjects pooled), and
RA-ILD (n=40, with 10 subjects pooled). The clinical
characteristics of the participants are summarized in Table I. Serum cytokine profiling was
performed using the Human Cytokine Antibody Array (cat. no.
GSH-CAA-440; RayBiotech, Inc.), a high-throughput platform with 11
non-overlapping arrays for simultaneous detection of 440 cytokines.
Peripheral blood serum samples were diluted 1:2 with blocking
buffer and incubated overnight in array chambers coated with
cytokine-specific capture antibodies. After washing away unbound
proteins, a biotin-conjugated anti-cytokine antibody cocktail was
added to form antibody-cytokine-antibody sandwich complexes.
Cy3-conjugated streptavidin was then applied to amplify fluorescent
signals via biotin-streptavidin binding. All incubation steps were
conducted with 100 µl of reagents per well. Fluorescence intensity
was measured with an InnoScan 300 Microarray Scanner (Innopsys) at
optimized 532 nm excitation/emission wavelengths for Cy3. Raw
signal values were normalized against internal positive and
negative controls for assay reproducibility.
 | Table IClinical information of subjects for
the antibody array assay. |
Table I
Clinical information of subjects for
the antibody array assay.
|
Characteristics | RA-ILD | RA | Healthy | P-value (RA-ILD vs.
RA) |
|---|
| n | 40 | 40 | 7 | |
| Age, years | 62.5±9.5 | 59.2±6.9 | 60.1±2.1 | 0.074 |
| Sex (F/M) | 30/10 | 30/10 | 4/3 | |
| KL-6 (U/ml) | 647.0±221.4 | 178.6±42.2 | 181.6±55.0 | <0.001 |
| DAS28 | 4.2±1.5 | 3.3±7.9 | - | 0.007 |
| CRP (mg/l) | 2.3±2.2 | 1.0±1.6 | - | 0.004 |
| VAS | 34.8±31.9 | 11.9±16.7 | - | <0.001 |
| ESR (mm/h) | 45.4±22.3 | 31.6±19.1 | - | 0.004 |
| D-dimer (mg/l) | 1.3±0.7 | 0.4±0.4 | 0.16±0.1 | <0.001 |
| FVC (%) | 60.3±13.4 | 93.2±9.4 | - | <0.001 |
| DLCO (%) | 57.2±15.7 | 91.9±7.3 | - | <0.001 |
| FEV1 (%) | 59.4±14.0 | 90.2±6.3 | - | <0.001 |
| SLS I | 2.6±1.1 | 0±0 | - | <0.001 |
Enzyme-linked immunosorbent assays
(ELISAs)
Insulin and IL-31 were validated by ELISAs using
commercially available kits (Human Insulin ELISA Kit; cat. no.
ELH-Insulin; Human IL-31 ELISA Kit; cat. no. ELH-IL31; RayBiotech,
Inc.) with an expanded cohort consisting of 64 patients with
RA-ILD, 64 patients with RA and 40 healthy controls (demographics
provided in Table II). Briefly,
serum samples were incubated in antibody-precoated wells overnight
at 4˚C. Following washing steps, biotin-conjugated detection
antibodies were added and incubated for 2 h. Horseradish
peroxidase-conjugated streptavidin was then applied to bind the
biotinylated complexes for 45 min. The enzymatic reaction was
developed using tetramethylbenzidine substrate form 30 min. After
the reaction was stopped, absorbance was measured at 450 nm using
an ELx800NB microplate reader (BioTek; Agilent Technologies,
Inc.).
 | Table IIClinical information of subjects for
ELISA. |
Table II
Clinical information of subjects for
ELISA.
|
Characteristics | RA-ILD | RA | Healthy | P-value (RA-ILD vs.
RA) |
|---|
| n | 64 | 64 | 40 | |
| Age, years | 65.6±9.6 | 62.1±8.0 | 57.8±9.9 | 0.081 |
| Sex (F/M) | 40/24 | 51/13 | 23/17 | |
| KL-6 (U/ml) | 457.3±260.2 | 208.0±127.6 | - | <0.001 |
| DAS28 | 4.1±1.5 | 3.2±1.3 | - | <0.001 |
| CRP (mg/l) | 2.2±2.0 | 0.9±1.5 | - | <0.001 |
| VAS | 32.4±29.1 | 12.9±16.5 | - | <0.001 |
| ESR (mm/h) | 43.3±19.9 | 33.1±20.9 | - | <0.001 |
| D-dimer (mg/l) | 1.2±0.8 | 0.4±0.4 | - | <0.001 |
| FVC (%) | 63.3±13.5 | 92.1±7.7 | - | <0.001 |
| DLCO (%) | 61.2±15.1 | 91.5±6.8 | - | <0.001 |
| FEV1 (%) | 62.7±13.2 | 90.9±6.0 | - | <0.001 |
| SLS I | 6.5±5.6 | 0±0 | - | <0.001 |
Statistical analysis
Statistical comparisons between experimental groups
were conducted using a one-way ANOVA followed by Bonferroni's post
hoc test in SPSS version 20.0 (IBM Corp.). Differences were
considered statistically significant when meeting two criteria: (i)
P<0.05; (ii) a fold change threshold of <0.83
(downregulation) or >1.2 (upregulation). Continuous data were
expressed as the mean ± standard deviation (SD). Correlation
analyses between serum biomarkers (IL-31 and insulin) and clinical
parameters of RA-ILD severity were performed using GraphPad Prism
9.0 (GraphPad Software; Dotmatics). Pearson's correlation
coefficient (r) was calculated to assess linear relationships
between biomarkers and clinical parameters. P<0.05 was
considered to indicate a statistically significant difference.
Results
Proteomic screening identifies 20
proteins associated with RA and RA-ILD progression
A panel of 20 proteins demonstrated sequential
upregulation, significantly elevated in patients with RA compared
with healthy controls (P<0.05), and showing even greater
increases in patients with RA-ILD compared with patients with RA
(P<0.05) (Fig. 1). These
proteins included carbonic anhydrase IX (CA9), IL-1F9,
ectodysplasin A2 (EDA-A2), growth arrest-specific gene 1 (Gas1),
IL-2Rb, IL-7, amphiregulin (AR), sonic hedgehog N-terminal (Shh-N),
IGF-1R, MMP-7, insulin, Eotaxin, EpCAM, tissue factor pathway
inhibitor (TFPI), MIP-3b, CRTAM, IL-31, tissue factor (TF),
syndecan-1 and cathepsin L. The information of these 20 proteins is
summarized in Table III.
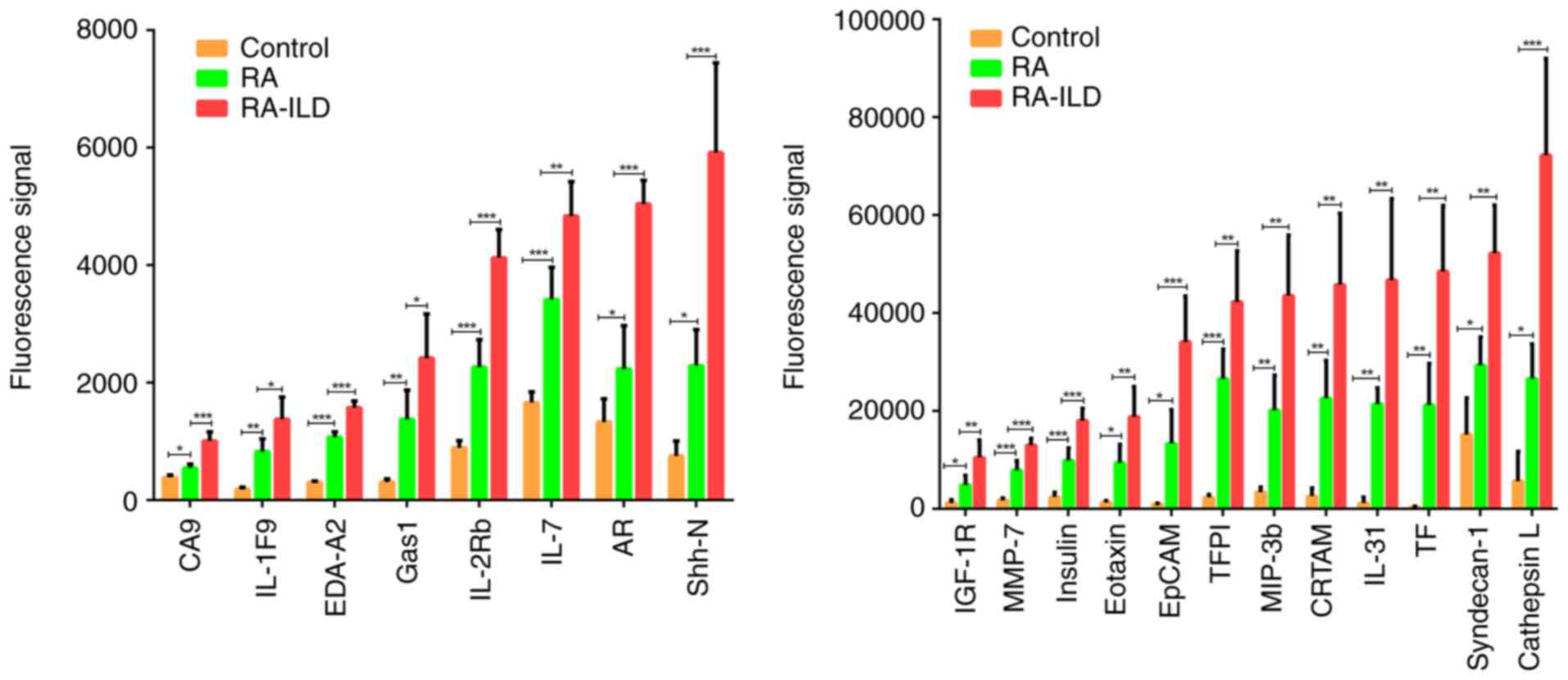 | Figure 1Histogram. Serum protein levels were
significantly higher in patients with RA compared with healthy
controls, and even more elevated in patients with RA-ILD compared
with patients with RA. The bar graph illustrates the relative
expression levels of 20 differentially expressed proteins among
healthy controls, patients with RA and patients with RA-ILD. The
data are presented as mean ± SD. The data from patients with RA and
patients with RA-ILD represent pooled samples.
*P<0.05, **P<0.01 and
***P<0.001. RA, rheumatoid arthritis; RA-ILD,
RA-associated interstitial lung disease; TFPI, tissue factor
pathway inhibitor; CA9, carbonic anhydrase IX; AR, amphiregulin;
Shh-N, sonic hedgehog N-terminal; Gas1, growth arrest-specific gene
1; EDA-A2, ectodysplasin A2; TF, tissue factor. |
 | Table IIIInformation on the 20 proteins. |
Table III
Information on the 20 proteins.
| Protein | Uniport ID | RA/Con | RA-ILD/RA | Known
functions | Novelty in
RA-ILD |
|---|
| CA9 | Q16790 | 1.4 | 1.8 | Response to
hypoxia | Induces hypoxic
fibrosis |
| IL-1F9 | Q9NZH8 | 4.5 | 1.7 | Interleukin-1
receptor binding | A proinflammatory
cytokine in the lung disease |
| EDA-A2 | Q92838 | 3.5 | 1.5 | A ligand activating
the DEATH-domain containing receptors EDAR and EDA2R | Activates the
inflammatory pathway NF-κB signaling |
| Gas1 | P54826 | 4.4 | 1.8 | Specific growth
arrest protein involved in growth suppression | Gas1/Axl
pathway |
| IL-2Rb | P14784 | 2.5 | 1.8 | Receptor for
interleukin-2 | Contributes to
immune microenvironment disorder |
| IL-7 | P13232 | 2.1 | 1.4 | Regulates B cell
proliferation | Exacerbates
IPF |
| AR | P15514 | 1.7 | 2.3 | Ligand of the EGF
receptor/EGFR | Involves into
inflammatory lung disease |
| Shh-N | Q15465 | 3.0 | 2.6 | Displays an
autoproteolysis and a cholesterol transferase activity | Promotes pulmonary
fibrosis |
| IGF-1R | P08069 | 3.9 | 2.2 | Receptor tyrosine
kinase which mediates actions of insulin-like growth factor 1 | Promotes IPF |
| MMP-7 | P09237 | 4.5 | 1.6 | Activates
pro-collagenase | A mediator of
extracellular matrix remodeling in IPF |
| Insulin | P01308 | 4.1 | 1.8 | Decreases blood
glucose concentration | Participates in
PI3K-Akt signaling |
| Eotaxin | P51671 | 7.0 | 2.0 | Promotes the
accumulation of eosinophils | Produces
profibrotic cytokines contributing to pulmonary fibrosis |
| EpCAM | P16422 | 14.0 | 2.6 | Plays a role in
embryonic stem cells proliferation and differentiation | Involves into
IPF |
| TFPI | P10646 | 11.3 | 1.6 | Inhibits
VIIa/tissue factor activity | Involves into
IPF |
| MIP-3b | Q99731 | 5.9 | 2.2 | Chemokine
activity | Involves into
IPF |
| CRTAM | O95727 | 8.8 | 2.0 | Mediates
heterophilic cell-cell adhesion | Activates STAT
signaling |
| IL-31 | Q6EBC2 | 18.3 | 2.2 | Activates
STAT3 | Activates JAK-STAT
signaling |
| TF | P13726 | 57.2 | 2.3 | Initiates blood
coagulation | Involves into
IPF |
| Syndecan-1 | P18827 | 1.9 | 1.8 | Cell surface
proteoglycan | Exacerbates
inflammation-to-fibrosis transitions in pulmonary fibrosis |
| Cathepsin L | P07711 | 4.7 | 2.7 | Regulates
CD4+ T cell positive selection | Involves into
IPF |
Classification capacity of the 20
proteins
To assess the discriminatory value of the 20
proteins, hierarchical clustering and principal component analysis
(PCA) were performed. Using normalized expression data from the
pooled analysis, hierarchical clustering of these proteins unveiled
unique expression patterns and demonstrated 100% accuracy in
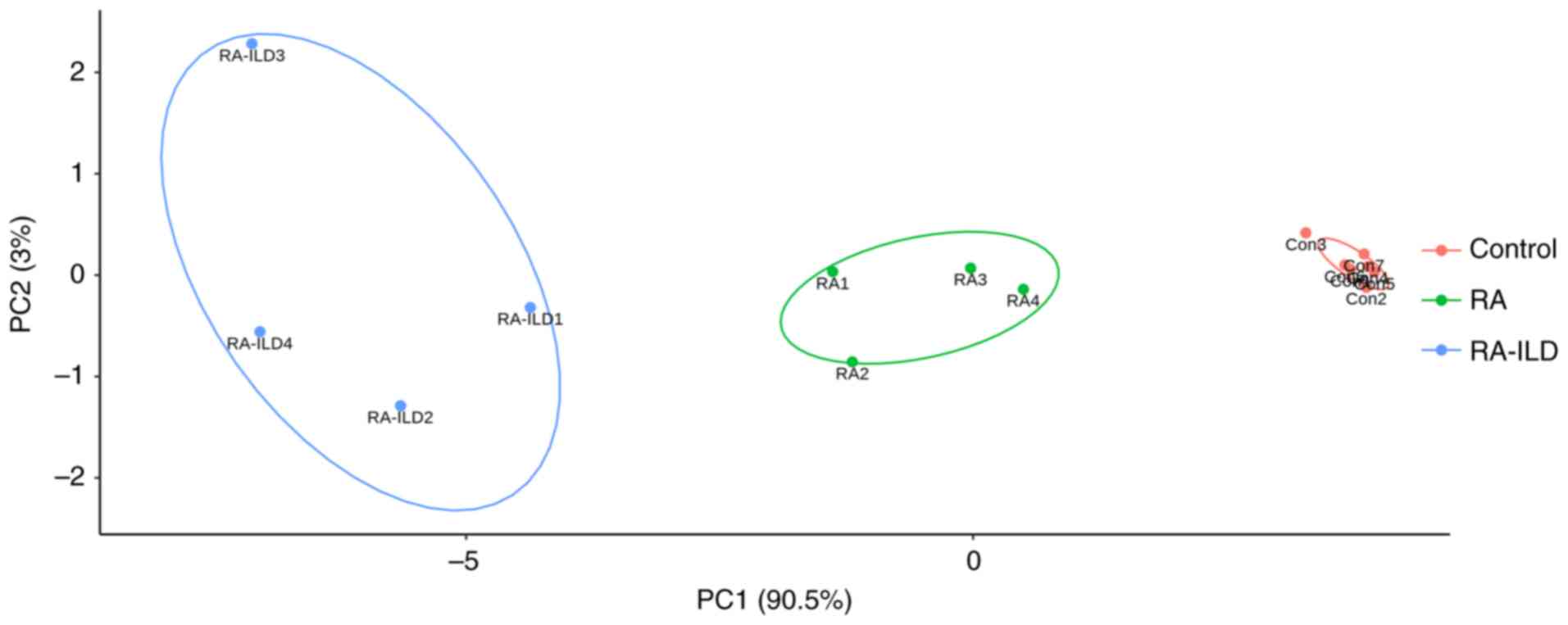
classifying RA-ILD from RA and the healthy groups (Fig. 2). PCA effectively segregated RA,
RA-ILD, and healthy controls into separate clusters within the
reduced-dimensional space (Fig.
3).
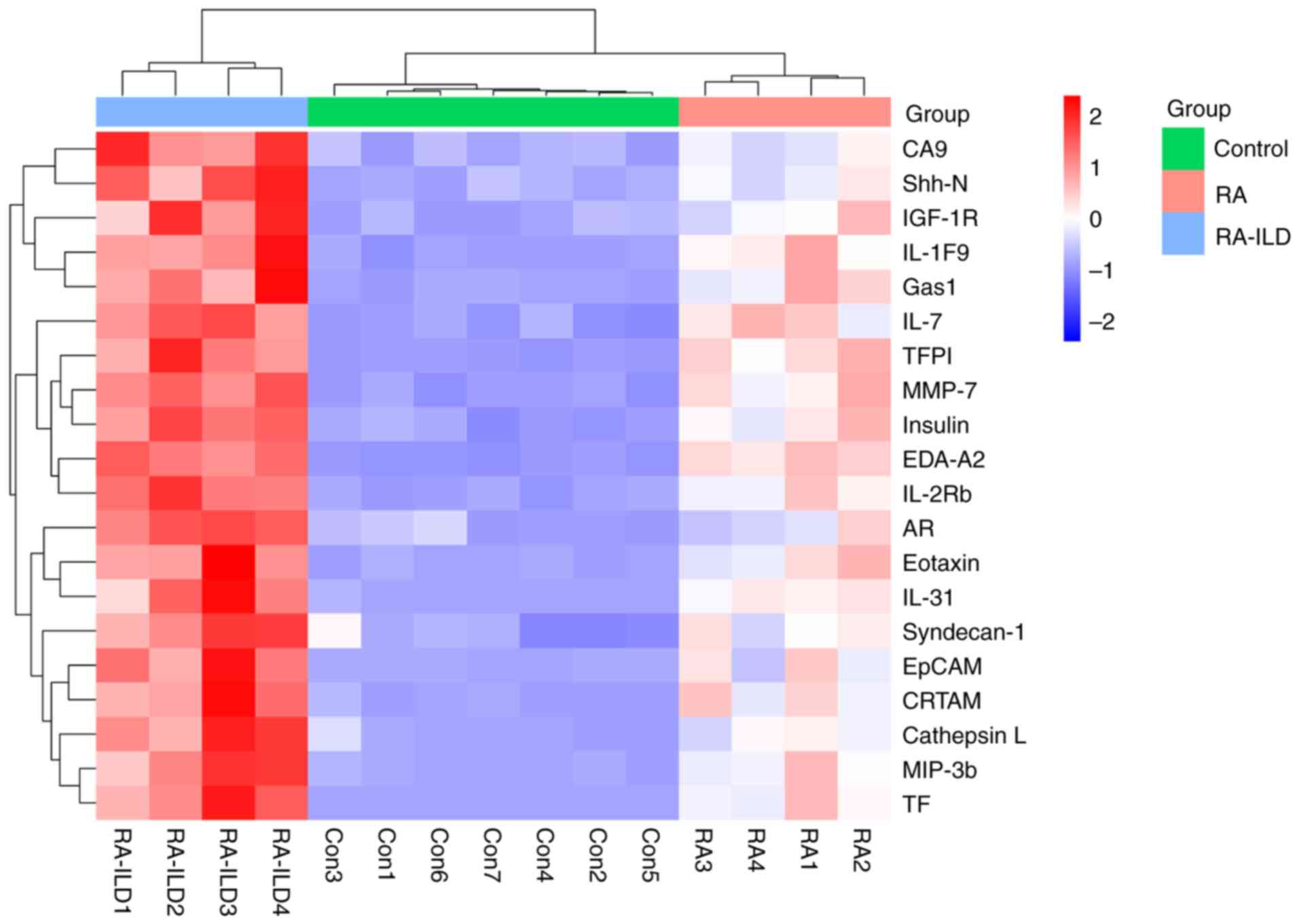 | Figure 2Heatmap. This heatmap represents
hierarchical clustering analysis based on the expression patterns
of 20 proteins, showing distinct molecular signatures among the
three groups. The color scale indicates normalized protein
expression levels (blue: low; red: high). The data from patients
with RA and patients with RA-ILD represent pooled samples. RA,
rheumatoid arthritis; RA-ILD, RA-associated interstitial lung
disease; TFPI, tissue factor pathway inhibitor; CA9, carbonic
anhydrase IX; AR, amphiregulin; Shh-N, sonic hedgehog N-terminal;
Gas1, growth arrest-specific gene 1; EDA-A2, ectodysplasin A2; TF,
tissue factor. |
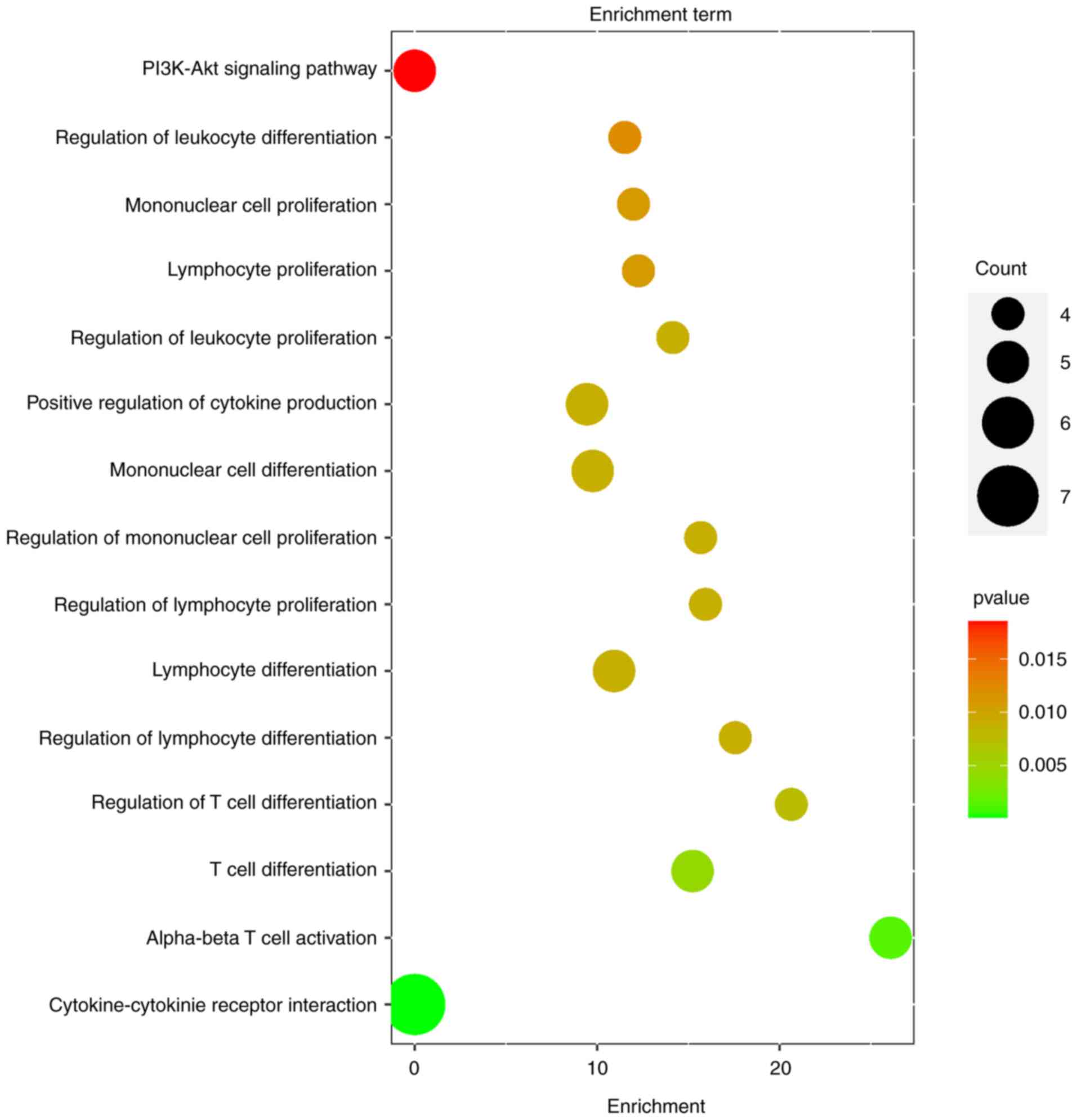
Functional enrichment analysis
Gene Ontology (GO) enrichment analysis revealed
critical biological processes associated with the 20 proteins,
including T cell differentiation, T cell activation, lymphocyte
differentiation, mononuclear cell proliferation, leukocyte
proliferation and positive regulation of cytokine production.
Further analysis using the Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway implicated two key pathways cytokine-cytokine
receptor interaction and the PI3K-Akt signaling pathway (Fig. 4).
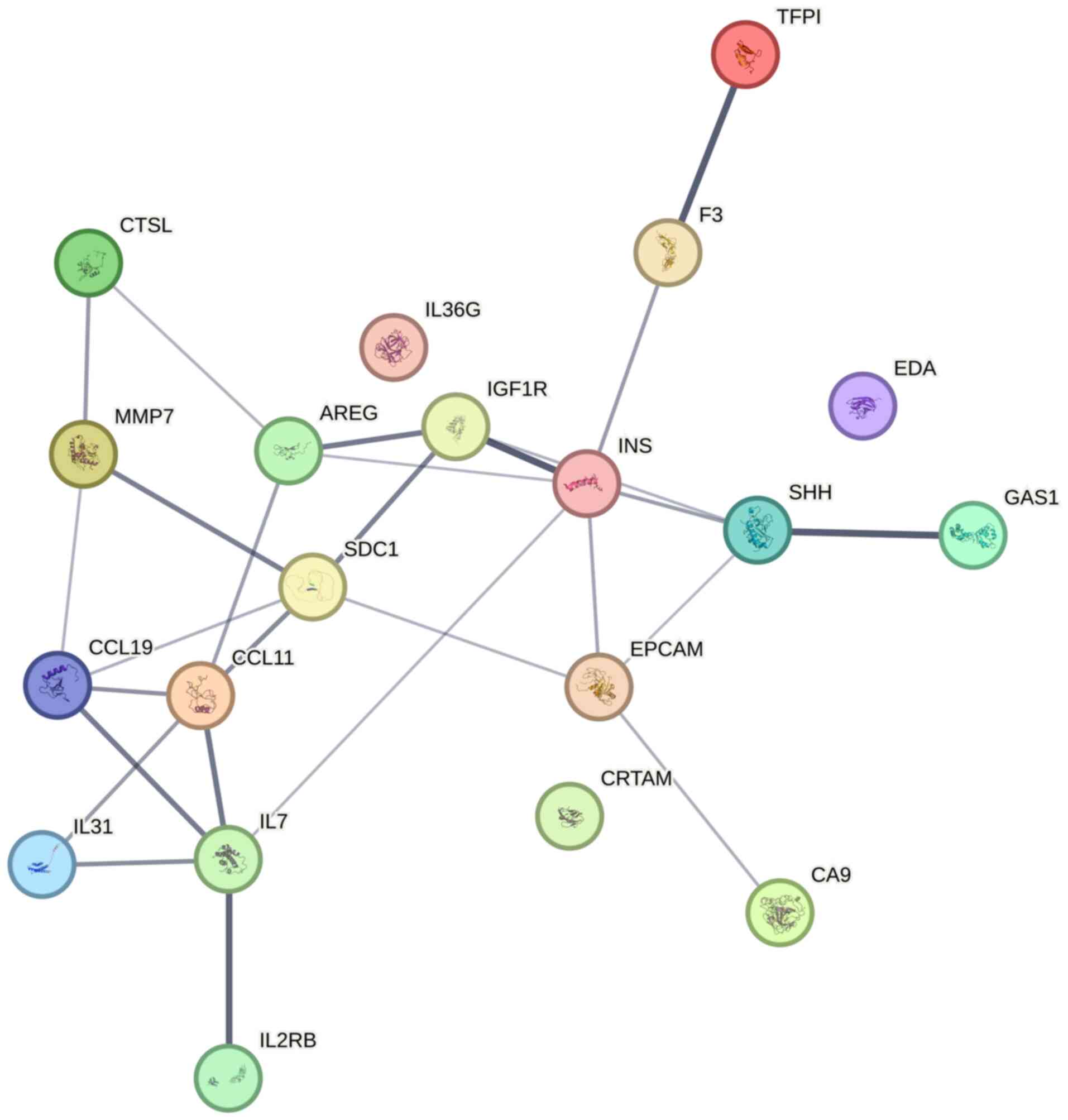
Protein-protein interaction (PPI)
analysis
PPI network analysis identified insulin as the
highest-degree hub protein (node degree=6), showing direct
interactions with multiple profibrotic and immunomodulatory
mediators: IGF-1R, Shh-N, AR, TF, IL-7 and EpCAM (Fig. 5). This indicates its central role in
coordinating molecular crosstalk within the RA-ILD proteomic
landscape.
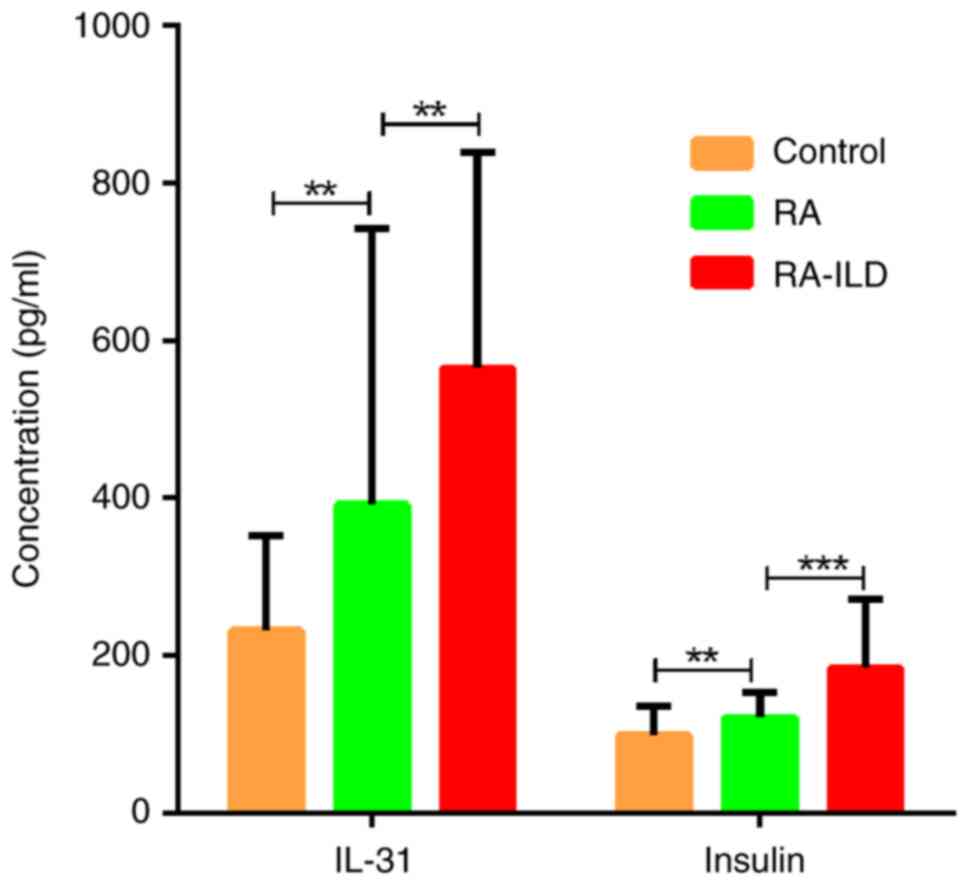
ELISA validation and clinical value
analysis
The independent validation of insulin and IL-31 with
an expanded cohort by ELISA demonstrated strong concordance with
the initial antibody array data (Fig.
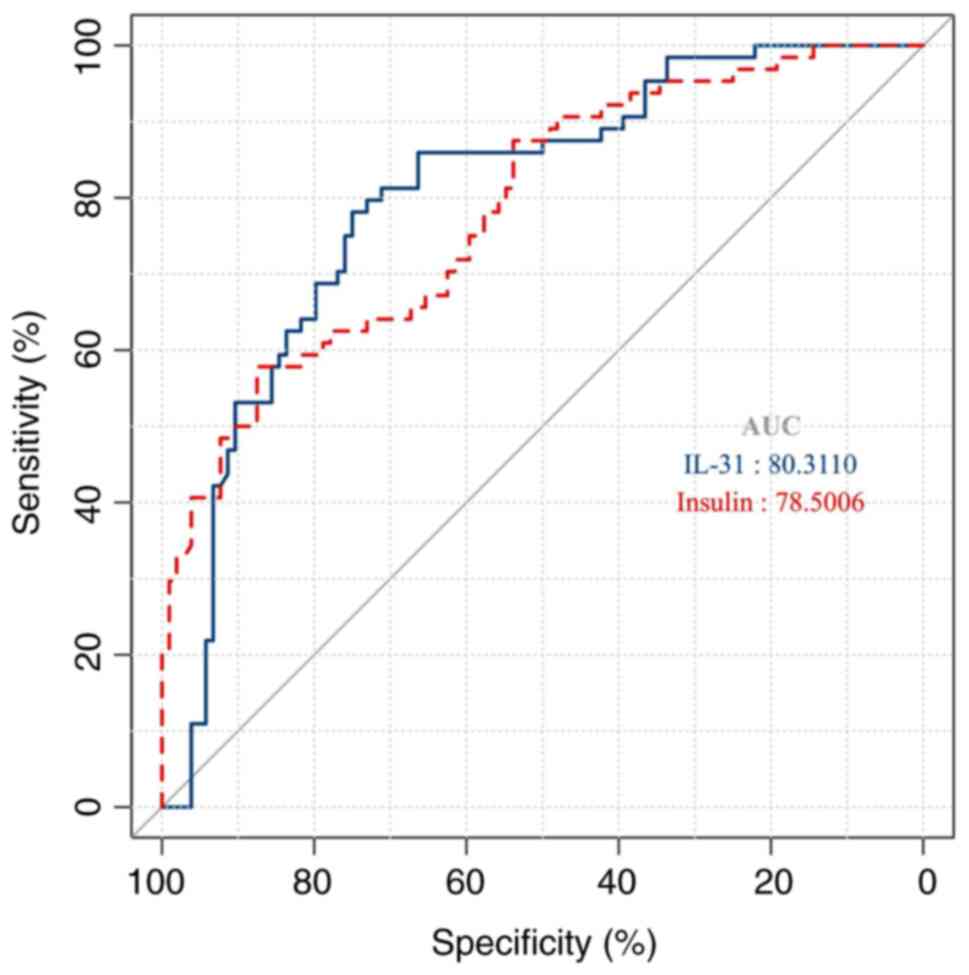
6). The diagnostic potential of these two biomarkers showed
excellent discriminatory capacity with area under the curve values
of >75% (Fig. 7). Notably, the
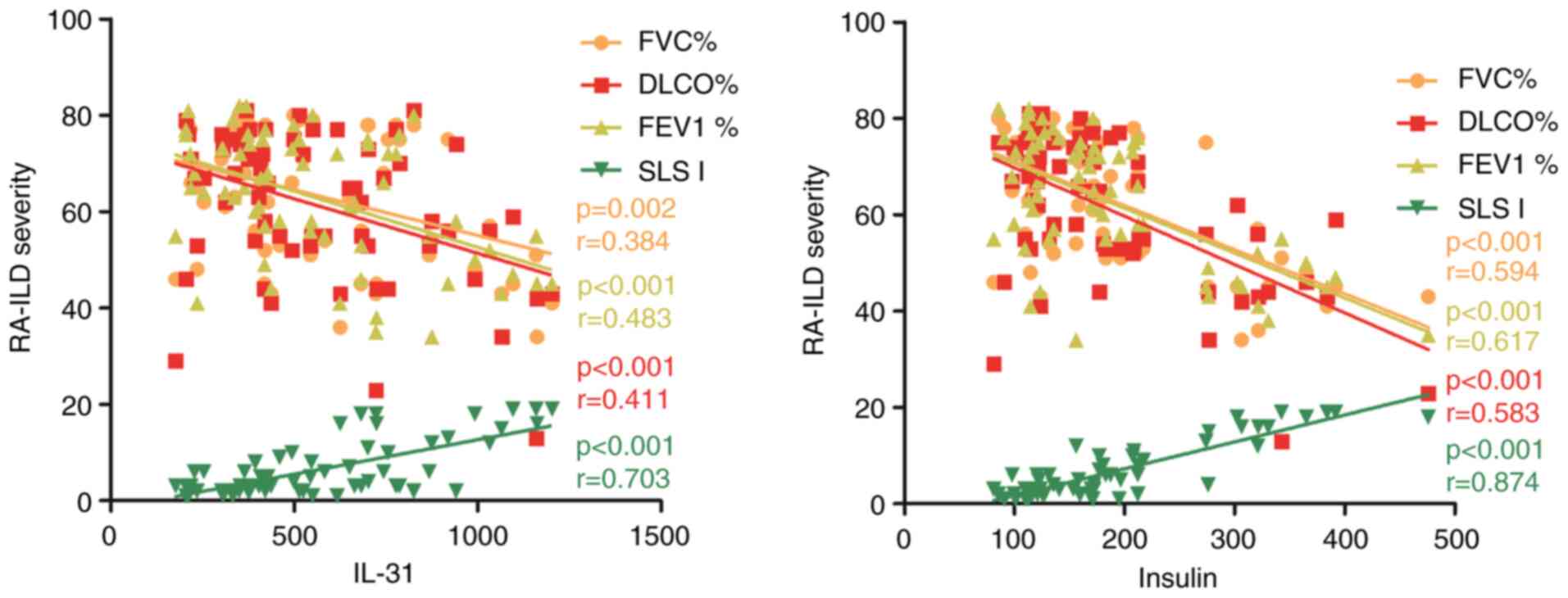
correlation analysis revealed a strong positive correlation between
these two biomarkers and the Scleroderma Lung Study I score of HRCT
images (Fig. 8, r>0.6). This
finding suggests that insulin and IL-31 may play a role in the
disease activity of RA-ILD.
Discussion
RA-ILD is a severe extra-articular manifestation of
RA, characterized by progressive pulmonary fibrosis and high
morbidity. Its etiology is multifactorial and complex, involving
genetic predisposition (for example, MUC5B promoter
variants), autoimmune-driven inflammation, aberrant fibroblast
activation, and environmental exposures such as smoking. The
interplay of these factors leads to dysregulated immune responses
and irreversible lung remodeling (10). However, the precise molecular
mechanisms triggering these immune responses remain elusive,
limiting the development of effective targeted therapies for
RA-ILD. In the present study, 20 proteins including CA9, IL-1F9,
EDA-A2, Gas1, IL-2Rb, IL-7, AR, Shh-N, IGF-1R, MMP-7, insulin,
Eotaxin, EpCAM, TFPI, MIP-3b, CRTAM, IL-31, TF, Syndecan-1 and
Cathepsin L were identified, which were significantly elevated in
patients with RA compared with healthy controls, with further
upregulation in RA-ILD relative to RA alone. The stepwise increase
in these proteins suggests their contribution to RA pathogenesis
and progression to ILD, the most common and serious pulmonary
complication of RA.
GO analysis revealed that these 20 proteins were
enriched in immune-related processes, including T cell
differentiation, T cell activation and cytokine production. These
findings align with prior studies implicating aberrant T cell
responses and cytokine dysregulation in RA-ILD pathogenesis
(24-26).
KEGG pathway analysis highlighted the cytokine-cytokine receptor
interaction pathway and PI3K-Akt signaling pathway, both of which
play key roles in immune cell proliferation and differentiation in
ILD (27). The PI3K-Akt pathway is
known to drive fibroblast activation and fibrosis in ILD (28). Moreover, cytokine-receptor
interactions -such as IL-7/IL-7R, IGF-1R/insulin-may perpetuate
chronic inflammation and fibrogenesis (29,30).
Notably, Wu et al (31)
reported that the AhR/IGF1R axis contributes to the development of
IPF through activation of the TGF-β/Smad/STAT signaling cascade. In
the present study, KEGG pathway analysis revealed that IGF-1R and
insulin were involved in the PI3K-Akt signaling pathway, suggesting
a potential role for the insulin/IGF-1R axis in the progression of
RA to ILD through this pathway. It was hypothesized that the
insulin/IGF1R axis could be a promising therapeutic target for the
treatment of RA-ILD. PPI analysis revealed that insulin is a
central hub protein that interacts with IGF-1R, Shh-N, AR, TF, IL-7
and EpCAM. These interactions suggest that insulin and its network
partners may collectively regulate immune and metabolic pathways in
RA-ILD, potentially exacerbating disease progression. While no
previous studies have directly linked insulin to RA-ILD, its
network prominence and functional interactions generate a strong
interest in investigating the modulation of the insulin pathway as
a potential therapeutic strategy. Further validation is needed to
confirm the potential involvement of insulin in RA-ILD.
Furthermore, among these 20 proteins, several have
well-established roles in RA or pulmonary fibrosis. For example,
IL-7 promotes T cell survival and Th17 differentiation (29), and its elevation has been linked to
the exacerbation of IPF (32).
MMP-7 serves as a predictive biomarker of disease progression and
mediates extracellular matrix remodeling in IPF (33). IL-1F9 acts as a proinflammatory
cytokine in lung disease by enhancing chemokine production and
inflammatory cell recruitment (34). Eotaxin is associated with increased
pulmonary infiltration of eosinophils and neutrophils, as well as
the production of profibrotic cytokines contributing to pulmonary
fibrosis (35). Both cathepsin L
and tissue factor have been implicated in the pathogenesis of IPF
and ILD (36,37). AR is elevated in inflammatory lung
disease associated with RA (38),
and EpCAM, TFPI and MIP-3b are upregulated in IPF (39-41).
Shh-N promotes pulmonary fibrosis through the hedgehog signaling
pathway (42), and Syndecan-1
shedding exacerbates the transition from inflammation to fibrosis
by releasing heparan sulfate-bound growth factors (43).
Notably, CA9, EDA-A2, Gas1, CRTAM, IL-2Rb and IL-31
have emerged as novel candidates, with no prior studies linking
them to RA-ILD or fibrotic diseases. However, chronic inflammation
is a well-established precursor to fibrotic tissue remodeling
(44), suggesting that these
proteins may contribute indirectly to fibrosis through sustained
inflammatory signaling. Among these novel targets, EDA-A2 activates
the inflammatory responses through NF-κB signaling by binding to
the EDA receptor (45). CRTAM
promotes STAT signaling via STAT1 phosphorylation (46). IL-2Rb contributes to immune
microenvironment disorder by disrupting the Th1/Th2 cell
differentiation balance (47), and
IL-31 drives inflammation primarily through JAK-STAT pathway
activation (48). Gas1 may play a
critical role in fibrotic diseases, including RA-ILD through the
Gas1/Axl axis, analogous to the pro-fibrotic Gas6/Axl signaling
pathway (49,50). In addition, CA9, a hypoxia-inducible
protein, may promote hypoxic-associated fibrosis (51). These findings indicate the potential
of these novel candidates to drive RA-ILD progression through their
roles in inflammatory and hypoxic signaling, warranting further
investigation.
However, the present study has several limitations.
First, the cohort included a relatively small number of healthy
controls (n=7), which limited the statistical power to detect
biologically relevant differences between healthy individuals and
patient groups. Second, the use of pooled samples for the antibody
array analysis-implemented due to cost constraints associated with
high-throughput proteomic screening-represents a significant
methodological limitation. Third, the present study only validated
insulin (the highest-degree hub protein) and IL-31 (a novel
candidate) using independent methods due to budget limitations.
Fourth, the functional roles of the identified protein signatures
were not experimentally validated. Future studies should prioritize
individual sample analysis, larger cohort sizes, and rigorous
validation of all these candidate proteins as potential biomarkers.
In addition, in vitro and in vivo studies are
warranted to elucidate the functional relevance of these proteins,
particularly those with novel associations, and to explore their
potential as therapeutic targets for RA-ILD.
In conclusion, our study highlights the complex
molecular interplay underlying RA-ILD pathogenesis. A total of 20
dysregulated proteins that collectively drive immune dysregulation
and fibrotic progression were identified. These proteins are
functionally enriched in critical pathways, including T cell
differentiation, cytokine-cytokine receptor interactions (such as
IL-7/IL-7R and IGF-1R/Insulin), and the PI3K-Akt signaling pathway.
The PPI network identifies insulin as a central hub, interacting
with multiple profibrotic mediators (IGF-1R, Shh-N and AR) and
immune modulators (IL-7, EpCAM), suggesting its pivotal role in
orchestrating profibrotic and inflammatory responses in RA-ILD.
While several proteins (MMP-7, IL-1F9 and Cathepsin L) have
established roles in pulmonary fibrosis, our identification of
novel candidates points to additional mechanisms involving
sustained inflammatory signaling and hypoxia-responsive pathways.
These findings provide new insight into RA-ILD pathogenesis and
suggest that targeting the insulin/IGF1R-PI3K-Akt axis may
represent a promising therapeutic strategy to disrupt the
immune-fibrotic cascade in this disease. Further validation and
mechanistic studies are warranted to explore these potential
targets.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the 2024 Haidian
Health Development Research, Cultivation Plan Project (grant no.
HP2024-30-101004) and the Beijing Shunyi District Hospital Research
and Development Special Fund (grant no. 2025Y01).
Availability of data and materials
The data generated in the present study may be found
in zenodo database under accession number 15852072 or at the
following URL: (https://zenodo.org/records/15852072).
Authors' contributions
WC conducted all experiments and wrote the first
draft of the manuscript. YZ and QY contributed to sample collection
and processing. BY and QY conducted statistical analyses. GZ
contributed to conception and design of the present study, and
revised the manuscript. WC and GZ confirm the authenticity of all
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Approval was obtained from the Research Ethics
Committees of Beijing Haidian Hospital (approval no. 2024-006;
Beijing, China) and Beijing Shunyi Hospital (approval no.
2023-k-021; Beijing, China). All participants provided informed
consent to participate in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang M, Yin J and Zhang X: Factors
associated with interstitial lung disease in patients with
rheumatoid arthritis: A systematic review and meta-analysis. PLoS
One. 18(e0286191)2023.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Farquhar HJ, Beckert L, Edwards AL,
Matteson EL, Frampton CMA, Ganly E, Yetton R, Thiessen R, Haslett
J, Bucknall D and Stamp LK: Rheumatoid interstitial lung disease in
Canterbury, Aotearoa New Zealand-A retrospective cohort study.
Semin Arthritis Rheum. 64(152359)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang HF, Wang YY, Li ZY, He PJ, Liu S and
Li QS: The prevalence and risk factors of rheumatoid
arthritis-associated interstitial lung disease: A systematic review
and meta-analysis. Ann Med. 56(2332406)2024.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Juge PA, Wemeau L, Ottaviani S, Desjeux G,
Zhuo J, Vannier-Moreau V, Flipo RM, Crestani B and Dieudé P:
Increased mortality in patients with RA-associated interstitial
lung disease: Data from a French administrative healthcare
database. RMD Open. 9(e003491)2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sullivan DI and Ascherman DP: Rheumatoid
arthritis-associated interstitial lung disease (RA-ILD): Update on
prevalence, risk factors, pathogenesis, and therapy. Curr Rheumatol
Rep. 26:431–449. 2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Narváez J: Moving forward in Rheumatoid
arthritis-associated interstitial lung disease screening. J Clin
Med. 13(5385)2024.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Koduri G and Solomon JJ: Identification,
monitoring, and management of rheumatoid arthritis-associated
interstitial lung disease. Arthritis Rheumatol. 75:2067–2077.
2023.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu M, Jiang Z, Liu M, Ni H, Li Y, Fang J,
Du Q and Dong Y: SLAMF1 as a novel molecule mediating the causal
association between rheumatoid arthritis and interstitial lung
disease: A Mendelian randomization study combined with
transcriptomics and in vivo validation. Int Immunopharmacol. 142
(Pt A)(113082)2024.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhong X, Wang X, Xu L, Zhang J, Yu W, Ji
L, Huang J, Zhong X, Zhang J and Long L: Alterations in gut
microbiota in Rheumatoid arthritis patients with interstitial lung
Disease: A Comparative study. Hum Immunol.
86(111239)2025.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kim Y, Yang HI and Kim KS: Etiology and
pathogenesis of rheumatoid arthritis-interstitial lung disease. Int
J Mol Sci. 24(14509)2023.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zheng W, Hu X, Zou M, Hu N, Song W, Wang
R, Liu Y, Hou Q, Liu Y, Chen X and Cheng Z: Plasma IL-36α and
IL-36γ as potential biomarkers in interstitial lung disease
associated with rheumatoid arthritis: A pilot study in the Chinese
population. Inflammation. 46:285–296. 2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bes C, Köybaşı G, İçaçan OC, Yalçın Mutlu
M and Yıldırım F: Antifibrotic therapies in rheumatoid arthritis
associated interstitial lung disease. Eur J Rheumatol. 9:176–179.
2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Liang B, Zhang Y, Ke D, Yan R, Jiang MN,
Li L, Zhang LX, Zhao XG, Yuan GP, Xu B and Liu XM: Serum YKL-40 and
Serum Krebs von den lungen-6 as potential predictive biomarkers for
rheumatoid arthritis-associated interstitial lung disease. Immunol
Invest. 53:989–1000. 2024.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Liu N, Fan X, Shao Y, Chen S, Wang T, Yao
T and Chen X: Resveratrol attenuates inflammation and fibrosis in
rheumatoid arthritis-associated interstitial lung disease via the
AKT/TMEM175 pathway. J Transl Med. 22(457)2024.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liu H, Yang Y, Zhang J and Li X:
Baricitinib improves pulmonary fibrosis in mice with rheumatoid
arthritis-associated interstitial lung disease by inhibiting the
Jak2/Stat3 signaling pathway. Adv Rheumatol. 63(45)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Venetsanopoulou AI, Ntinopoulou M,
Papagianni E, Koletsos N, Voulgari PV and Chrysanthopoulou A:
Neutrophil extracellular traps as immunofibrotic mediators in
RA-ILD; pilot evaluation of the nintedanib therapy. Front Immunol.
15(1480594)2024.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Shoda T, Kotani T, Mitsuhiro K, Yoshikawa
A, Wada Y, Makino H, Osuga K and Takeuchi T: The therapeutic
efficacy of abatacept for rheumatoid arthritis-associated
interstitial lung disease: Insights from a 12-month trial using
semi-quantitative chest high-resolution computed tomography
imaging. J Clin Med. 13(5871)2024.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang S, Liu M, Li X, Zhang J, Wang F,
Zhang C, Roden A, Ryu JH, Warrington KJ, Sun J, et al: Canonical
and noncanonical regulatory roles for JAK2 in the pathogenesis of
rheumatoid arthritis-associated interstitial lung disease and
idiopathic pulmonary fibrosis. FASEB J. 36(e22336)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Rodríguez Portal JA, Brito García N, Díaz
Del Campo Fontecha P, Valenzuela C, Ortiz AM, Nieto MA,
Mena-Vázquez N, Cano-Jiménez E, Castellví I, Aburto M, et al:
SER-SEPAR recommendations for the management of rheumatoid
arthritis-related interstitial lung disease. Part 1: Epidemiology,
risk factors and prognosis. Reumatol Clin (Engl Ed). 18:443–452.
2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Albrecht K, Strangfeld A, Marschall U and
Callhoff J: Interstitial lung disease in rheumatoid arthritis:
Incidence, prevalence and related drug prescriptions between 2007
and 2020. RMD Open. 9(e002777)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Huang Z, Wu T, Lu R, Zhou H, Zhang Y,
Huang L, Gan Y and He H: Prevalence and clinical significance of
anti-neutrophil cytoplasmic antibodies in rheumatoid
arthritis-associated interstitial lung disease. BMC Pulm Med.
25(177)2025.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Aletaha D, Neogi T, Silman AJ, Funovits J,
Felson DT, Bingham CO III, Birnbaum NS, Burmester GR, Bykerk VP,
Cohen MD, et al: 2010 Rheumatoid arthritis classification criteria:
An American college of rheumatology/European league against
rheumatism collaborative initiative. Arthritis Rheum. 62:2569–2581.
2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Raghu G, Remy-Jardin M, Myers JL, Richeldi
L, Ryerson CJ, Lederer DJ, Behr J, Cottin V, Danoff SK, Morell F,
et al: Diagnosis of idiopathic pulmonary fibrosis. An Official
ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Crit Care
Med. 198:e44–e68. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kono M: New insights into the metabolism
of Th17 cells. Immunol Med. 46:15–24. 2023.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang Y, Zhu J, Xiao K, Liu H, Du K, Wu D
and Zou Q: Single-cell sequencing of PBMC characterizes the
transformation of T cell subsets in the inflammatory
microenvironment of RA-ILD. Research Square: https://doi.org/10.21203/rs.3.rs-3990097/v1.
|
|
26
|
Jeong E, Hong H, Lee YA and Kim KS:
Potential rheumatoid arthritis-associated interstitial lung disease
treatment and computational approach for future drug development.
Int J Mol Sci. 25(2682)2024.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Liu C and Ge Y: Immune-related genes
associated with interstitial lung disease in dermatomyositis. Int J
Gen Med. 17:5261–5271. 2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yang Z, Han R, Yin H, Li J, Cao Y, Guo R,
Sheng Y, Song L and Zhang Y: Mechanism of Lycopodii herba for
RA-ILD using integrated metabolomics and network pharmacology. Anal
Biochem. 648(114679)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang C, Kong L, Kim S, Lee S, Oh S, Jo S,
Jang I and Kim TD: The role of IL-7 and IL-7R in cancer
pathophysiology and immunotherapy. Int J Mol Sci.
23(10412)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Knuever J, Willenborg S, Ding X, Akyüz MD,
Partridge L, Niessen CM, Brüning JC and Eming SA: Myeloid
cell-restricted insulin/IGF-1 receptor deficiency protects against
skin inflammation. J Immunol. 195:5296–5308. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wu SM, Tsai JJ, Pan HC, Arbiser JL, Elia L
and Sheu ML: Aggravation of pulmonary fibrosis after knocking down
the aryl hydrocarbon receptor in the insulin-like growth factor 1
receptor pathway. Br J Pharmacol. 179:3430–3451. 2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Liu Z, Peng Z, Lin H, Zhou K, Liang L, Cao
J, Huang Z and Mei J: Identifying potential drug targets for
idiopathic pulmonary fibrosis: A mendelian randomization study
based on the druggable genes. Respir Res. 25(217)2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Bauer Y, White ES, de Bernard S,
Cornelisse P, Leconte I, Morganti A, Roux S and Nayler O: MMP-7 is
a predictive biomarker of disease progression in patients with
idiopathic pulmonary fibrosis. ERJ Open Res. 3:00074–2016.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ramadas RA, Ewart SL, Medoff BD and LeVine
AM: Interleukin-1 family member 9 stimulates chemokine production
and neutrophil influx in mouse lungs. Am J Respir Cell Mol Biol.
44:134–145. 2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Huaux F, Gharaee-Kermani M, Liu T, Morel
V, McGarry B, Ullenbruch M, Kunkel SL, Wang J, Xing Z and Phan SH:
Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in
bleomycin-induced lung injury and fibrosis. Am J Pathol.
167:1485–1496. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yuan L, Zou C, Ge W, Liu Y, Hu B, Wang J,
Lin B, Li Y and Ma E: A novel cathepsin L inhibitor prevents the
progression of idiopathic pulmonary fibrosis. Bioorg Chem.
94(103417)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Novelli F, Neri T, Tavanti L, Armani C,
Noce C, Falaschi F, Bartoli ML, Martino F, Palla A, Celi A and
Paggiaro P: Procoagulant, tissue factor-bearing microparticles in
bronchoalveolar lavage of interstitial lung disease patients: An
observational study. PLoS One. 9(e95013)2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Poole JA, Thiele GM, Janike K, Nelson AJ,
Duryee MJ, Rentfro K, England BR, Romberger DJ, Carrington JM, Wang
D, et al: Combined collagen-induced arthritis and organic
dust-induced airway inflammation to model inflammatory lung disease
in rheumatoid arthritis. J Bone Miner Res. 34:1733–1743.
2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Schuliga M, Kanwal A, Read J, Blokland
KEC, Burgess JK, Prêle CM, Mutsaers SE, Grainge C, Thomson C, James
A, et al: A cGAS-dependent response links DNA damage and senescence
in alveolar epithelial cells: A potential drug target in IPF. Am J
Physiol Lung Cell Mol Physiol. 321:L859–L871. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cella G, Cipriani A, Tommasini A, Rampin
E, Sbarai A, Rocconi R, Mazzaro G and Luzzatto G: Tissue factor
pathway inhibitor (TFPI) antigen plasma level in patients with
interstitial lung disease before and after heparin administration.
Semin Thromb Hemost. 23:45–49. 1997.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Russo RC and Ryffel B: The chemokine
system as a key regulator of pulmonary fibrosis: Converging
pathways in human idiopathic pulmonary fibrosis (IPF) and the
bleomycin-induced lung fibrosis model in mice. Cells.
13(2058)2024.PubMed/NCBI View Article : Google Scholar
|
|
42
|
He CH, Lv JM, Khan GJ, Duan H, Wang W,
Zhai KF, Zou GA and Aisa HA: Total flavonoid extract from
Dracocephalum moldavica L. improves pulmonary fibrosis by reducing
inflammation and inhibiting the hedgehog signaling pathway.
Phytother Res. 37:2745–2758. 2023.PubMed/NCBI View
Article : Google Scholar
|
|
43
|
Feng F, Wang LJ, Li JC, Chen TT and Liu L:
Role of heparanase in ARDS through autophagy and exosome pathway
(review). Front Pharmacol. 14(1200782)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li Y, Zhao J, Yin Y, Li K, Zhang C and
Zheng Y: The role of IL-6 in fibrotic diseases: Molecular and
cellular mechanisms. Int J Biol Sci. 18:5405–5414. 2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cai Z, Deng X, Jia J, Wang D and Yuan G:
Ectodysplasin A/Ectodysplasin A receptor system and their roles in
multiple diseases. Front Physiol. 12(788411)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zheng S, Yang B, Li L, Chen M, Zhang L,
Chi W, Shao ZM, Xiu B, Chi Y and Wu J: RTAM promotes antitumor
immune response in triple negative breast cancer by enhancing CD8+
T cell infiltration. Int Immunopharmacol.
129(111625)2024.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liao Y, Ke B, Long X, Xu J and Wu Y:
Upregulated expression of IL2RB causes disorder of immune
microenvironment in patients with Kawasaki disease. Biomed Res Int.
2022(2114699)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Pastor Bandeira I, de Almeida Franzoi AE,
Murillo Wollmann G, de Medeiros Junior WLG, Nogueira Brandão W,
Schatzmann Peron JP, Becker J, Nascimento OJM and Magno Gonçalves
MV: Interleukin-31 and soluble CD40L: New candidate serum
biomarkers that predict therapeutic response in multiple sclerosis.
Neurol Sci. 43:6271–6278. 2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chen CC, Chen CY, Yeh CT, Liu YT, Leu YL,
Chuang WY, Shih YH, Chou LF, Shieh TM and Wang TH: Corylin
attenuates CCl4-induced liver fibrosis in mice by regulating the
GAS6/AXL signaling pathway in hepatic stellate cells. Int J Mol
Sci. 24(16936)2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Fiebeler A, Park JK, Muller DN, Lindschau
C, Mengel M, Merkel S, Banas B, Luft FC and Haller H: Growth arrest
specific protein 6/Axl signaling in human inflammatory renal
diseases. Am J Kidney Dis. 43:286–295. 2004.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Cao Z, Li J, Hu W, Xu J, Zhao F, Wang Y,
Qin S, Liu M, Wang P, Duan J, et al: Near-infrared imaging agent
ABSi-148 alleviates CA IX-mediated hypoxic fibrosis in
inflammation-cancer transition. Adv Healthc Mater.
18(e2404935)2025.PubMed/NCBI View Article : Google Scholar
|